Abstract
Recent genome-wide association studies have identified multiple genetic loci and single nucleotide polymorphisms (SNPs) associated with either increased or decreased risk of colorectal cancer (CRC). In the present study, our objective was to determine whether 11 of the new susceptibility CRC loci are associated with tumor morphology and to confirm these loci as distinct and etiologically different risk factors in the development of CRC. The following clinical and morphological parameters were analyzed in 1572 samples: tumor size, T-stage, lymph node metastases, degree of differentiation, mucin production, Crohn-like peritumoral lymphocytic infiltration, tumor-infiltrating lymphocytes, desmoplastic reaction, necrosis, invasion of blood or lymph vessels, perineural growth, medullary type, budding, and tumor margin. One SNP from each of the 11 loci (rs6983267 on 8q24.21, rs16892766 on 8q23.3, rs719725 on 9p24.1, rs10795668 on 10p14, rs3802842 on 11q23.1, rs4444235 on 14q22.2, rs4779584 on 15q13.3, rs9929218 on 16q22.1, rs4939827 on 18q21.1, rs10411210 on 19q13.11, and rs961253 on 20p12.3) was genotyped for all cases. Odds ratios, 95% confidence intervals, and the corresponding P values were calculated for the 11 SNPs identified above. A cross tabulation between SNPs and morphology was performed. Several loci showed statistically significant associations with specific phenotypes. The findings are consistent with pathogenic variants in several loci that act in distinct CRC and morphogenetic pathways. Further large-scale studies are required to validate these findings.
Numerous genes are involved in colorectal carcinogenesis either as somatic or epigenetic changes, for example, KRAS and BRAF, but also susceptibility genes such as those involved in familial adenomatous polyposis or hereditary non-polyposis colorectal cancer (HNPCC).1 Most of these genes belong to morphogenetic signaling pathways, conserved from fruit fly to man, such as wingless-type mouse mammary tumor virus integration site family member (WNT), sonic hedgehog homolog, transforming growth factor beta receptor 1, and other growth factor families including fibroblast growth factor, epidermal growth factor, platelet-derived growth factor, and vascular endothelial growth factor.1 Morphogens are important cellular signaling molecules with major influences in cell and tissue morphology. Thus, tumor phenotypes can reflect an underlying genetic contribution. Tumors originating in the hereditary colorectal cancer (CRC) syndromes familial adenomatous polyposis and HNPCC caused by germline mutations in the Adenomatous Polyposis Coli gene (WNT signaling pathway) and DNA mismatch repair genes (with effects on WNT/transforming growth factor beta receptor 1 signaling pathways), respectively, show clear morphological patterns. Microsatellite instability-positive tumors in HNPCC and sporadic microsatellite instability-positive tumors express different phenotypes as a result of different means of morphogenesis, although the DNA mismatch repair pathway is involved in both. Lymphocytic infiltration and tumor budding are clearly associated with HNPCC and mucin secretion, poor differentiation, tumor heterogeneity, and glandular serration are associated with microsatellite instability-positive tumors in the serrated pathway.2 Different, but equivalent pathways involved in HNPCC (KRAS) and serrated pathway (BRAF) demonstrate a possible genetic background for these differences.3
Recent genome-wide association studies have identified multiple genetic loci and single nucleotide polymorphisms (SNPs) associated with increased or decreased CRC risk.4,5,6,7,8,9,10,11 The first was a susceptibility region on 8q24.21,4,6,9 where the most significant SNP showed an OR of 1.19. During the last 2 years, another 10 susceptibility loci have been published, at 8q23.3, 9p24.1, 10p14, 11q23.1, 14q22.2, 15q13.3, 16q22.1, 18q21.1, 19q13.11, and 20p12.34,5,7,8,10,11.
Our aim was to determine whether the 11 new susceptibility loci were associated with tumor morphology to confirm them as distinct and etiologically different risk factors in colorectal carcinogenesis. To this end, we selected one published SNP in each region and analyzed them in consecutive CRC cases to demonstrate their role in morphogenetic pathways.
