Abstract
The accumulation of intracellular storage vesicles is a hallmark of lysosomal storage diseases. Neither the identity nor origin of these implicated storage vesicles have yet been established. The vesicles are often considered as lysosomes, endosomes, and/or autophagosomes that are engorged with undigested materials. Our studies in the mouse model of mucopolysaccharidosis type IIIB, a lysosomal storage disease that induces neurodegeneration, showed that large storage vesicles in cortical neurons did not receive material from either the endocytic or autophagy pathway, which functioned normally. Storage vesicles expressed GM130, a Golgi matrix protein, which mediates vesicle tethering in both pre- and cis-Golgi compartments. However, other components of the tethering/fusion complex were not associated with GM130 on storage vesicles, likely accounting for both the resistance of the vesicles to brefeldin A and the alteration of Golgi ribbon architecture, which comprised distended cisterna connected to LAMP1-positive storage vesicles. We propose that alteration in the GM130-mediated control of vesicle trafficking in pre-Golgi and Golgi compartments affects Golgi biogenesis and gives rise to a dead-end storage compartment. Vesicle accumulation, Golgi disorganization, and alterations of other GM130 functions may account for neuron dysfunction and death.
Lysosomal storage disorders (LSDs) represent the most common cause of neurodegeneration in pre-adulthood. In these inherited metabolic diseases, a biochemical disorder interrupts the degradation of macromolecules in lysosomes or the recycling of degradation products. This primary defect triggers cascades of intricate events, which frequently results in neuronal dysfunction and death. A hallmark of LSD pathology is the intracellular accumulation of storage vesicles with characteristics of lysosomes. They predominate in specific cell types depending on the disease. Observations in animal models show progressively increasing numbers and size of storage vesicles with age in affected tissues, which is evocative of a slow and irreversible process.
Storage vesicles in cells with LSDs are commonly identified as crippled lysosomes engorged with undigested material resulting form the genetic defect.1 The activation of a genetic program controlling lysosome proliferation could compensate deficient lysosomal functions.2 Storage vesicles are however not normal lysosomes and might be distinct from lysosomes. Whereas lysosomes are usually small, highly dynamic, and contain homogeneous dense material, storage vesicles are large, polymorphic, poorly mobile,3 and contain highly heterogeneous materials, frequently consisting of internal vesicles and membranes. These morphological features are reminiscent of multivesicular bodies (MVBs), which could accumulate due to impaired capacity for lysosomal degradation. Activation of macroautophagy, or impairment of autophagosomal-lysosomal fusion, or a combination of both, were reported in several LSDs.4,5,6,7,8,9 Activation and/or interruption of the macroautophagy flux can increase the number and size of autophagosomes, leading to imbalance between production and elimination, and subsequent accumulation. It is therefore conceivable that storage vesicles are MVBs, autophagosomes, a combination of both, or amphisomes resulting from merging of these two types of vesicles. Alterations of the endo-lysosomal and/or macroautophagy pathways are intimately associated to neurodegeneration, as shown in Alzheimer and Parkinson diseases.10,11,12 There are additional cell disorders in LSDs that are not necessarily linked to these pathways. They include defects in calcium homeostasis,13,14,15 alterations in phospholipid synthesis,16,17 oxidative and endoplasmic reticulum (ER) stress.18,19,20
We report here the results of a comprehensive analysis of storage vesicles in the neurons of a mouse model of mucopolysaccharidosis IIIB mice (MPSIIIB, Sanfilippo syndrome type B). This LSD is caused by α-N-acetylglucosaminidase (NaGlu, EC 3.2.1.50) deficiency, a lysosomal hydrolase necessary for heparan sulfate (HS) degradation. The enzyme defect induces the production of partially digested HS oligosaccharides. The associated accumulation of substances like GM3 gangliosides,21 subunit c of mitochondrial ATP synthase (ScMAS),22 ubiquitin, or lysozyme23 is not currently understood. Mechanisms leading to the development of neuroinflammation24 and loss of synaptophysin expression,25 which both predominates in the rostral cortex, and neuropathology, which is spread throughout the central nervous system, are also ignored. Neurological manifestations are predominant in children with MPSIIIB, evolving over few years from behavioral changes to profound mental retardation.26 Cortical neurons are severely affected and presumably play a major role in disease clinical expression.27,28 In the mouse model of MPSIIIB, storage vesicle accumulation is conspicuous in the central nervous system and contrasts with limited amounts of undigested HS accumulating in this tissue.21 Genetic correction at early stage of disease progression rescues HS degradation, clears other accumulating substances, prevents storage vesicle accumulation, and improves animal behavior.29 Our results provide evidence that storage vesicles in MPSIIIB mouse neurons are not related to endosomes or autophagosomes but more likely to the Golgi complex.
