Abstract
The Sox4 transcription factor is involved in various cellular processes, such as embryonic development and differentiation. Deregulated expression of Sox4 in several human cancers has been reported to date, but its role in melanoma is unknown. We explored the role of Sox4 in melanoma pathogenesis in vivo and in vitro. Using tissue microarray, we evaluated Sox4 expression in 180 melanocytic lesions and investigated its role in melanoma cell migration and invasion. Sox4 expression was remarkably reduced in metastatic melanoma compared with dysplastic nevi (P < 0.05) and primary melanoma (P < 0.01). This reduction was correlated with a poorer disease-specific survival of melanoma patients (P = 0.039). Multivariate Cox regression analysis revealed that reduced Sox4 expression is an independent prognostic factor (P = 0.049). Knockdown of Sox4 enhanced melanoma cell invasion, migration, and stress fiber formation. The increased migration and invasion on Sox4 knockdown depends on the presence of nuclear factor (NF)-κB p50 and is abrogated when p50 is knocked down. We further observed inhibition of NF-κB p50 transcription by Sox4, in addition to a reverse pattern of expression of Sox4 and NF-κB p50 in different stages of melanocytic lesions. Our results suggest that Sox4 regulates melanoma cell migration and invasion in an NF-κB p50-dependent manner and may serve as a prognostic marker and potential therapeutic target for human melanoma.
Melanoma is a type of skin cancer arising from uncontrolled proliferation of melanocytes.1 It is a highly aggressive disease, due to its notorious potential to invade and metastasize to other organs, as well as its resistance to conventional radiotherapy and chemotherapy.1,2,3 Metastatic spread has been accounted as the main reason for the observed mortality in melanoma.4 Accordingly, patients with metastatic melanoma have a very poor survival of only 6 to 8 months, and a 5-year survival rate of less than 5%.2 Therefore, it is essential to reveal the factors involved in the progression and metastasis of melanoma to design novel therapeutic strategies to overcome this disease.
The family of Sry-related high-mobility group box (Sox) transcription factors encode a set of proteins with a common feature of the presence of a high-mobility group DNA binding domain.5,6 Members of this family have been shown to play important roles in embryonic development, cell fate decision, and differentiation.7,8,9 On the other hand, aberrant expression or function of members of Sox family has also been reported in various cancers.10,11,12 Sox4 is a 47-kDa protein member of this family encoded by a single exon gene.13 Sox4 preferentially binds the AACAAAG sequence motif through its high-mobility group domain and regulates transcription of target genes.14,15
Aberrant expression of Sox4 has also been found in several human cancers. It has been shown that Sox4 mRNA is up-regulated in cancers of lung,16 salivary gland,17 and brain.18 Elevated expression of Sox4 protein has also been reported in prostate cancer,19 adenoid carcinoma,20 and endometrial cancer.21 Nevertheless, higher Sox4 expression is reported to be positively correlated with better 5-year survival in bladder carcinoma patients.22 Moreover, others have shown that expression of Sox4 induces apoptosis in HEK293, as well as human bladder and hepatocellular carcinoma cells.22,23,24 Sox4 can also promote cell cycle arrest and apoptosis to inhibit tumorigenesis in human colon cancer cell lines,25 suggesting that Sox4 has a potential tumor-suppressive function in certain tissues. However, so far no study has been conducted on Sox4 expression in different stages of human melanoma, its possible correlation with tumor progression or patient survival, and its biological functions in melanoma cells.
In the present study we examined the expression of Sox4 protein in different stages of melanocytic lesions by tissue microarray (TMA) technology and immunohistochemistry. We observed a significant correlation between reduced expression of Sox4 and melanoma metastasis. Moreover, we observed a significant positive and independent correlation between expression of Sox4 protein and disease-specific 5-year survival of primary and metastatic melanoma patients. In addition, we showed that depletion of Sox4 expression by RNA interference in melanoma cell lines increases cell migration and invasion in a nuclear factor (NF)-κB p50−dependent manner. Furthermore, we observed a reverse expression pattern of Sox4 and NF-κB p50 in different stages of melanoma.
Materials and Methods
Construction of TMA
Formalin-fixed paraffin-embedded tissues from 43 dysplastic nevi, 89 primary melanomas, and 48 metastatic melanomas were used in the present study. All specimens were obtained from the 1990 to 1998 archives of the Department of Pathology, Vancouver General Hospital. The use of human skin tissues in this study was approved by the Clinical Research Ethics Board of the University of British Columbia. The most representative tumor area from each sample was carefully selected and marked on the H&E-stained slide. The TMAs were assembled using a tissue-array instrument (Beecher Instruments, Silver Spring, MD). Duplicate 0.6-mm−thick tissue cores were taken from each biopsy specimen and spotted on two high-density TMA blocks. Multiple 4-μm sections were cut with a Leica microtome and transferred to adhesive-coated slides.
Immunohistochemistry
TMA slides or single tumor slides were dewaxed at 55°C for 30 minutes and three consequent washes with xylene. Tissues were rehydrated by a series of washes in 100%, 95%, and 80% ethanol, followed by two washes in distilled water. Antigen retrieval was done by heating the samples at 95°C for 30 minutes in 10 mmol/L sodium citrate (pH 6.0). After inactivating the endogenous peroxidase by incubating in 3% H2O2 for 30 minutes and blocking with universal blocking serum for 30 minutes, slides were incubated with a polyclonal rabbit anti-Sox4 antibody (1:25 dilution, Abcam, Cambridge, MA) or rabbit anti-NF-κB p50 antibody (1:150; Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C overnight. The slides were then incubated with biotin-labeled secondary antibody and streptavidin-peroxidase for 30 minutes each, followed by developing with diaminobenzidine substrate kit (Vector Laboratories, Burlington, Ontario, Canada) and counterstained with hematoxylin.
Evaluation of Immunostaining
The evaluation of Sox4 nuclear expression was made blinded by three independent observers (including one dermatopathologist) simultaneously, and a consensus score was reached for each core. The nuclear Sox4 staining was scored into four grades according to the following staining intensities: 0, 1+, 2+, and 3+. Percentages of Sox4-positive cells were also scored into five categories: 0 (0%), 1 (1 to 25%), 2 (26 to 49%), 3 (51 to 75%), and 4 (76 to 100%). In cases with a discrepancy between duplicate cores, the higher score from the duplicate tissue cores was taken as the final score. The immunoreactive score, which is calculated by multiplying the scores of staining intensity and the percentage of positive cells, was used as the final staining score, defined as follows: 0, negative; 1 to 3, weak; 4 to 6, moderate; and 8 to 12, strong.
Cell Culture, Plasmid Construction and Small Interfering RNA Transfection
Two human melanoma cell lines; MMRU (derived from a patient with metastatic melanoma26) and RPEP (derived from a recurrent primary melanoma patient26) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum in 5% CO2 atmosphere at 37°C. The Sox4 open reading frame was subcloned from pOTB7-Sox4 (ImaGegen, Berlin, Germany) into 3×Flag-CMV-7.1 using PCR primers harboring EcoRI and BamHI restriction sites. Cells were grown to 50% confluency before transfection of small interfering (si)RNAs. For siRNA transfection, cells were incubated with either nonspecific control siRNA (siCTR), Sox4 specific siRNA (siSox4, Qiagen SI00047880) or NF-κB p105/p50 siRNA (sip50, Qiagen SI00047880) using Silenfect transfection reagent (Bio-Rad, Mississauga, Ontario, Canada). The cells were harvested at 72 hours after transfection and lysed for Western blot assay or further processed for indicated analysis.