Materials and Methods
Study Sample
Patients were recruited within a national study conducted by the Swedish Low-Risk Colorectal Cancer Study Group. Samples were obtained during 2004 to 2006 from 14 different surgical clinics in the middle of Sweden. Patients who were too ill or too old to be invited were excluded; otherwise, all patients were eligible. By December 2006, 2175 patients had agreed to participate. Informed consent was obtained from each subject in accordance with the Swedish law concerning the ethical approval of research on human subjects (2002:409, 2003:198). Blood was obtained from 1786 patients, and DNA was extracted using routine methods. Of the 1786 surgical specimens, all underwent evaluation directly after operation by a local pathologist. Within the study, 1599 samples were available for re-examination, and 1583 had a known family history. After exclusion of cases of familial adenomatous polyposis (n = 2) and HNPCC according to Amsterdam criteria (n = 9), 1572 cases were included in the analysis. The initial 18 cases (1.1%) were evaluated by two experienced gastrointestinal pathologists (S.G. and N.P.) to discuss and establish consensus criteria regarding morphological parameters such as necrosis, budding, desmoplasia, and tumor-infiltrating lymphocytes, which are not part of routine pathology reporting in Sweden. The rest of the cases were then evaluated by one of the pathologists (S.G.).
Morphology Coding
A specific protocol was submitted for each tumor. The following data were analyzed: tumor size in three dimensions, TNM classification, total number of lymph nodes and number with metastases, degree of differentiation in the major part of the tumor, mucin production, Crohn-like peritumoral lymphocytic infiltration, tumor-infiltrating lymphocytes, desmoplastic reaction, necrosis, invasion of blood or lymph vessels, perineural growth, medullary type, budding, and tumor margin.
For patients with more than one tumor, the largest tumor was chosen for study. If size was not assessable, the tumor with the most informative morphological parameters was chosen. Tumors were divided into two groups depending on the largest diameter, 6 cm or more (large) and smaller. For tumor stage, T1 and T2 tumors were grouped together, while T3 and T4 tumors formed one group. Lymph node metastases were categorized as present or not, that is, N1 and N2 cases were grouped together. Tumor grade was determined according to the World Health Organization classification.12 Tumors were assigned two grades: poor (including poor and undifferentiated) and other (moderate and well differentiated) because the distinction between poor and moderate is clinically most relevant when it comes to prognosis and management13 and because of the general difficulty in distinguishing moderate and well differentiated tumors. Mucin production was divided into two categories: 0 to 50% and greater than 50% of the tumor area. Tumors containing more than 50% extracellular mucin were classified as mucinous and, by definition, low-grade, according to World Health Organization and Swedish consensus criteria.12,14 Crohn-like peritumoral lymphocytic reaction was recorded if there were at least four nodular aggregates of lymphocytes deep to the advancing tumor margin in low power (×4) field.15 Number of tumor-infiltrating lymphocytes were categorized in two groups, ≤ 30 /10 high-power fields and >30/10 high-power fields from H&E-stained sections, avoiding adenomas, intramucosal carcinomas, and early invasive tumor components.16 Desmoplasia was defined as a hypocellular intense fibrous reaction around infiltrating tumor tissue and was scored as present or absent. Tumor necrosis was defined as presence of cell detritus and inflammatory cells within glandular lumina, and was scored as present or absent.17 Vascular invasion was recorded when unequivocal tumor aggregates were found in preformed spaces lined by endothelium indicating lymphatic or blood vessels. Perineural growth was defined as tumor cells infiltrating underneath the perineurium at the invasive margin of the tumor or deep to it. Medullary carcinoma was defined according to World Health Organization criteria.12 The presence or absence of budding, defined as detachment of single isolated cancer cells or a cluster of up to four cells by H&E staining was recorded as this has been shown to be an adverse prognostic factor.18 Quantification of buds was, however, not performed. Tumor margin was classified as dominantly circumscribed (pushing with even, rounded infiltration) or infiltrative (invading foci identified).19 In the analysis of necrosis, desmoplastic reaction, and budding, tumors located in the rectum were excluded due to the fact that most cases of rectal cancer in Sweden receive pre-operative radiotherapy, which might alter the morphology and affect the results.