Materials and Methods
Antibodies and Reagents
Cell culture reagents, media, and Dulbecco’s PBS (DPBS) were from Invitrogen (Cergy-Pontoise, France), fetal calf serum (FCS) was from PAA Laboratories (Les Mureaux, France). Other products were from Sigma (Lyon, France) if not specified otherwise. Antibodies (dilutions for immunofluorescence): mouse monoclonal (mAb) IgG1 anti-α-synuclein (clone 42, 1:500, BD Biosciences, Erembodegem, Belgium), rabbit polyclonal anti-β-COP (COPI, 1:1000, Novus Biologicals, Cambridge, UK), rabbit polyclonal anti-Calnexin (1:1000, Stressgen, Victoria, Canada), rabbit polyclonal anti-EEA1 (1:200, Affinity BioReagents, Golden, US), rabbit polyclonal anti-ERGIC53 (1:200, Sigma), rabbit polyclonal anti-Giantin (1:1000, Covance, Emeryville, CA), biotinylated goat polyclonal anti-GFP/YFP (1:3000, Rockland, Gilbertsville, PA), mouse mAb IgM anti-GM3 (clone GMR6, 1:100, Sheikagaku, Tokyo, Japan), mouse mAb IgG1 anti-GM130 (clone 35, 1:200, BD Biosciences), mouse mAb IgG1 anti-Golgin97 (clone CDF4, 1:200, Molecular Probes, Eugene, OR), rabbit polyclonal anti-GRASP65 (1:500, Novus Biologicals), mouse mAb IgG1 anti-GS15 (clone 19/GS15, 1:200, BD Biosciences), mouse mAb IgM anti-heparan sulfate (clones HepSS-1 and F58-10E4, 1:200, Sheigagaku), rat mAb IgG2b anti-LAMP1 (clone 1D4B, 1:500, BD Biosciences), mouse mAb IgG1 anti-LC3-II (clone 4E12, 1:100, MBL, Naka-ku Nagoya, Japan), rabbit polyclonal anti-LC3B (1:250, Novus Biologicals), mouse mAb IgG2a anti-M6PR (clone 2G11, 1:400, Abcam, Cambridge, UK), mouse mAb IgG1 anti-Neurofilament 200kD (clone RT97, 1:1000, Hybrydoma Bank, Iowa City, US), mouse mAb IgG1 anti-p115 (clone 15/p115, 1:200, BD Biosciences), mouse mAb IgG1 anti-p62 (clone 3/p62, 1:200, BD Biosciences), rabbit polyclonal anti-ScMAS (1:1000, a gift from Pr D. Palmer, Centre for Molecular Biology, Lincoln University, Canterbury, New Zealand),30 rabbit polyclonal anti-Sec23 (COPII, 1:300, Affinity BioReagents), rabbit polyclonal anti-Syntaxin 5 (1:100, Synaptic Systems, Göttingen, Germany), rabbit polyclonal anti-Ubiquitin (1:500, Dako, Glostrup, Denmark). LysoTracker Red DND-99 (50 nmol/L), streptavidin (1 μg/ml) and secondary antibodies conjugated to Alexafluor 488, 555, or 647 were from Molecular Probes (Invitrogen). Dextran-biotin was from Sigma.