Western Blot
Cells were harvested and washed with PBS twice and proteins were extracted as described previously.27 After protein concentration was determined by protein assay (Bio-Rad), Western blot analysis was performed. The following antibodies were used for Western blot: rabbit anti-Sox4 (1:250), rabbit anti-NF-κB p50 (1:1000; Santa Cruz), mouse anti-D-tag (1:1000; Applied Biological Materials, Richmond, BC, Canada), and mouse anti-β-actin (1:10000; Sigma, St Louis, MO). Infrared-dye−labeled secondary antibody was applied to the blot for 1 hour at room temperature and then signals were detected with Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln, NE).
Real-Time Reverse-Transcription PCR
Total RNA was prepared by Trizol extraction (Invitrogen, Carlsbad, CA) and reverse transcribed into cDNA with the SuperScript First-Strand Synthesis System (Invitrogen) according to the manufacturer’s protocol. Real-time reverse transcription-PCR was performed with SYBR Green Master mix system (Applied Biosystem, Carlsbad, CA). The sequences of human Sox4 primers were 5′-GGTCTCTAGTTCTTGCACGCTC-3′ (forward) and 5′-CGGAATCGGCACTAAGGAG-3′ (reverse). The primers for human NF-κB p50 were 5′-GGATTTCGTTTCCGTTATGT-3′ (forward) and 5′- TGTCCTTGGGTCCAGCAGTT-3′ (reverse). The primers for human β-actin were 5′- GCTCTTTTCCAGCCTTCCTT-3′ (forward) and 5′-CGGATGTCAACTTCACACTT-3′ (reverse).
Sulforhodamine B Cell Growth Assay
To compare cell growth rates, cells were seeded in 24-well plates 24 hours after the transfection with siRNA. At each time point, cells were fixed with 10% trichloroacetic acid, stained with 0.4% sulforhodamine B in 1% acetic acid, and then destained with 1% acetic acid. Cell density was quantified by dissolving bound dye in 10 mmol/L Tris (pH 10.5) followed by colorimetric determination at 550 nm. The initial time point (0 hours) was measured by fixing cells immediately after they had attached to the tissue culture plate, 6 hours after seeding. Subsequent time points were measured by fixing cells 24, 48, and 72 hours later. Relative rates of cell growth were calculated as a ratio of the cell density at each time point over the cell density at 0 hours.
Chromatin Immunoprecipitation Assay
MMRU cells were transfected with 3×Flag-Sox4 construct using the Effectene transfection reagent (Qiagen, Valencia, CA). Twenty four hours after transfection, formaldehyde-fixed cells were immunoprecipitated overnight with rabbit anti-Sox4 (Santa Cruz) and the associated genomic DNA was analyzed by PCR and agarose electrophoresis. We designed four sets of primers, spanning a distance from −2.5 kb to + 850bp of the NF-κB p50 transcription start site. We also used four sets of primers designed for Dicer1 promoter region. Immunoprecipitation of Dicer1 promoter sequence by Sox4 was done as a positive control. A list of the sequences of primers and the site of their corresponding amplicon is shown in Table 1.
Table 1.
Sequences of the Primers Used for Chromatin Immunoprecipitation (ChIP) Assay and the Distance of the Amplicon to the Corresponding Transcription Start Site
| Forward | Reverse | Distance* | |
|---|---|---|---|
| P50-A | 5′-AGGAATGGAGGAGGGTTAATC-3′ | 5′-GACACCAGTCATTGGATTTAGAG-3′ | −2181 |
| P50-B | 5′-ACTGGAGGAGGAGGATGGAG-3′ | 5′-TCATTGTTTCACTACTGGGTGG-3′ | −936 |
| P50-C | 5′-CTGTGAAGAGATGTGAATGTAACTG-3′ | 5′-GTGCCGCTGATAGAGTCATG-3′ | −392 |
| P50-D | 5′-TCATTTCTCTTCACGTCCCTC-3′ | 5′-ACCACCTTCCCTCTCTCCTC-3′ | +701 |
| Dicer-A | 5′-TAACACGGGCTGAAATATAGG-3′ | 5′-CTGTATCCGTTCTAATGGTCTATC-3′ | −3019 |
| Dicer-B | 5′-ACTAGGACAGGTGTGAGGGAC-3′ | 5′-TCAGTAGAGACGGGGTTTCAC-3′ | 2378 |
| Dicer-C | 5′-AAATTAGCTGGGTGTGGTGG-3′ | 5′-GGTGCTGAAACTGCTTCCTG-3′ | 1269 |
| Dicer-D | 5′-CAGAGAGTCTGCCAAACTTAGC-3′ | 5′-CTGGCGGTGAAAGGTTAATC-3′ | 208 |
Sequences upstream of the transcription start site are marked as (−) and those downstream of the transcription start site are marked as (+).
Wound-Healing Assay
Forty-eight hours after transfection with siRNAs, cells were washed with PBS and a standard 200 μl pipette tip was used to draw across the well to produce a wound at the center of each well. The monolayers were then washed three times with PBS to remove floating cells and incubated in fresh complete medium for another 18 hours. Photographs were taken at the same position of the wounds at 0 and 18 hours time points. The starting wound edges were defined in each photo by a black lines based on the scratch at 0 hours time point and the numbers of migrating cells across these lines were counted to quantify the rate of cell migration.28
Immunofluorescence
Cells were transfected with siCTR or siSox4 and subcultured onto coverslips in six-well plates in complete medium. Forty-eight hours after transfection, the medium was removed, cells were washed with PBS and a new serum-free medium applied for overnight. Then, cells were collected after stimulation with complete medium containing 10% fetal bovine serum for 30 minutes. In the case of ROCK inhibitor Y27632 (10 μmol/L) treatment, the inhibitor was added to the serum free medium and applied overnight. Then cells were collected after stimulation with complete medium containing 10% fetal bovine serum and Y27632 for 30 minutes. The cells were fixed with 2 ml of fixative (1% paraformaldehyde and 0.5% Triton X-100 in PBS) for 20 minutes at room temperature. After washing with PBS, the cells were incubated with bovine serum albumin for 1 hour followed by staining with phalloidin-rhodamine (1 unit/coverslip; Invitrogen, Carlsbad, CA) for 30 minutes, then incubated with rabit-anti-Sox4 primary antibody for 2 hours and Cy2-conjugated goat anti-rabbit secondary antibody (1:2000, Jackson ImmunoResearch, West Grove, PA) for 1 hour. Finally, the coverslips were incubated with 1:3000 dilution of stock Hoechst 33258 (20 mmol/L) for 5 minutes and the cells were visualized under a fluorescent microscope. Photos were taken with a cooled mono 12-bit Retiga-Ex camera equipped with Northern Eclipse imaging software. To quantify the intensity of F-actin staining, 10 images per slide were taken under a fluorescent microscope. Images were analyzed using ImageJ software (NIH, Bethesda, MD), and the mean of relative cellular fluorescent intensity was measured.
Cell Invasion Assay
Cell invasion analysis was done using the Boyden chamber assay as previously described.29 In brief, 20 μl of 5 mg/ml Matrigel (BD Biosciences, Mississauga, Canada) in serum-free medium was added to the upper compartment of 24-well Transwell culture chambers (with 8.0 μm pore size polycarbonate membrane). MMRU cells (4 × 104) suspended in 250 μl of serum-free medium were seeded on the upper compartment, and 750 μl of complete medium was added to the lower compartment. After 24 hours incubation, cells were fixed with 10% trichloroacetic acid at 4°C for 1 hour. Non-invaded cells were removed from the upper surface of the filter carefully with a cotton swab. Invaded cells on the lower side of the filter were stained with 0.5% crystal violet for 2 hours at room temperature and the retained dye on the filters was extracted by 30% acetic acid followed by reading the absorbance at 590 nm.