Genotyping
All cases were genotyped for one SNP from each CRC risk locus—rs16892766 on 8q23.3, rs6983267 on 8q24.21, rs719725 on 9p24, rs10795668 on 10p14, rs3802842 on 11q23.1, rs4444235 on 14q22.2, rs4779584 on 15q13.3, rs9929218 on 16q22.1, rs4939827 on 18q21.1, rs10411210 on 19q13.1, and rs961253 on 20p12.3. Exploiting linkage disequilibrium between SNPs, we excluded the following published SNPs: rs355527 on 20p12.3 (tagged by rs961253), and rs7259371 on 19q13.1 (tagged by rs10411210). Six of the SNPs (rs719725, rs4444235, rs4779584, rs9929218, rs10411210, and rs961253) were genotyped using TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA). Genotyping of the remaining five SNPs (rs6983267, rs16892766, rs10795668, rs4939827, and rs3802842) were performed using a technology developed by Nanogen, at deCode Genetics, Reykjavik, Iceland.
Statistical Analysis
The cross tabulation between SNP data and morphology was done using Statistica version 8, (Statsoft Inc., Tulsa, OK). Pearson χ2 test was used for calculating the P value (see Supplemental Appendix at http://ajp.amjpathol.org). The significant results from these genotype–phenotype analyses were studied further by using the DeFinetti program (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl).20 Deviations of genotype frequencies in cases and controls from those expected under Hardy-Weinberg equilibrium were calculated by χ2 tests (one degree of freedom). The significance level for statistical tests was set at 0.05. Odds ratios, 95% confidence intervals, and the corresponding P values were calculated using the same program. The results were validated using Statistica version 8. Results are presented without correction for multiple testing to avoid the loss of valuable information due to the limited number of patients.
Results
Of the enrolled participants, 53% were men and 47% women. The average age of CRC diagnosis was 69 years, with a median of 70 years and a range of 27–95 years. With regard to tumor pathology, 7.3% of the cases had T1 tumors, 16.3% had T2, 62.2% had T3, and 12.6% had T4. A small group (1.7%) had unknown T status or were not verified as cancers (T0); 56.2% were N0, 21.3% were N1, and 18.4% were N2. A small percentage (4.1%) had unknown N status. The majority of cases (96.1%) had inadequate information about the M status. Tumors were located as follows: 0.5% in appendix, 14.7% in cecum, 10.9% in the ascending colon, 4.2% in the right flexure, 5.1% in the transverse colon, 2.6% in the left flexure, 3.6% in the descending colon, 22.0% in the sigmoid colon, and 35.2% in rectum; 1.3% had an unknown location. Cases missing information on pathological features were excluded from further analysis regarding these features.
Genotyping success rate was between 97 to 99%. The genotypes for the 11 SNPs were then tested for association with the fourteen morphological parameters (see Supplemental Appendix at http://ajp.amjpathol.org). Five of the gene variants (on 8q24.21, 10p14, 16q22.1, 18q21.1, and 19q13.11) have been published to have a protective effect, meaning that the most common variant (the wild-type allele) constitutes the risk allele. For consistency, we will address the results associated with the variant allele, regardless of whether this exerts a risk or a protective effect. Cross tabulation analysis addressed if there was any genotype-phenotype correlation. For five loci, the variants, rs16892766 on 8q23.3, rs3802842 on 11q23.1, rs4779584 on 15q13.3, rs9929218 on 16q22.1, and rs4939827 on 18q21.1, no statistically significant phenotype was found (see Supplemental Appendix at http://ajp.amjpathol.org); however, six individual loci showed statistically significant results and a total of 10 genotype-phenotype associations were significant (see Supplemental Appendix at http://ajp.amjpathol.org). Table 1 presents the results of the DeFinetti analysis to obtain the ORs and confidence intervals for the 10 tests with statistically significant results) and demonstrates that five SNPs are significantly associated with morphological parameters.
Table 1.