Animals
Mouse experiments were approved by the Institut Pasteur ethical committee for animal research and performed by authorized investigators (authorization no. 75-268, Ministère de l’Agriculture et de la Pêche). C57Bl/6NaGlu+/− mice were obtained from Pr. E. Neufeld (UCLA, Los Angeles, CA)21 and bred. C57Bl/6NaGlu−/− mutant embryos were identified by PCR on DNA from tail-clipped.
Viral Vectors
The structure, preparation, and use of AAV-hNaGlu, HIV-hNaGlu, and HIV-IDUA-GFP vectors were previously described.29,31
Cortical Neuron Cultures
Cerebral cortices from 4 to 6 C57Bl/6NaGlu−/− or C57Bl/6NaGlu+/+ E17 embryos were pooled. Cells were prepared, plated, and cultured as previously described.25 Neuron cultures were exposed to lentivirus vectors coding for the missing enzyme NaGlu or for the lysosomal hydrolase α-L-iduronidase fused to GFP (IDUA-GFP) at day 3 or day 7, respectively (10 ng of p24 protein/coverslip). Cultures were incubated with DMSO, rapamycin (250 nmol/L), and/or leupeptin (50 μmol/L) at day 6 for 48 hours, or with BFA (1.25 μg/ml) at day 7.
ts045 VSV-G-YFP Trafficking
Neurospheres from wild-type or MPSIIIB mouse E17 ganglionic eminences were prepared and grown as previously described.32 Nucleofection was performed according to the manufacturer protocol (Mouse NSC Nucleofector Kit, LONZA, Levallois-Perret, France) with 4 μg of ts045VSV-G-YFP plasmid DNA.33 Cell differentiation was induced by growth factor withdrawal and adhesion in the presence of 2% FCS onto coverslips coated with 50 μg/ml polyornithine, 5 μg/ml laminine. Neurospheres were incubated 24 hours at 40°C, then switched to 32°C to allow VSV-G trafficking. VSV-G-YFP was revealed on fixed cells with anti-GFP antibodies.
Fluorescence Labeling and Imaging
Mouse were killed with ketamine injection and perfused intracardiacally with PBS followed by PBS, 4% paraformaldehyde (PFA, 50 ml). Brains were removed, postfixed in the same fixative for 1 hour at 4°C, then embedded in 20% gelatin and sliced sagittally with vibratome (60-μm-thick sections). Sections were delipidated for 3 minutes with pure ethanol, rinsed 3 times in PBS before overnight incubation at 4°C with primary antibodies diluted in PBS, 0.3% triton X100, 1% bovin serum albumin (BSA), 2% normal goat serum (NGS). Bound antibodies were revealed by one-hour incubation at room temperature with fluorescent secondary antibodies. Coverslips were mounted in FluoromountG (Southern Biotech, Birmingham, AL).
Cultured cells were washed in DPBS and fixed with DPBS, 4% PFA for 15 minutes. After blocking with PBS, 0.1% saponin, 1% BSA, 2% NGS, cells were incubated overnight at 4°C with primary antibodies diluted in blocking buffer. Bound antibodies were revealed after one-hour incubation at room temperature with fluorescent secondary antibodies. Coverslips were mounted in FluoromountG. Living neuron cultures were treated with biotinylated dextran for 5 minutes at 37°C in Neurobasal medium and washed three times in Dulbecco’s PBS before incubation in Neurobasal supplemented with 5% FCS. Cells were then processed for immunostaining.
Quantification of Immunolabeling
Images were acquired either with an Axioplan 2 imaging optic microscope equipped with Apotome and AxioCam TR camera controlled by the AxioVision software (Zeiss, Le Pecq, France), or with a SP5 confocal system (Leica, Reuil-Malmaison, France). Time exposure was kept constant for all analyses in the same experiment. Fluorescent signal was binarised, keeping threshold constant for all acquisitions in the same experiment. Quantification in neuronal processes was performed by scoring stained individual objects (vesicles), using the Axiovision or ImageJ softwares. Quantification in neurons soma, where individual objects could hardly been distinguished from each other, was performed by measuring specific signal pixel numbers relative to total cell surface, using the Axiovision colocalization module. Confocal images were deconvoluted with the Huygens 2 software (SVi, Hilversum, The Netherlands). Deconvoluted 3D images were constructed with the Imaris x64 software (Bitplan, Zurich, Switzerland).