Statistical Analyses
The Kruskal-Wallis test was applied to compare the Sox4 nuclear staining between dysplastic nevi, primary melanoma, and metastatic melanoma using the GraphPad prism4 software. Other statistical analyses were performed with the SPSS 11.5 software (SPSS, Chicago, IL). The correlations between nuclear Sox4 expression and clinicopathologic variables, including age, gender, tumor thickness, location, and ulceration were analyzed by χ2 test. The Kaplan-Meier method and log-rank test were used to evaluate the correlations between Sox4 staining and patient survival. The Cox regression model was used for multivariate analysis. A P value of less than 0.05 was considered significant.
Results
Clinicopathologic Features of TMAs
Forty-three dysplastic nevi, 89 primary melanomas, and 48 metastatic melanomas were included in our analysis. For 89 cases of primary melanoma with age ranging from 21 to 93 (median 58), there were 56 male and 33 female cases (Table 2). For melanoma staging, we used Breslow thickness and American Joint Committee on Cancer stages as our criteria. Forty-five cases of primary melanomas were ≤1.5 mm thick (low-risk), and 44 were thicker than 1.5 mm (high-risk). Nineteen cases were located in sun-exposed area (head and neck) while 70 were located in sun-protected sites (other locations). Eighteen cases had tumor ulceration at diagnosis (Table 2).
Table 2.
Sox4 Nuclear Staining and Clinicopathological Characteristics of 89 Cases of Primary Melanoma
| Variables | Sox4 staining
|
|||
|---|---|---|---|---|
| Negative-weak | Moderate-strong | Total | P value* | |
| Age | ||||
| ≤58 | 14 (30.4%) | 32 (69.6%) | 46 | >0.05 |
| >58 | 5 (11.6%) | 38 (88.4%) | 43 | |
| Sex | ||||
| Male | 10 (17.9%) | 46 (82.1%) | 56 | >0.05 |
| Female | 9 (27.3%) | 24 (72.7%) | 33 | |
| Tumor thickness (mm) | ||||
| ≤1.5 | 12 (26.7%) | 33 (73.3%) | 45 | >0.05 |
| >1.5 | 7 (15.9%) | 37 (84.1%) | 44 | |
| Ulceration | ||||
| Present | 1 (5.5%) | 17 (94.5%) | 18 | >0.05 |
| Absent | 18 (25.3%) | 53 (74.7%) | 71 | |
| Tumor subtype | ||||
| Superficial spreading | 12 (31.6%) | 26 (68.4%) | 38 | >0.05 |
| Lentigo maligna | 1 (5.9%) | 16 (94.1%) | 17 | |
| Other† | 6 (17.6%) | 28 (82.4) | 34 | |
| Site‡ | ||||
| Sun-protected | 17 (24.3%) | 53 (75.7%) | 70 | >0.05 |
| Sun-exposed | 2 (10.5%) | 17 (89.5%) | 19 | |
χ2 test.
Other: unspecified subtype.
Sun-protected sites: trunk, arm, leg and feet; Sun-exposed sites: head and neck.
Reduced Sox4 Expression Is Correlated with Melanoma Progression
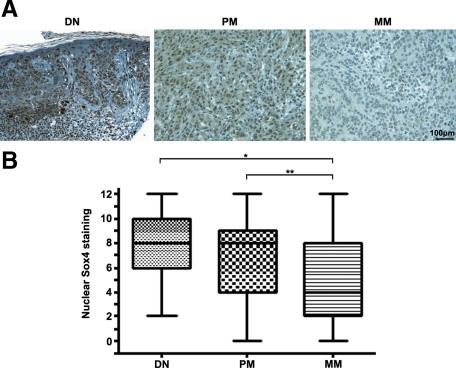
Microscopic analysis of immunostained cores revealed a distinct nuclear staining of Sox4 in dysplastic nevi (Figure 1A) with faint cytoplasmic expression in few samples. Notably, significant differences in nuclear Sox4 staining were observed between dysplastic nevi and metastatic melanomas and between primary melanomas and melanoma metastases (P < 0.05 and P < 0.01, respectively, Kruskal-Wallis test; Figure 1B). Nevertheless, there is no significant difference in Sox4 staining between primary melanoma and dysplastic nevi (P > 0.05, Kruskal-Wallis test; Figure 1B). Similarly, when we grouped the samples into negative-weak (0 to 3) or moderate-strong (4 to 12), a significant reduction of nuclear Sox4 expression was observed between metastatic melanoma and dysplastic nevi (P = 0.012, χ2 test) and also between metastatic melanoma and primary melanoma (P = 0.005, χ2 test), but not between the primary melanoma and dysplastic nevi (P = 0.708, χ2 test) (see Supplemental Figure S1 at http://ajp.amjpathol.org). We did not observe any significant correlation between nuclear expression of Sox4 and patient sex, age, tumor thickness, location, subtype, or ulceration status (Table 2).
Figure 1.
Expression of Sox4 protein in cutaneous melanoma. A: Representative images of dysplastic nevi (DN) with strong nuclear staining, primary melanoma (PM) with moderate staining, and metastatic melanoma (MM) with negative staining. Scale bar = 100 μm. B: Kruskal-Wallis test for differences in Sox4 staining among DN, PM, and MM. The median is depicted as a horizontal line inside each box. *P < 0.05; **P < 0.01.
Stronger Nuclear Sox4 Staining Correlates with Better 5-Year Survival of Melanoma Patients
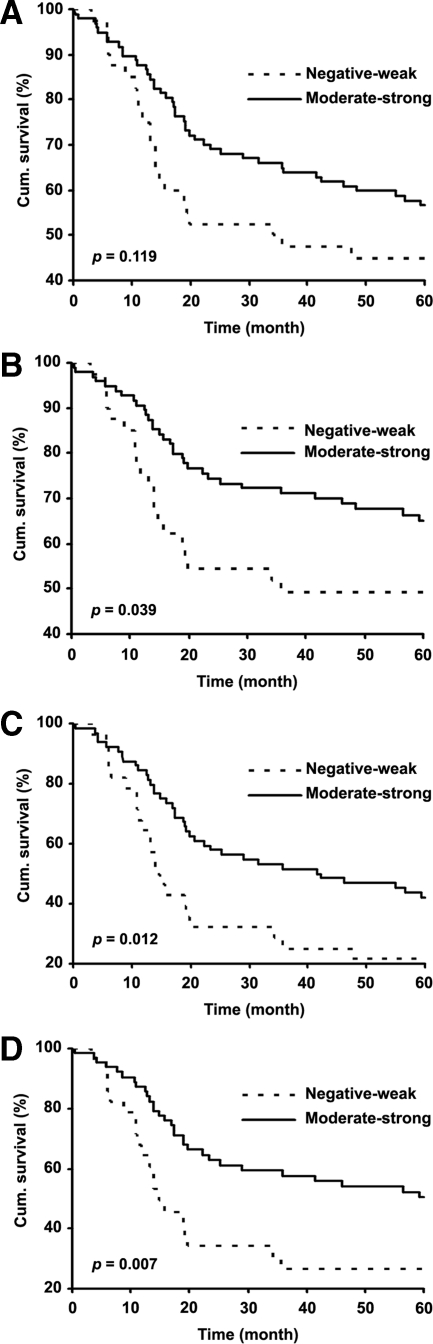
To evaluate the correlation between nuclear Sox4 expression and 5-year survival of melanoma patients, we constructed Kaplan-Meier survival curves using overall or disease-specific 5-year patient survival data to analyze the biopsies stained negative-weak versus moderate-strong Sox4 expression. Our data revealed that overall survival in moderate-strong Sox4 staining group was 55.7% compared to 45% in negative-weak group. However, the log rank analysis indicates that this difference is not significant (P = 0.119; Figure 2A). On the other hand, the disease-specific 5-year survival of the patients was significantly reduced from 65.2% in moderate-strong Sox4 group to 49.2% in negative-weak Sox4 group (P = 0.039; Figure 2B). We also performed multivariate Cox regression analysis including Sox4 nuclear staining, age, and gender to examine whether the nuclear Sox4 expression is an independent prognostic marker for melanoma. The results showed that nuclear Sox4 expression is an independent factor for predicting the disease-specific, but not the overall patient survival (P = 0.049 and 0.095, respectively; Table 3) when we combined all primary and metastatic melanoma cases for the analysis.