Correlation Analysis between SNPs and Morphological Profiles
| Locus/SNP | Morphology | Genotypes | No | Yes | OR (95% CI) | P values |
|---|---|---|---|---|---|---|
| 8q24.21 | Crohn-like lymphocytic reaction | GG | 137 | 318 | 1 | |
| rs6983267 | GT | 284 | 492 | 0.75 (0.58–0.96) | 0.021 | |
| TT | 90 | 207 | 0.99 (0.72–1.36) | 0.955 | ||
| 9p24 | Necrosis | AA | 102 | 291 | 1 | |
| rs719725 | AC | 112 | 346 | 1.08 (0.79–1.48) | 0.615 | |
| CC | 47 | 83 | 0.62 (0.41–0.95) | 0.026 | ||
| 9p24 | Desmoplastic reaction | AA | 58 | 335 | 1 | |
| rs719725 | AC | 42 | 415 | 1.71 (1.12–2.61) | 0.012 | |
| CC | 29 | 102 | 0.61 (0.37–1.00) | 0.049 | ||
| 9p24 | Tumor infiltrating lymphocytes >30/10 high-power fields | AA | 459 | 146 | 1 | |
| rs719725 | AC | 576 | 146 | 0.89 (0.61–1.03) | 0.087 | |
| CC | 152 | 59 | 1.22 (0.86–1.74) | 0.269 | ||
| 9p24 | Budding | AA | 292 | 226 | 1 | |
| rs719725 | AC | 327 | 285 | 1.13 (0.89–1.43) | 0.323 | |
| CC | 120 | 58 | 0.62 (0.44–0.89) | 0.010 | ||
| 10p14 | Moderate or well differentiated | GG | 159 | 608 | 1 | |
| rs10795668 | GA | 179 | 506 | 0.74 (0.58–0.94) | 0.015 | |
| AA | 34 | 98 | 0.75 (0.49–1.16) | 0.194 | ||
| 14q22.2 | Crohn-like lymphocytic reaction | TT | 163 | 347 | 1 | |
| rs4444235 | TC | 229 | 495 | 1.02 (0.80–1.30) | 0.902 | |
| CC | 125 | 190 | 0.71 (0.53–0.96) | 0.024 | ||
| 19q13.1 | Desmoplastic reaction | CC | 105 | 741 | 1 | |
| rs10411210 | CT | 23 | 123 | 0.76 (0.46–1.24) | 0.266 | |
| TT | 4 | 5 | 0.18 (0.05–0.67) | 0.004 | ||
| 20p12.3 | Mucin production in >50% of the tumor | CC | 558 | 75 | 1 | |
| rs961253 | CA | 585 | 119 | 1.51 (1.11–2.07) | 0.009 | |
| AA | 192 | 44 | 1.71 (1.14–2.56) | 0.010 | ||
| 20p12.3 | Infiltrative | CC | 320 | 285 | 1 | |
| rs961253 | tumor margin | CA | 330 | 350 | 1.19 (0.96–1.48) | 0.118 |
| AA | 137 | 87 | 0.71 (0.52–0.98) | 0.034 |
Genotypes for each morphological parameter that was significant (P value <0.05; see Supplemental Appendix at http://ajp.amjpathol.org). Significant OR and P values are presented in bold. Yes: support for the morphological parameter; No: no support.
Heterozygous carriers of the T allele of rs6983267 (8q24.21) had decreased Crohn-like peritumoral lymphocytic reaction: ORhet = 0.75 (P value = 0.021). The SNP rs719725 (9p24.1) initially displayed four statistically significant phenotypic characteristics (see Supplemental Appendix at http://ajp.amjpathol.org), tumor-infiltrating lymphocytes (P = 0.038), necrosis (P = 0.027), desmoplastic reaction (P = 0.0003), and budding (P = 0.0003). However, the results for tumor-infiltrating lymphocytes did not remain significant after the DeFinetti analysis. Also, the results for desmoplastic reaction, budding, and necrosis were inconsistent in homo- and heterozygous carriers and were considered false positives (Table 1). For rs10795668 (10p14), the heterozygote genotype was associated with poor differentiation ORhet = 0.74 (P = 0.015). Homozygosity for the C allele of rs4444235 (14q22.2) showed a decreased Crohn-like peritumoral reaction ORhom = 0.71 (P = 0.024). The T allele of rs10411210 (19q13.11) was negatively associated with desmoplastic response ORhom = 0.18 (P = 0.004) (Figure 1). For rs961253 (20p12.3), the A allele was associated with mucin producing tumors ORhom = 1.71 (P = 0.010) and ORhet = 1.51 (P = 0.009) respectively (Figure 2) and homozygous carriers of the A allele more commonly had tumors with circumscribed margins ORhom = 0.71 (P = 0.034). However, for heterozygous carriers the results suggested an opposite effect (ORhet = 1.19).