Electron Microscopy and Immunogold Labeling
Cultured cells were washed in DPBS before fixation in 0.1M cacodylated, 2.5% glutaraldehyde, 1% sucrose and postfixed in 0.1M cacodylate, 0.1% osmium tetroxide, 1.5% potassium ferricyanide. Samples were then dehydrated and embedded in epon 812. Anesthetized mice were perfused with 0.1 mol/L Sorensen buffer, 20 U/ml heparin, followed by 0.1 mol/L Sorensen, 2% PFA. Coronal cortical sections (2 mm) were postfixed with Karnovsky fixative (0.08M cacodylate, 2% PFA, 2.5% glutaraldehyde, 5 mmol/L CaCl2). Fragments (1 mm3) were prepared and postfixed in 1% aqueous osmium tetroxide and embedded in epon 812. Ultrathin sections (70 nm) were prepared using a Reichert Ultracut S Wild M3z microtome (Leica) and contrasted with uranyl acetate and lead citrate.
For immunogold ultrastructural microscopy, anesthetized mice were perfused with 0.1 mol/L Sorensen buffer containing 20U/ml heparin followed by 0.1 mol/L Sorensen, 4% PFA. Coronal cortical sections were postfixed with 0.1 mol/L Sorensen, 4% PFA, 0.05% glutaraldehyde. Fragments were prepared according to the Tokuyasu cryo-ultramicrotomy method. Fragments were quenched in 50 mmol/L NH4Cl, cryoprotected in 2.1 mol/L sucrose and frozen in liquid nitrogen. Ultrathin cryosections were prepared using a cryo-ultramicrotome EM FC6 (Leica). Double immunogold labeling was made sequentially at room temperature. Briefly, for the first labeling, sections were quenched with 50 mmol/L NH4Cl and saturated with PBS, 1% BSA, 0.1% NGS, then incubated with anti-GM130 (1:25) revealed with a goat anti-mouse IgG conjugated to 10 nm gold particles (1:25, BBI, Cardiff, UK). After postfixation in PBS, 1% glutaraldehyde, sections were similarly processed for the second labeling, using anti-LAMP1 (1:50) revealed with a goat anti-rat IgG conjugated to 15 nm gold particles (1:25, BBI). Sections were contrasted with uranyl acetate mixed with methylcellulose.
Analyses were performed on a JEOL 1200EXII transmission electron microscope (JEOL, Croissy-sur-Seine, France) equipped with an Eloise Megaview camera controlled by Analysis Pro 3.1 software (Eloise, Roissy-CDG, France).
Turnover of Long-Lived Proteins
Neuron cultures were incubated with L-leucine-free medium supplemented with 2% FCS for 1 hour to deplete the endogenous L-leucine pool before the addition of L-leucine-free medium/FCS containing 5μCi/ml 3H-labeled L-leucine (GE Health care, Aulnay-sous-Bois, France). After 24 hours of incubation, the medium was replaced with the normal medium containing excess of L-leucine or with medium containing 250 nmol/L rapamycin. Aliquots of medium were collected at the indicated times (8 hours, 24 hours, 48 hours). Proteins were precipitated in 20% TCA and stored at 4°C. Upon acquisition, all samples were centrifuged at 15,000g for 5 minutes at 4°C. Supernatants were collected, the pellets were washed twice with cold 20% TCA, then all supernatants were pooled and the total radioactivity was measured by scintillation counting. Cells were then washed with PBS and solubilized in extraction buffer. An aliquot of solubilized cells was used to determine total protein concentration. Relative proteolysis was determined by normalizing TCA soluble radioactivity in the medium to protein concentration from the solubilized cells.