Figure 2.
Kaplan-Meier analysis of correlation between Sox4 expression and 5-year survival. A and B: Overall and disease-specific 5-year survival of all primary and metastatic melanoma patients. C and D: Overall and disease-specific 5-year survival of metastatic melanoma and high-risk primary melanoma patients (thickness >1.5 mm).
Table 3.
Multivariate Cox Regression Analysis of Sox4 Nuclear Expression in 48 Cases of Metastatic Melanoma and 89 Cases of Primary Melanoma
| Variable* | All primary and metastatic melanomas
|
High-risk melanomas (thickness > 1.5 mm)
|
||||
|---|---|---|---|---|---|---|
| Relative risk | 95% CI† | P value | Relative risk | 95% CI† | P value | |
| Overall survival | ||||||
| Sox4 | 1.571 | 0.924–2.673 | 0.095 | 1.904 | 1.078–3.362 | 0.026 |
| Age | 0.676 | 0.407–1.122 | 0.122 | 0.974 | 0.574–1.650 | 0.922 |
| Sex | 1.238 | 0.736–2.084 | 0.419 | 1.075 | 0.606–1.906 | 0.802 |
| Disease-specific survival | ||||||
| Sox4 | 1.780 | 1.001–3.163 | 0.049 | 1.994 | 1.087–3.658 | 0.025 |
| Age | 1.780 | 0.540–1.662 | 0.850 | 1.337 | 0.753–2.373 | 0.320 |
| Sex | 1.100 | 0.618–1.958 | 0.744 | 1.011 | 0.547–1.870 | 0.970 |
Coding of variables: Sox4 nuclear staining was coded as 1 (negative-weak) and 2 (moderate-strong). Age was coded as 1 (≤58 years), and 2 (>58 years). Gender was coded as 1 (female) and 2 (male).
CI: confidence interval.
As shown in Figure 2A, the nuclear Sox4 staining is not significantly correlated with overall 5-year patient survival, although a trend toward a poorer overall patient survival with negative-weak Sox4 staining is evident. This may be due to the fact that a small number of deaths occurred in the low-risk (≤1.5 mm) melanoma patients within 5 years. When we combined the metastatic melanoma with high risk primary melanoma cases (thickness >1.5 mm) and excluded the low risk (≤1.5 mm) cases of primary melanoma, we found that nuclear Sox4 expression has a significant positive correlation with both overall and disease-specific 5-year survival (P = 0.012 and 0.007, respectively; Figure 2, C and D). Accordingly, overall survival reduces from 42.2% in moderate-strong group to 21.4% in negative-weak group and disease-specific survival declines from 50.3% in moderate-strong group to 26.5% in negative-weak group (Figure 2, C and D). Furthermore, the multivariate Cox regression analysis revealed that nuclear Sox4 staining is an independent factor for predicting both overall and disease-specific 5-year patient survival (P = 0.026 and 0.025, respectively; Table 3) of metastatic melanoma and high-risk primary melanoma.
Sox4 Knockdown Enhances Melanoma Cell Migration
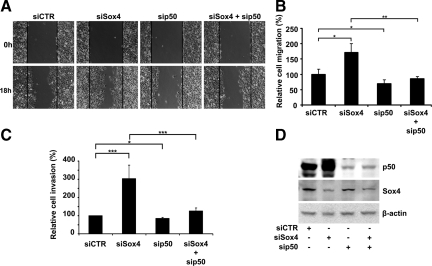
Increased ability of the cells to migrate is one of the critical steps of cancer cell invasion, resulting in higher metastasis potential and shorter survival of melanoma patients.30,31 To investigate whether the declined expression of Sox4 can affect the migration of melanoma cells, we used Sox4 specific siRNA to knockdown (KD) its expression (Figure 3D) and examined the rate of cell migration by wound healing assay. Our results revealed that Sox4-KD MMRU cells have a marked increase in cell migration compared with their control counterparts (Figure 3A). Accordingly, Sox4-KD resulted in a 70% increase of migrated cells into wound area compared with the control siRNA-transfected cells (Figure 3B). We also observed a 37% increase in cell migration of another melanoma cell line, RPEP, after Sox4 knockdown compared with the control siRNA (see Supplemental Figure S2, A-C at http://ajp.amjpathol.org). It is noteworthy that although diminished expression of Sox4 enhances melanoma cells migration, it does not significantly affect the growth rate of these cells based on the cell proliferation assay (see Supplemental Figure S3 at http://ajp.amjpathol.org).
Figure 3.
Enhancement of melanoma cell migration and invasion on Sox4 knockdown in a NF-κB p50-dependent manner. MMRU melanoma cells were transfected with either siCTR, siSox4, sip50, or co-transfected with siSox4/sip50. A: Wound-healing assay was performed on monolayers of MMRU melanoma cells 48 hours after transfection. Original magnification, ×100. B: Quantitation of A, *P < 0.05; **P < 0.01. C: For Boyden chamber assay, cells were suspended in serum-free medium and seeded on Matrigel, incubated at 37°C for 24 hours, stained by crystal violet, and quantified. *P < 0.05; **P < 0.01, ***P < 0.001. D: Protein extracts were prepared at 72 hours after transfection and analyzed for the expression of Sox4 and NF-κB p50 Western blot analysis. β-actin was used as a loading control.
Expression of NF-κB p50 Is Required for Sox4-KD-Induced Migration of Melanoma Cells
Our previous studies revealed that NF-κB p50 enhances melanoma cell migration.28 Therefore, we sought the possible role of NF-κB p50 in Sox4 KD-mediated melanoma cell migration. For this purpose we used specific siRNAs to concomitantly knockdown Sox4 and p50 and measured cell migration rate by wound healing assay (Figure 3A). Our results showed that while Sox4-KD induced a 70% increase in cell migration, p50 knockdown reduced cell migration by 30% comparing to the control group. Strikingly, when we co-knocked down Sox4 and p50 in MMRU cells, we observed a rate of cell migration comparable to the control siRNA treatment (Figure 3, A and B). These results indicate that Sox4-KD mediated increase in cell migration requires the expression of NF-κB p50 in melanoma cells.
Sox4 Regulates Melanoma Cell Invasion in an NF-κB p50 Dependent Manner
Elevated potential of the cells to invade through basal membrane is another pivotal characteristic of metastatic melanoma.30 To study the role of Sox4 in melanoma cell invasion, we examine the ability of MMRU cells to invade through the Matrigel matrix using the Boyden chamber assay. We found that cell invasion was induced by threefold in Sox4-KD cells, compared with the control (Figure 3C). On the other hand, p50 knockdown reduced cell invasion by 18% comparing with the control group (Figure 3C). Similar to the migration assay, when we co-knocked down Sox4 and p50, we observed a rate of cell invasion comparable to the control siRNA treatment (Figure 3C), indicating the requirement of NF-κB p50 in Sox4-KD−mediated increases melanoma cell invasion. Interestingly, we also observed a remarkable increase in the expression of NF-κB p50 protein (Figure 3D) on Sox4 knockdown, which further explains the observed increased cell migration and invasion on depletion of Sox4 protein and abrogation of these effects by co-knockdown of p50 and Sox4.