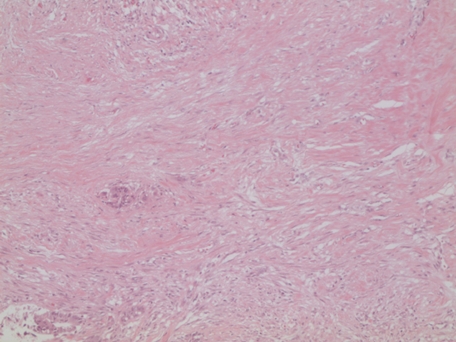
Figure 1.
Desmoplasia. Intense hypocellular fibrous reaction around infiltrating tumor glands from a patient homozygous for the C allele of SNP rs10411210 (19q13.1) H&E magnification original ×100.
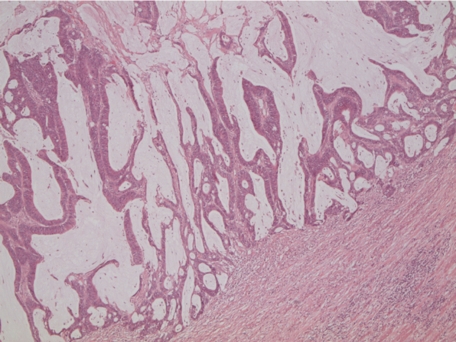
Figure 2.
Mucinous tumor. Presence of greater than 50% extracellular mucin in a tumor from a patient homozygous for the A allele of SNP rs961253 (20p12.3) H&E magnification original ×25.
Discussion
Our study demonstrates a unique pattern of morphological parameters associated with recently published low-risk gene variants of CRC, with loci on chromosomes 19 and 20 displaying the greatest and most significant effect on CRC risk. Consistent with other reports, we found an overdominant effect, such that the most significant associations were observed in heterozygotes.21 The first published susceptibility region on 8q24.214,9 (rs6983267) is associated with an elevated risk of adenoma development and is a known risk locus for prostate cancer9. Here, this variant was demonstrated to be negatively associated with Crohn-like peritumoral lymphocytic infiltration. Several studies have published genotype-phenotype analysis for this locus, 8q24.21, sex, tumor site, age at diagnosis, and family history.6,21,22 However, none of these did report association between this SNP and sex, tumor site, or age.6,21,22 In contrast, Tuupanen et al22 reported a positive association to family history for the same SNP. In addition, rs6983267 on 8q24.21 has been associated missmatch repair status, tumor site, and tumor stage.23 Baeten et al24 studied leukocyte infiltration in tumors as a prognostic marker and reported that the number of inflammatory cells in the tumor stroma and tumor cell nests, but not the peritumoral leukocytes, predicted patient survival. This is consistent with our result that suggests that Crohn-like peritumoral lymphocytic infiltration is a separate entity from intratumoral lymphocytes. The results for the rs719725 (9p24) variant were inconsistent and therefore unlikely to be associated with any of the suggested phenotypes. The region on 19q13.11 was negatively associated with desmoplastic reaction, which is generally considered favorable.25 However there are conflicting reports regarding the role of the fibrotic stromal response in cancer development and whether it favors the host or the tumor.26 The region on 20p12.3 harbors a risk allele that is associated with mucin-producing tumors and with tumors predominantly showing defined margins. Mucin production is typical for villous tumors; in HNPCC, where villous tumors are over-represented, there is generally an association with better prognosis than sporadic CRC even though HNPCC tumors show poor differentiation. The protective variant at the 10p14 locus was associated with poorly differentiated tumors but with no other HNPCC like morphology. Similar to the locus on 8q24.21, the 14q22.2 locus harbors an allele negatively correlated to Crohn-like peritumoral lymphocytic reaction However, for the variant on 8q24.21, it is the allele providing a protective effect that is associated with tumor phenotype, while for the variant on 14.q22.2, it is the risk allele.