Western Blots
After intracardiac perfusion with PBS, mouse brains were removed and cortical fragments (3 mm3) were collected. Total proteins were extracted from brain fragments or cultured cortical neurons homogenized in lysis buffer (0.1% SDS,1% NP-40, 0.2% deoxycholate, 0.15 M NaCl, 50 mmol/L Tris pH7.8, protease inhibitors). Membrane proteins were extracted with a CNMCS Compartmental extraction kit (Biochain, Hayward, US) according to manufacturer recommendations. Endo-H (Biolabs, Beverly, US) and peptide N-glycosidase F (PNGase F) were used according to manufacturer recommendations. Proteins were analyzed by Western blot, as previously described.25 Signals were revealed with anti-LC3B (1:2000), anti-LAMP1 (1:5000), mouse mAb anti-IDUA (clone ID1A, 1:100, a gift from Dr. D. Brooks, Women’s and Children’s Hospital, Adelaide, Australia), anti-GM130 (1:500), rabbit polyclonal anti-actin (1:500, Abcam), mouse mAb anti-actin (1:5000, Sigma) or goat polyclonal anti-CD56 (1:5000, AbCys, Paris, France) antibodies followed by appropriate horseradish peroxydase-coupled secondary antibodies (AbCys, 1:10000) and SuperSignal West Dura chemiluminescent substrate (Pierce, Rockford, US). Signal intensities were measured with the LAS-1000CH Luminescent photofilm LTD system, piloted by the IR-LAS-Pro software (Fujifilm Life Science, Courbevoie, France). Specific signal value is relative to actin signal in the same lane.
Statistical Analysis
Statistics were performed using the SPSS software (SPSS). The assumption that the values follow normal distribution was verified by the Shapiro-Wilk’s test. Nonparametric tests were used when normal distribution was not assumed.
Results
Storage Vesicles Are Distinct from Lysosomes
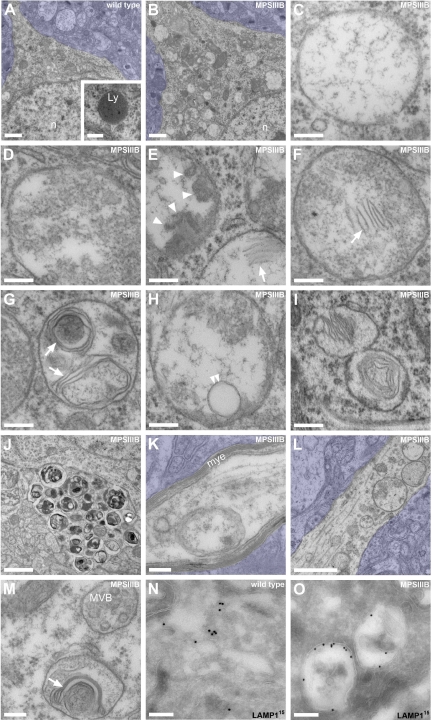
We and others previously reported behavioral manifestations reminiscent of symptoms in children with Sanfilippo syndrome in MPSIIIB mice from the age of 4–5 months.21,29,34 In our colony, life expectancy is 12 ± 2 months (n = 91). Cortical atrophy, neuronal loss and decreased synaptic density are absent until end-stage disease.35 Vesicular distension in brain cells is prominent at 4–5 months and worsens progressively with age. Electron microscopy showed that the morphology and size of storage vesicles accumulating in neuronal soma and processes of 8-month-old MPSIIIB mouse cortical neurons differed from normal lysosomes (Figure 1, A–N). Individual cells accumulated numerous vesicles with highly heterogeneous content, ranging from clear amorphous material, internal debris, internal vesicles, isolated membranes fragments, dense aggregates, multilamellar structures, or even densely packed stacks of membranes, called zebra bodies. Storage vesicles diameter varied from less than 0.1 μm to several micrometers. Immunogold electron page 10.microscopy detected the lysosomal marker LAMP1 in storage vesicle limiting membranes (Figure 1O).
Figure 1.