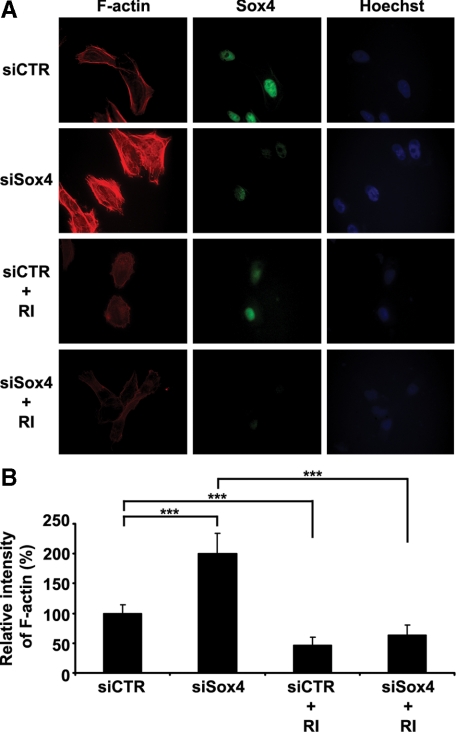
Sox4 Knockdown Requires ROCK Activity to Increase the Stress Fiber Formation in Melanoma Cells
We previously reported that NF-κB p50 induces cell migration through increased RhoA/ROCK activity and formation of F-actin stress fiber formation.28 To further investigate the mechanisms involved in Sox4-KD induced cell migration, we starved the Sox4-KD and control MMRU cells overnight with serum depletion and stained the cells with rhodamine-conjugated phalloidin (Figure 4A). On average, cells transfected with siSox4 had a 99% increase in stress fiber formation compared with control siRNA after 30 minutes of serum stimulation (Figure 4B). Similarly, knockdown of Sox4 elevates formation of stress fiber in RPEP melanoma cell line by over 62% (see Supplemental Figure S4, A and B, at http://ajp.amjpathol.org). Interestingly, when treated with Rock inhibitor, Y27632, the formation of stress fiber was effectively abrogated in both control and Sox4-KD cells, indicating the requirement of ROCK activity in Sox4-KD induced stress fiber formation (Figure 4, A and B).
Figure 4.
Sox4 knockdown induces stress fiber formation. MMRU cells were transfected with siSox4 or siCTR, followed by serum starvation overnight and serum stimulation for 30 minutes. For Rock inhibitor treatment, cells were treated with serum-free medium containing 10 ìM Y27632 overnight, and then incubated with complete medium containing 10% fetal bovine serum and 10 ìM Y27632 for 30 minutes. A: Increased formation of stress fiber on Sox4 knockdown compared to the control. Treatment with ROCK inhibitor Y27632 (RI) abrogates the Sox4-KD effect. Original magnification ×630. B: Quantitation of A, ***P < 0.001.
Inhibition of NF-κB p50 Transcription by Sox4 in Melanoma Cells
A shown in Figure 3D, Sox4-KD induced the expression of NF-κB p50 protein, implying an inhibitory role of Sox4 in regulation of NF-κB p50 expression. However, it is not clear whether this regulation happens at the transcription level or at the levels of translation and/or protein stability. To further investigate this observation, we performed a real-time quantitative PCR assay measuring the expression of NF-κB p50 mRNA after Sox4-KD in MMRU cells. Our results revealed a marked overexpression of NF-κB p50 mRNA after Sox4-KD (Figure 5A), implying that Sox4 can regulate the expression of NF-κB p50 at least in part at the transcript level. To further study the mechanistic details of this effect, we conducted a chromatin immunoprecipitation assay using MMRU cells transiently transfected with a construct encoding 3×Flag-Sox4 (Figure 5B). Using the specific polyclonal anti-Sox4 antibody, we were able to pull down nucleoprotein complexes containing Sox4 protein. By using specific primers designed for various regions of NF-κB p50 promoter area, we were able to detect precipitation of a genomic fragment corresponding to the immediate upstream sequence of NF-κB p50 promoter (Figure 5C). Since binding of Sox4 to the promoter sequence of Dicer1 has been reported before,32 we used precipitation of this sequence as a positive control for this assay.
Figure 5.
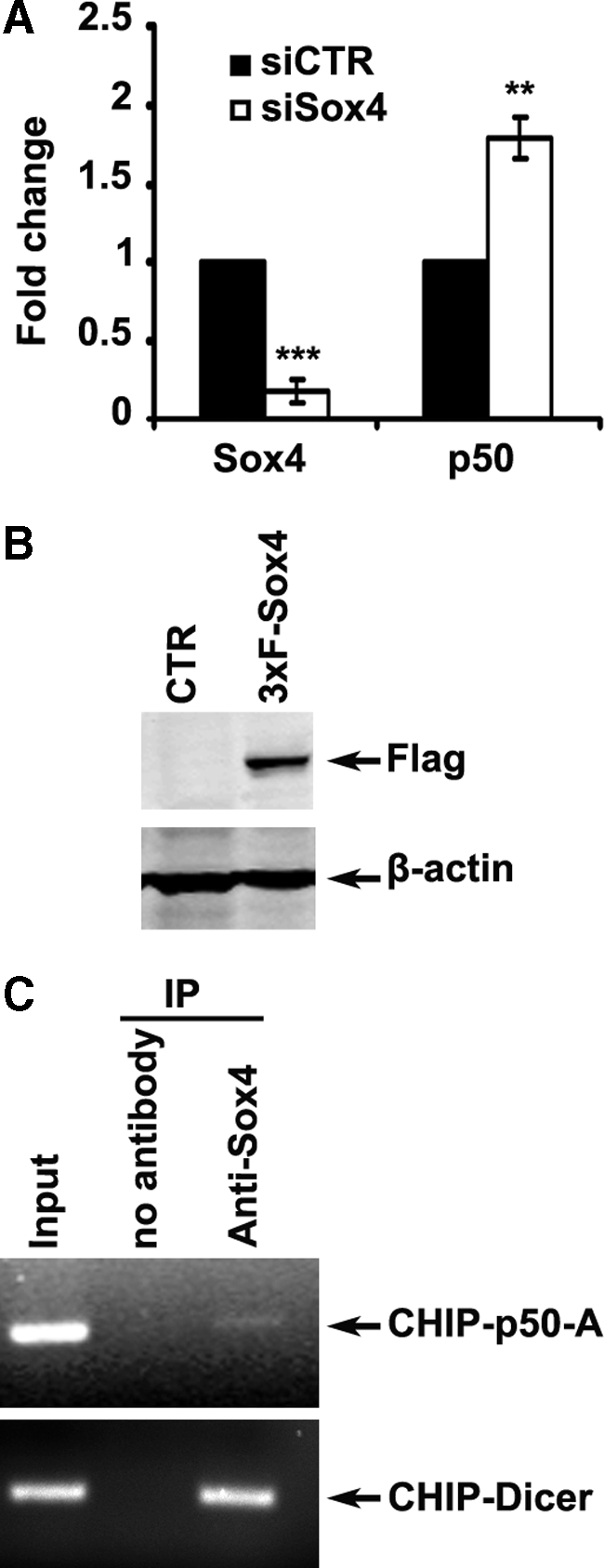
Inhibition of NF-κB p50 transcription by Sox4. A: For quantitative reverse transcription-PCR analysis, cells were transfected with siCTR or siSox4, and lysed for total RNA extraction and reverse transcription 72 hours after transfection. Expression of NF-κB p50 and Sox4 mRNAs was measured by real-time quantitative PCR and normalized with β-actin as loading control. **P < 0.01, ***P < 0.001. B: Protein extracts were prepared at 24 hours after transfection and analyzed for the expression of 3×Flag-Sox4. β-actin was used as a loading control. C: Sox4 protein binds to the NF-κB p50 gene promoter sequence in MMRU cells in vivo, demonstrated by chromatin immunoprecipitation assay. Binding of Sox4 protein to Dicer1 promoter sequence was used as a positive control.