It is difficult to discuss how some of these correlations might be interpreted in their biological context as the exact pathogenic SNP is still not known for all risk loci. However, one study has recently demonstrated that the 8q24.21 locus affects the last nucleotide of a binding site for transcription factor 4 that could explain some of the risk for carriers of this allele.27 The closest gene mapping telomeric to 20p12.3 is bone morphogenic protein 2, and similarly, bone morphogenic protein 4 maps close to the 14q22.2 locus.28 Both genes belong to the transforming growth factor-β family, which is already known as a morphogenetic pathway involved in CRC carcinogenesis. For the SNP on 10p14, there is no coding transcript or predicted gene within 0.4 Mb of sequence from the SNP.29 The 19q13.1 locus maps to a 96-kb block of linkage disequilibrium that contains the gene RHPN2, suggested to be involved in the biology of invasiveness of CRC.30
As this is the first study of detailed morphology associated to CRC low-risk alleles, we thought it was important to show all possible results for future comparisons and therefore chose not to correct for multiple testing. However, it cannot therefore be ruled out that some, or even all, of our results are false positives.
In the study of cancer as a complex disease, it is expected that numerous genes and pathways will act together and that this will influence risk effects. The effect of each individual genetic variant has been demonstrated to be extremely small with relative risks only just above 1. Hence, understanding the genetic effects on function as seen by clinical parameters such as tumor phenotype is important. That cancer-causing genes do influence morphology was indicated from high-risk genes,1 and here we have investigated 11 published CRC risk loci for 14 different morphological parameters and detected genotype-phenotype correlations for five of them. The findings are consistent with the pathogenic variants in these loci acting in distinct different CRC and morphogenetic pathways. The knowledge of genes or genetic variants involved in cancer development has future clinical potential in prevention, diagnosis, and prognosis and even for decisions regarding therapeutic strategies. However, our results are preliminary, and more studies are required to confirm these findings, and in particular, a long-term follow-up would be important to evaluate the survival implications related to these risk alleles.
Acknowledgments
We thank all patients and their spouses, Berith Wejderot for excellent technical assistance, and deCode Genetics for genotyping.
Footnotes
Address reprint requests to Annika Lindblom, M.D., Ph.D., Department of Molecular Medicine and Surgery, CMM02, Karolinska University Hospital, S 17176 Stockholm, Sweden. E-mail: Annika.Lindblom@ki.se.
Supported by the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and the Karolinska Institute, The Swedish Cancer Society, The Stockholm Cancer Foundation, and The Swedish Research Council. Work done in Edinburgh was funded by grants from Cancer Research UK (C348/A8896); a Centre grant from CORE as part of the Digestive Cancer Campaign; Scottish Government Chief Scientist Office (K/OPR/2/2/D333); Medical Research Council (G0000657–53203).
Author contributions: Study concept and design: S.G., N.P., A.L.; Acquisition of data: S.G., N.P., S.V.H., S.P., U.L.; Analysis and interpretation of data: S.G., N.P., S.V.H., S.P., A.T., S.M.F., H.C., M.G.D., A.L.; Statistical analysis: S.V.H., S.P.; Obtained funding: A.L.; Drafting of the manuscript: S.G., N.P., S.V.H., S.P., A.L.; Recruiting patients: The Low-Risk Colorectal Cancer Study Group: David Edler, Karolinska Universitetssjukhuset/Solna (Stockholm); Claes Lenander, Mag-tarm-centrum, Ersta sjukhus (Stockholm); Johan Dalén, S:t Görans sjukhus (Stockholm); Fredrik Hjern, Danderyds sjukhus (Danderyd); Nils Lundqvist, Norrtälje sjukhus (Norrtälje); Ulrik Lindforss, Södertälje sjukhus (Södertälje); Lars Påhlman, Akademiska sjukhuset (Uppsala); Kennet Smedh, Centrallasarettet (Västerås); Anders Törnqvist, Centralsjukhuset (Karlstad); Jörn Holm, Länssjukhuset Gävle-Sandviken (Gävle); Martin Janson, Karolinska Universitetssjukhuset/Huddinge (Huddinge); Magnus Andersson, Universitetssjukhuset (Örebro); Susanne Ekelund, Södersjukhuset (Stockholm); Louise Olsson, Mälarsjukhuset (Eskilstuna).