Vesicles accumulating in the MPSIIIB mouse rostral cortex are distinct from lysosomes. A–M: Rostral cortex fragments of 8-month-old wild-type (A) or MPSIIIB mice (B–M) were processed for electron microscopy on ultrathin sections. Low magnification images of wild-type (A) and MPSIIIB neurons (B) show polymorphic distended vesicles in the affected cell. A normal lysosome is shown in A (inset). High magnifications of vesicles present in MPSIIIB neuron soma (C–I) show intraluminal electro-dense fibrillar (C), granular (D), globular (arrowheads in E), or multilamellar (arrows in E–G) materials. A sequestered vesicle is visible in H (double arrowhead). Images consistent with vesicle fusion or scission were observed (I). Vesicles with zebra bodies are shown in I. Enlargement of a process filled with dense vesicular bodies is visible in J. Vesicles also accumulate in myelinated axons (K) and in dendrites (L–M). A dendritic multivesicular body with normal aspect juxtaposed to a vesicle loaded with multilamellar material (arrow) is shown (M). Azure stains tissue beyond cell limits. Ly, lysosome; mye, myelin; MVB, multivesicular body; n, nucleus. Scale bars, 1 μm (A, B, J, and L); 0.2 μm (C–H, K, and M); 0.4 μm (I). N and O: Ultrathin cryosections from wild-type (N) or MPSIIIB (O) rostral cortex were immunostained for LAMP1 using 15 nm gold particles, revealing LAMP1 in small vesicles (N) or in limiting membranes of most distended vesicles (O). Scale bars, 0.2 μm.
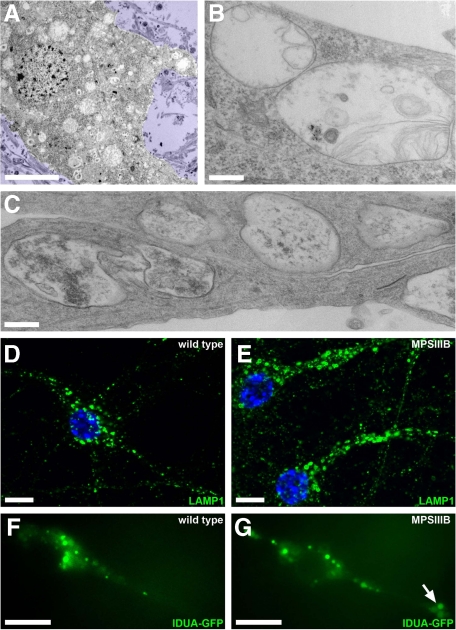
The accumulation of LAMP1 vesicles was also observed in primary cultures of embryonic MPSIIIB cortical neurons. The ultrastructure of storage vesicles accumulating in cultured cell bodies and neurites after 7 days in vitro was reminiscent of brain pathology (Figure 2A–C). In neurites, LAMP1 vesicles were easily distinguished from each other by fluorescent microscopy (Figure 2, D and E), allowing reliable scoring according to size. Increased vesicle density in MPSIIIB neurites, when compared with wild type, concerned all size categories (see Supplemental Figure 1A at http://ajp.amjpathol.org). Clearance after genetic correction with a lentiviral vector coding for the missing enzyme indicated that vesicle accumulation was related to the enzyme defect (see Supplemental Figure 1B at http://ajp.amjpathol.org). Storage vesicles were similarly observed in living neurons stained with LysoTracker, a marker of acidic vesicles (not shown), or with IDUA-GFP, a marker of late endosomes and lysosomes.31 Combined labeling showed that LAMP1 vesicles were also stained with Lysotracker and contained IDUA-GFP. The movements of vesicles stained with IDUA-GFP were recorded in living neuron processes (see Supplemental Videos 1, 2, and 3 at http://ajp.amjpathol.org). These vesicles were larger in MPSIIIB (0.71 ± 0.03 μm2, n = 295) than in wild type (0.54 ± 0.02 μm2, n = 185, P < 0.01) neurons, and more frequently immobile (72.2% versus 49.7%). The dynamics of vesicles accumulating in MPSIIIB neuron processes therefore differed from normal lysosomes. Large vesicles (>1 μm2, 16.2% of total) were always static in MPSIIIB neurons and often formed clusters in neurites (38% of vesicles located in MPSIIIB neurites belong to clusters, Figure 2F and G). Clusters hampered displacement of smaller mobile vesicles (see Supplemental Video 3 at http://ajp.amjpathol.org), which marked longer arrests than in wild type neurons (16.52 ± 1.30 seconds versus 11.65 ± 1.25 seconds, P < 0.001). Thus, storage vesicles clogged in MPSIIIB neuron processes affected vesicular transport. Velocity, direction changes and arrest frequency of mobile vesicles were otherwise comparable in MPSIIIB and wild type neurites, suggesting that the disease did not affect the transport machinery involved in normal lysosome dynamics.