Inverse Correlation between Expression of Sox4 and NF-κB p50 in Melanoma
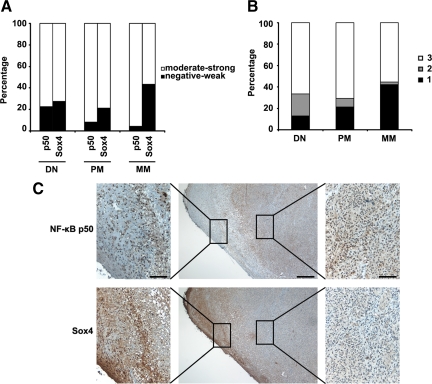
Based on the observed upregulation of NF-κB p50 on Sox4-KD and its critical requirement for enhancement of melanoma cell migration and invasion on Sox4-KD, we sought to analysis the protein expression of both Sox4 and NF-κB p50 in the same 169 melanocytic lesions. When all of the cases were analyzed, we found that whereas Sox4 staining is significantly decreased in metastatic melanoma comparing to dysplastic nevi and primary melanoma (P = 0.0260, χ2 test; Figure 6A), the expression of NF-κB p50 increases in primary melanoma and metastatic melanoma cases comparing to dysplastic nevi (P = 0.0147, χ2 test; Figure 6A). Next, we categorized each sample based on the concurrent expression level of Sox4 and NF-κB p50 into three groups: 1, low (negative-weak) Sox4/high (moderate-strong) NF-κB p50; 2, high Sox4/low NF-κB p50; and 3, high Sox4/high NF-κB p50. No sample with low Sox4 and low NF-κB p50 staining was available. Interestingly, we found a remarkable increase in the number of samples with low Sox4 and high NF-κB p50 in metastatic melanoma comparing to the primary melanoma and dysplastic nevi (P = 0.0001, χ2 test; Figure 6B). On the other hand, the percentage of samples with high Sox4 and low NF-κB p50 significantly declines in metastatic melanoma comparing with the primary melanoma and dysplastic nevi (P = 0.0002, χ2 test; Figure 6B). However, no statistically significant changed were observed in the number of samples with high Sox4 and high NF-κB p50 staining among different stages of melanocytic lesions.
Figure 6.
Inverse correlation between expression of Sox4 and NF-κB p50 in melanoma. A: Inverse correlation between Sox4 and NF-κB p50 expression in 169 melanocytic lesions at different stages. A significant reduced expression of Sox4 (P = 0.0260, χ2 test) and elevated expression of NF-κB p50 (P = 0.0147, χ2 test) in metastatic melanoma compared with the primary melanoma and dysplastic nevi is observed. B: Combined analysis of Sox4 and NF-κB p50 staining. Each sample was categorized based on the expression of Sox4 and NF-κB p50 as follows: 1, low (negative-weak) Sox4/high (moderate-strong) NF-κB p50; 2, high Sox4/low NF-κB p50; 3, high Sox4/high NF-κB p50. C: Representative images of serial sections of a high-risk nodular primary melanoma stained for NF-κB p50 and Sox4. Scale bar = 100 μm for left and right panels and 400 μm for the middle panels.
We further assessed a number of single tissue slides belonging to high risk nodular melanoma and superficial spreading melanoma patients for the expression of Sox4 and NF-κB p50 proteins. As shown in Figure 6C, in the samples which were positively stained for Sox4, the strongest staining was contained to the epidermis region as opposed to the deeper tumor mass which is mostly negative for Sox4 staining. On the other hand, the expression of NF-κB p50 was mainly restricted to the tumor area with very weak or negative staining in the epidermis. These data further indicate a mutually exclusive expression of Sox4 and NF-κB p50 in melanoma samples.
Discussion
Multiple members of Sox gene family are involved in various human malignancies.10,11,33,34 Sox4 has also been shown to be aberrantly expressed at either transcript or protein levels in different cancers.16,17,18,19,20,21,22 However, to our knowledge, no study on the expression and possible functions of Sox4 in human melanoma and its correlation with tumor progression or patient survival has been conducted so far. In the current study we analyzed Sox4 expression in 43 cases of dysplastic nevi, 89 cases of primary melanoma, and 48 cases of melanoma metastases by TMA technology and also investigated its role in regulating the migration and invasion abilities of melanoma cells as critical steps in cancer metastasis.
Microscopic analysis revealed that Sox4 is mainly a nuclear protein (Figure 1A) which is consistent with previous reports in addition to its function as a transcription factor.35,36 Statistical analysis of the tissue microarray immunoreactivity scores revealed a significant reduction in nuclear Sox4 expression in metastatic melanoma compared with primary melanoma and dysplastic nevi. However, we were not able to detect any significant difference between dysplastic nevi and primary melanoma (Figure 1B and see Supplemental Figure S1 at http://ajp.amjpathol.org). These results suggest that reduced or lost expression of Sox4 may be relevant to the process of melanoma metastasis.
Despite previous reports on elevated expression of Sox4 in cancers of lung,16 salivary gland,17 brain,18 and prostate,19 we observed a reduced expression of Sox4 in metastatic melanoma compared with primary melanoma and dysplastic nevi. This discrepancy could be due to intrinsic differences in the biology of various types of tissues. Accordingly, Sox4 is reported to have anti-apoptotic function in prostate cancer cells which may explain its elevated expression in tumor biopsies.19 It is noteworthy that similar contradictory expression pattern and functions have been also reported for other members of the Sox family. For instance, Sox9 is suggested to be an oncogene in prostate cancer,37 while it can suppress formation of melanoma.38 On the other hand, other studies have shown elevated apoptosis rate on overexpression of Sox4 in HEK293, bladder, and colon cancer cells.22,24,25 Furthermore, Ahn and colleagues23 showed that up-regulation of Sox4 on treatment with Delta12-prostaglandin J2 induced apoptosis and antisense-mediated knockdown of Sox4 abrogated apoptosis in hepatocellular carcinoma. The p53 up-regulated modulator of apoptosis (PUMA) has also been reported as a direct transcription target of Sox4,19 indicating the role of Sox4 in the tumor suppression pathways. Nevertheless, there has been no study dealing with the possible role of Sox4 in cancer metastasis so far. In this report we revealed that Sox4 expression is reduced or lost in the course of progression of primary melanoma to metastatic melanoma (Figure 1). Moreover, we demonstrated a positive correlation between nuclear Sox4 expression and 5-year disease-specific survival of melanoma patients as well as 5-year overall and disease-specific survival of high-risk melanoma cases (Figure 2). Sox4 knockdown significantly stimulated melanoma cell migration (Figure 3, A and B; see Supplemental Figure S2 at http://ajp.amjpathol.org) and invasion (Figure 3C), which are two of the most critical events in the process of cancer progression toward metastasis.39 These observations suggest that Sox4 functions as an inhibitor of melanoma metastasis, which by far is the most important cause for the high rate of mortality of melanoma,4 explaining the observed positive correlation between Sox4 expression and higher survival rate in patients. This observation is also in line with the previous report of positive correlation between Sox4 expression and survival of patients with bladder cancer22 and hepatocellular carcinoma.40
NF-κB has been regarded as one of the main regulator in several cellular pathways including cell proliferation, inflammation, cell survival, migration, and metastasis.41,42,43 We previously reported that expression of NF-κB p50 is elevated during melanoma progression and that abrogation of its expression by RNA interference reduces cell migration in vitro.28 A positive role of NF-κB p50 in facilitating the invasion of melanoma has also been reported by others.44,45 Here we showed that RNA interference-mediated knockdown of NF-κB p50 in melanoma cells reduces the cell migration and invasion (Figure 3). Strikingly, co-knockdown of NF-κB p50 abolished both Sox4-KD-induced cell migration and invasion (Figure 3). Furthermore, we observed a marked up-regulation of NF-κB p50 expression on Sox4-KD (Figure 3D), implying that Sox4 functions upstream of NF-κB p50 and can suppress the expression of NF-κB p50 and hence the activity of NF-κB pathway. Moreover, we observed a remarkable increase in the formation of F-actin stress fibers on Sox4 knockdown in both MMRU and RPEP cells (Figure 4, A and B; see Supplemental Figure S4, A and B, at http://ajp.amjpathol.org). Formation of stress fiber is commonly considered as a critical event for cell motility and metastasis.46 We previously reported induced RhoA activity and ROCK-mediated formation of stress fiber on overexpression of NF-κB p50 in melanoma cells. Here we showed that Sox4 knockdown has a stimulatory effect on stress fiber formation and treatment with ROCK-inhibitor, Y27632, abrogates this effect, providing further evidence for the involvement of NF-κB p50-RhoA/ROCK-stress fiber formation pathway in the Sox4-KD-induced cell migration.