The authors disclose no conflicts.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- van den Brink GR, Offerhaus GJ. The morphogenetic code and colon cancer development. Cancer Cell. 2007;11:109–117. doi: 10.1016/j.ccr.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Jass JR. HNPCC and sporadic MSI-H colorectal cancer: a review of the morphological similarities and differences. Fam Cancer. 2004;3:93–100. doi: 10.1023/B:FAME.0000039849.86008.b7. [DOI] [PubMed] [Google Scholar]
- Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O'Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, Jaeger E, Vijayakrishnan J, Kemp Z, Gorman M, Chandler I, Papaemmanuil E, Penegar S, Wood W, Sellick G, Qureshi M, Teixeira A, Domingo E, Barclay E, Martin L, Sieber O, Kerr D, Gray R, Peto J, Cazier JB, Tomlinson I, Houlston RS. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, Carvajal-Carmona L, Howarth K, Jaeger E, Spain SL, Walther A, Barclay E, Martin L, Gorman M, Domingo E, Teixeira AS, Kerr D, Cazier JB, Niittymaki I, Tuupanen S, Karhu A, Aaltonen LA, Tomlinson IP, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Cetnarskyj R, Porteous ME, Pharoah PD, Koessler T, Hampe J, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Chang-Claude J, Hoffmeister M, Brenner H, Zanke BW, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, Reid FJ, Smith LA, Kavoussanakis K, Koessler T, Pharoah PD, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Schmidt CO, Hampe J, Chang-Claude J, Hoffmeister M, Brenner H, Wilkening S, Canzian F, Capella G, Moreno V, Deary IJ, Starr JM, Tomlinson IP, Kemp Z, Howarth K, Carvajal-Carmona L, Webb E, Broderick P, Vijayakrishnan J, Houlston RS, Rennert G, Ballinger D, Rozek L, Gruber SB, Matsuda K, Kidokoro T, Nakamura Y, Zanke BW, Greenwood CM, Rangrej J, Kustra R, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Lipton L, Gibbs P, Chapman M, Desai J, Jones IT, Yeatman TJ, East P, Tomlinson IP, Verspaget HW, Aaltonen LA, Kruhoffer M, Orntoft TF, Andersen CL, Sieber OM. DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin Cancer Res. 2008;14:8061–8069. doi: 10.1158/1078-0432.CCR-08-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, Walther A, Spain S, Pittman A, Kemp Z, Sullivan K, Heinimann K, Lubbe S, Domingo E, Barclay E, Martin L, Gorman M, Chandler I, Vijayakrishnan J, Wood W, Papaemmanuil E, Penegar S, Qureshi M, Farrington S, Tenesa A, Cazier JB, Kerr D, Gray R, Peto J, Dunlop M, Campbell H, Thomas H, Houlston R, Tomlinson I. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Aaltonen LA. World Health Organization classification of tumours. Lyon: IARC Press,; Pathology and Genetics of Tumours of the Digestive System. 2000:pp 103–119. [Google Scholar]
- Chandler I, Houlston RS. Interobserver agreement in grading of colorectal cancers-findings from a nationwide web-based survey of histopathologists. Histopathology. 2008;52:494–499. doi: 10.1111/j.1365-2559.2008.02976.x. [DOI] [PubMed] [Google Scholar]
- Stewart PM, Stears A, Tomlinson JW, Brown MJ. Regulation–the real threat to clinical research. BMJ. 2008;337:a1732. doi: 10.1136/bmj.a1732. [DOI] [PubMed] [Google Scholar]
- Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA, Borten MM, Stitz R, Searle J, McKeone D, Fraser L, Purdie DR, Podger K, Price R, Buttenshaw R, Walsh MD, Barker M, Leggett BA, Jass JR. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrk TC, Watson P, Kaul K, Lynch HT. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–2422. [PubMed] [Google Scholar]