Despite the observed overexpression of NF-κB p50 protein after Sox4-KD, it was not understood at what step of the gene expression this regulation occurs. To address this question, we performed quantitative PCR assay after Sox4-KD and revealed a significant overexpression of NF-κB p50 mRNA after Sox4-KD (Figure 5A), implying that Sox4 regulates the expression of NF-κB p50 at least in part at the transcript level. Since the Sox4 protein is a DNA binding factor, we sought to further examine its ability to bind to NF-κB p50 promoter sequence by chromatin immunoprecipitation assay. Interestingly, we were able to pull down a genomic fragment upstream of NF-κB p50 promoter (Figure 5C). The promoter region of the human NF-κB p50 has been previously defined to be located at the first 1 kb nucleotides of the transcription start site as well as part of the first exon.47 We were not able to detect binding of Sox4 to this promoter area; nevertheless, our PCR results showed binding of Sox4 to a genomic sequence adjacent to this promoter region (approximately 2 kb from start site). Interestingly, this fragment contains a distinctive Sox4 binding site AACAAAG,14 which can explain the observed binding of Sox4 to this specific sequence. It is worth noting that although Sox4 has been initially identified as a transcription factor, it is possible that it can also inhibit transcription of its target genes depending on the binding partners present. This mechanism has been previously described for members of the Sox family.48,49 It can also explain the observed contradictory functions of Sox4 in different cancer types. However, the exact mechanistic details of Sox4-mediated inhibition of NF-κB p50 transcription and also its potential binding partner(s) to the promoter region is yet to be revealed. Collectively, these data suggest a significant influence of Sox4 on the NF-κB p50 pathway in melanoma cells.
In an interesting report, Pan et al25 recently showed a direct role of Sox4 in stabilization of p53 protein on UV-irradiation or application of genotoxic agents and subsequent induction of apoptosis, cell cycle arrest, and suppression of tumorigenesis. p53 has long been considered as a main hub in the cellular stress response to UV radiation,50,51 which is the most prominent environmental factor for melanoma development.52,53 It is also known that p53 can inhibit NF-κB−mediated tumorigenesis54 and absence or aberrant function of p53 may result in activation of NF-κB pathway, hence formation or promotion of cancer. Nevertheless, the frequency of p53 mutation in melanoma is significantly lower than other types of human malignancy.55,56 It has been suggested that melanoma cells compensate for lack of p53 mutation by mutation or reduced expression of other factors involved in the p53-mediated tumor suppression, such as Apaf-1.1,55,57,58 Therefore, reduced expression of Sox4, as an upstream stabilizer of p53, in metastatic melanoma, may indeed help the tumor cells to activate the NF-κB pathway and acquire further ability to invade other tissues. This idea may provide another explanation for our observed reverse pattern of Sox4 and NF-κB p50 expression (Figure 6). The possible relationship between the expression of Sox4, stability and activity of p53, and NF-κB pathway is currently under investigation in our group.
Sinner and colleagues59 have previously observed that Sox4 protein may interact and stabilize β-catenin protein which is a main hub of wnt signaling pathway in colon carcinoma cells. A similar observation has been made in prostate cancer cells.32 Wnt signaling is a prominent signaling pathway involved in various features of tumorigenesis including epidermal−mesenchymal transition and metastasis,60 cell attachment, cell motility, and matrix metalloproteinase expression and activity.61 Considering the metastasis suppressor function of Sox4 in melanoma and its reported interaction with β-catenin protein, it would also be of interest to investigate the effect of Sox4 on the wnt signaling pathway and its possible influence on the cell detachment as a main feature of epithelial mesenchymal transition and metastasis.
In conclusion, our data indicate that Sox4 expression is significantly reduced in metastatic melanoma and reduced Sox4 is correlated with a poorer 5-year patient survival. Furthermore, we demonstrated the inhibitory effect of Sox4 on NF-κB pathway and melanoma cell migration and invasion in vitro. We also reported a reverse pattern of expression of Sox4 and NF-κB p50 in different stages of melanocytic lesions, as well as mutually exclusive expression pattern of Sox4 and NF-κB p50 in high-risk primary melanoma. These data suggest that Sox4 plays an important inhibitory role in melanoma progression and metastasis, and may be used as a promising prognostic marker and therapeutic target for malignant melanoma.
Acknowledgments
We thank Ronald Wong, Jun Li, Jin Bai, Yabin Cheng, and Anousheh Zardan for technical assistance and Robert H. Bell for helpful discussions.
Footnotes
Address reprint requests to Gang Li, M.D., Ph.D., Jack Bell Research Centre, 2660 Oak Street, Vancouver, BC, Canada V6H 3Z6. E-mail: gangli@interchange.ubc.ca.
Supported by grants from Canadian Institutes of Health Research (MOP-84559, MOP-93810) and Canadian Dermatology Foundation (G.L.). S.M.J. is a recipient of Canadian Institutes of Health Research Skin Research Training Centre Trainee Award and University of British Columbia Graduate Fellowship.