- Greenson JK, Bonner JD, Ben-Yzhak O, Cohen HI, Miselevich I, Resnick MB, Trougouboff P, Tomsho LD, Kim E, Low M, Almog R, Rennert G, Gruber SB. Phenotype of microsatellite unstable colorectal carcinomas: well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am J Surg Pathol. 2003;27:563–570. doi: 10.1097/00000478-200305000-00001. [DOI] [PubMed] [Google Scholar]
- Ueno H, Murphy J, Jass JR, Mochizuki H, Talbot IC. Tumour ‘budding’ as an index to estimate the potential of aggressiveness in rectal cancer. Histopathology. 2002;40:127–132. doi: 10.1046/j.1365-2559.2002.01324.x. [DOI] [PubMed] [Google Scholar]
- Jass JR, Ajioka Y, Allen JP, Chan YF, Cohen RJ, Nixon JM, Radojkovic M, Restall AP, Stables SR, Zwi LJ. Assessment of invasive growth pattern and lymphocytic infiltration in colorectal cancer. Histopathology. 1996;28:543–548. doi: 10.1046/j.1365-2559.1996.d01-467.x. [DOI] [PubMed] [Google Scholar]
- Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–1261. [PubMed] [Google Scholar]
- Poynter JN, Figueiredo JC, Conti DV, Kennedy K, Gallinger S, Siegmund KD, Casey G, Thibodeau SN, Jenkins MA, Hopper JL, Byrnes GB, Baron JA, Goode EL, Tiirikainen M, Lindor N, Grove J, Newcomb P, Jass J, Young J, Potter JD, Haile RW, Duggan DJ, Le Marchand L. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Niittymaki I, Nousiainen K, Vanharanta S, Mecklin JP, Nuorva K, Jarvinen H, Hautaniemi S, Karhu A, Aaltonen LA. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res. 2008;68:14–17. doi: 10.1158/0008-5472.CAN-07-5766. [DOI] [PubMed] [Google Scholar]
- Cicek MS, Slager SL, Achenbach SJ, French AJ, Blair HE, Fink SR, Foster NR, Kabat BF, Halling KC, Cunningham JM, Cerhan JR, Jenkins RB, Boardman LA, Petersen GM, Sargent DJ, Alberts SR, Limburg PJ, Thibodeau SN. Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2492–2500. doi: 10.1158/1055-9965.EPI-09-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeten CI, Castermans K, Hillen HF, Griffioen AW. Proliferating endothelial cells and leukocyte infiltration as prognostic markers in colorectal cancer. Clin Gastroenterol Hepatol. 2006;4:1351–1357. doi: 10.1016/j.cgh.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Caporale A, Vestri AR, Benvenuto E, Mariotti M, Cosenza UM, Scarpini M, Giuliani A, Mingazzini P, Angelico F. Is desmoplasia a protective factor for survival in patients with colorectal carcinoma? Clin Gastroenterol Hepatol. 2005;3:370–375. doi: 10.1016/s1542-3565(04)00674-3. [DOI] [PubMed] [Google Scholar]
- Ueno H, Jones AM, Wilkinson KH, Jass JR, Talbot IC. Histological categorisation of fibrotic cancer stroma in advanced rectal cancer. Gut. 2004;53:581–586. doi: 10.1136/gut.2003.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, Mecklin JP, Jarvinen H, Ristimaki A, Di-Bernardo M, East P, Carvajal-Carmona L, Houlston RS, Tomlinson I, Palin K, Ukkonen E, Karhu A, Taipale J, Aaltonen LA. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- Kim JS, Crooks H, Dracheva T, Nishanian TG, Singh B, Jen J, Waldman T. Oncogenic beta-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62:2744–2748. [PubMed] [Google Scholar]
- Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- Bellovin DI, Simpson KJ, Danilov T, Maynard E, Rimm DL, Oettgen P, Mercurio AM. Reciprocal regulation of RhoA and RhoC characterizes the EMT and identifies RhoC as a prognostic marker of colon carcinoma. Oncogene. 2006;25:6959–6967. doi: 10.1038/sj.onc.1209682. [DOI] [PubMed] [Google Scholar]