The authors declare no conflict of interest.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- Cummins DL, Cummins JM, Pantle H, Silverman MA, Leonard AL, Chanmugam A. Cutaneous malignant melanoma. Mayo Clin Proc. 2006;81:500–507. doi: 10.4065/81.4.500. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JC. Back to basics: sox genes. Dev Dyn. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, He W, Lu C, Wang Z, Wang J, Giercksky KE, Nesland JM, Suo Z. Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res. 2009;29:1233–1241. [PubMed] [Google Scholar]
- Nonaka D. Differential expression of SOX2 and SOX17 in testicular germ cell tumors. Am J Clin Pathol. 2009;131:731–736. doi: 10.1309/AJCP7MNCNBCRN8NO. [DOI] [PubMed] [Google Scholar]
- Lu B, Fang Y, Xu J, Wang L, Xu F, Xu E, Huang Q, Lai M. Analysis of SOX9 expression in colorectal cancer. Am J Clin Pathol. 2008;130:897–904. doi: 10.1309/AJCPW1W8GJBQGCNI. [DOI] [PubMed] [Google Scholar]
- Farr CJ, Easty DJ, Ragoussis J, Collignon J, Lovell-Badge R, Goodfellow PN. Characterization and mapping of the human SOX4 gene. Mamm Genome. 1993;4:577–584. doi: 10.1007/BF00361388. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangur CS, Switzer A, Fan L, Marton MJ, Meyer MR, Wang T. Identification of genes over-expressed in small cell lung carcinoma using suppression subtractive hybridization and cDNA microarray expression analysis. Oncogene. 2002;21:3814–3825. doi: 10.1038/sj.onc.1205480. [DOI] [PubMed] [Google Scholar]
- Frierson HF, Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002;161:1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, Luider TM, den Boer ML, Pieters R. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10:648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH. Epigenetic repression of microRNA-129–2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- Ahn SG, Kim HS, Jeong SW, Kim BE, Rhim H, Shim JY, Kim JW, Lee JH, Kim IK. Sox-4 is a positive regulator of Hep3B and HepG2 cells’ apoptosis induced by prostaglandin (PG)A(2) and delta(12)-PGJ(2). Exp Mol Med. 2002;34:243–249. doi: 10.1038/emm.2002.34. [DOI] [PubMed] [Google Scholar]
- Hur EH, Hur W, Choi JY, Kim IK, Kim HY, Yoon SK, Rhim H. Functional identification of the pro-apoptotic effector domain in human Sox4. Biochem Biophys Res Commun. 2004;325:59–67. doi: 10.1016/j.bbrc.2004.09.215. [DOI] [PubMed] [Google Scholar]
- Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, Zhang PJ, Li AL, Zhang XM. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci USA. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers HR, Etoh T, Doherty JR, Sober AJ, Mihm MC., Jr Cell migration and actin organization in cultured human primary, recurrent cutaneous and metastatic melanoma. Time-lapse and image analysis. Am J Pathol. 1991;139:423–435. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li G. ING3 promotes UV-induced apoptosis via Fas/caspase-8 pathway in melanoma cells. J Biol Chem. 2006;281:11887–11893. doi: 10.1074/jbc.M511309200. [DOI] [PubMed] [Google Scholar]
- Gao K, Dai DL, Martinka M, Li G. Prognostic significance of nuclear factor-kappaB p105/p50 in human melanoma and its role in cell migration. Cancer Res. 2006;66:8382–8388. doi: 10.1158/0008-5472.CAN-05-4402. [DOI] [PubMed] [Google Scholar]
- Wong RP, Ng P, Dedhar S, Li G. The role of integrin-linked kinase in melanoma cell migration, invasion, and tumor growth. Mol Cancer Ther. 2007;6:1692–1700. doi: 10.1158/1535-7163.MCT-07-0134. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Huang DY, Lin YT, Jan PS, Hwang YC, Liang ST, Peng Y, Huang CY, Wu HC, Lin CT. Transcription factor SOX-5 enhances nasopharyngeal carcinoma progression by down-regulating SPARC gene expression. J Pathol. 2008;214:445–455. doi: 10.1002/path.2299. [DOI] [PubMed] [Google Scholar]
- Pan X, Li H, Zhang P, Jin B, Man J, Tian L, Su G, Zhao J, Li W, Liu H, Gong W, Zhou T, Zhang X. Ubc9 interacts with SOX4 and represses its transcriptional activity. Biochem Biophys Res Commun. 2006;344:727–734. doi: 10.1016/j.bbrc.2006.03.194. [DOI] [PubMed] [Google Scholar]
- Boyd KE, Xiao YY, Fan K, Poholek A, Copeland NG, Jenkins NA, Perkins AS. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2006;107:733–741. doi: 10.1182/blood-2003-05-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Leav I, Ibaragi S, Wegner M, Hu GF, Lu ML, Balk SP, Yuan X. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68:1625–1630. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, Hearing VJ. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J Clin Invest. 2009;119:954–963. doi: 10.1172/JCI34015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, Lee SB, Yoon SK. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro, Carcinogenesis. 31:1298–1307. doi: 10.1093/carcin/bgq072. [DOI] [PubMed] [Google Scholar]
- Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314–1319. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Tergaonkar V. NFkappaB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis. 2009;14:348–363. doi: 10.1007/s10495-009-0315-0. [DOI] [PubMed] [Google Scholar]
- Perkins ND. NF-kappaB: tumor promoter or suppressor? Trends Cell Biol. 2004;14:64–69. doi: 10.1016/j.tcb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Amiri KI, Richmond A. Role of nuclear factor-kappa B in melanoma. Cancer Metastasis Rev. 2005;24:301–313. doi: 10.1007/s10555-005-1579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Stolpe A, Caldenhoven E, Stade BG, Koenderman L, Raaijmakers JA, Johnson JP, van der Saag PT. 12-O-tetradecanoylphorbol-13-acetate- and tumor necrosis factor alpha-mediated induction of intercellular adhesion molecule-1 is inhibited by dexamethasone. Functional analysis of the human intercellular adhesion molecular-1 promoter. J Biol Chem. 1994;269:6185–6192. [PubMed] [Google Scholar]
- Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell PC, Scheinman RI, Baldwin AS., Jr Promoter of the human NF-kappa B p50/p105 gene. Regulation by NF-kappa B subunits and by c-REL. J Immunol. 1993;150:2794–2804. [PubMed] [Google Scholar]
- Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Latonen L, Laiho M. Cellular UV damage responses–functions of tumor suppressor p53. Biochim Biophys Acta. 2005;1755:71–89. doi: 10.1016/j.bbcan.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Park HY, Kosmadaki M, Yaar M, Gilchrest BA. Cellular mechanisms regulating human melanogenesis. Cell Mol Life Sci. 2009;66:1493–1506. doi: 10.1007/s00018-009-8703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340:1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- Atillasoy ES, Seykora JT, Soballe PW, Elenitsas R, Nesbit M, Elder DE, Montone KT, Sauter E, Herlyn M. UVB induces atypical melanocytic lesions and melanoma in human skin. Am J Pathol. 1998;152:1179–1186. [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Merlino G, DePinho RA. Malignant melanoma: modern black plague and genetic black box. Genes Dev. 1998;12:3467–3481. doi: 10.1101/gad.12.22.3467. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Blaszyk H, Cunningham JS, McGovern RM, Schroeder JS, Helander SD, Pittelkow MR, Sommer SS, Kovach JS. Overexpression and mutations of p53 in metastatic malignant melanomas. Int J Cancer. 1996;67:313–317. doi: 10.1002/(SICI)1097-0215(19960729)67:3<313::AID-IJC1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, Cordon-Cardo C, Lowe SW. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Monzon J, Liu L, Brill H, Goldstein AM, Tucker MA, From L, McLaughlin J, Hogg D, Lassam NJ. CDKN2A mutations in multiple primary melanomas. N Engl J Med. 1998;338:879–887. doi: 10.1056/NEJM199803263381305. [DOI] [PubMed] [Google Scholar]
- Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Delmas V. The WNT/Beta-catenin pathway in melanoma. Front Biosci. 2006;11:733–742. doi: 10.2741/1831. [DOI] [PubMed] [Google Scholar]
- Crampton SP, Wu B, Park EJ, Kim JH, Solomon C, Waterman ML, Hughes CC. Integration of the beta-catenin-dependent Wnt pathway with integrin signaling through the adaptor molecule Grb2. PLoS One. 2009;4:e7841. doi: 10.1371/journal.pone.0007841. [DOI] [PMC free article] [PubMed] [Google Scholar]