Figure 2.
Storage vesicles accumulate in MPSIIIB mouse cortical neuron cultures. A–C: MPSIIIB embryonic cortical neuron cultures were fixed at day 7 to 16 and processed for electron microscopy. Many polymorphic vesicles are visible in the cell soma (A and B) and prolongations (C). Scale bars, 5 μm (A); 0.3 μm (B and C). D and E: Cultures of wild-type or MPSIIIB embryonic cortical neurons were fixed at day 12 and immunolabeled with anti-LAMP1 antibodies (green). Nuclei were counterstained in blue. Abundant and often distended LAMP1-positive vesicles are visible in MPSIIIB neuronal processes. Scale bars, 10 μm. F and G: Cultures of wild-type or MPSIIIB embryonic cortical neurons were exposed to a lentivirus vector coding for the fluorescent lysosomal hydrolase IDUA-GFP at day 7 and examined at day 12 by fluorescent time-lapse microscopy (see Supplemental Videos 1, 2, and 3 at http://ajp.amjpathol.org). A cluster of IDUA-GFP–positive vesicles is visible in a MPSIIIB neuronal process (arrow). Scale bars, 10 μm.
Storage Vesicles Are Not Related to the Endocytic Pathway
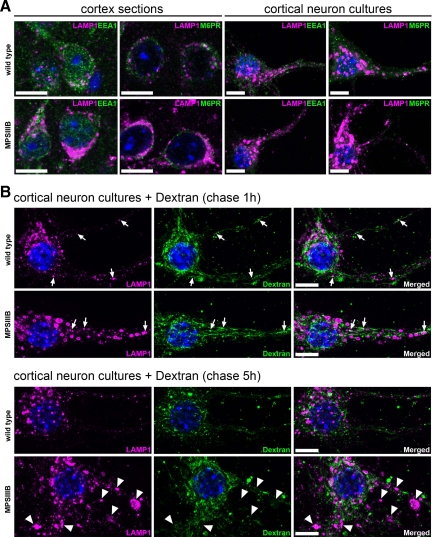
In the MPSIIIB mouse brain, we did not observed LAMP1-positive vesicles expressing the early endosomal antigen 1 (EEA1), or the mannose-6-phosphate receptor (M6PR), a marker of late endosomes and MVBs (Table 1, Figure 3A, see Supplemental Figure 2 at http://ajp.amjpathol.org). In the neurites of cultured MPSIIIB cortical neurons, only few LAMP1 vesicles reacted with anti-EEA1, and none reacted with anti-M6PR antibodies.
Table 1.
Summary of Investigated Markers
| Rostral cortex sections
|
Cortical neuron cultures
|
|||
|---|---|---|---|---|
| Similar staining patterns in MPSIIIB and wild type | Colocalization with LAMP1 in MPSIIIB* | Similar staining patterns in MPSIIIB and wild type | More frequent colocalization with LAMP1 in MPSIIIB than in wild type† | |
| ER markers | ||||
| Calnexin | ND | ND | Yes | No |
| ER-to-Golgi markers | ||||
| ERGIC53 | Yes | No | Yes | No |
| COPII (sec23) | Yes | Yes‡ | Yes | Yes (×1.6) |
| betaCOP | Yes | No | Yes | No |
| Golgi markers | ||||
| GM130 | No§ | Yes | No§ | Yes (×2.3) |
| Syntaxin 5 | ND | ND | Yes¶ | No |
| Giantin | Yes | No | Yes¶ | No |
| p115 | Yes | No | Yes¶ | No |
| GRASP65 | Yes | No | Yes¶ | No |
| GS15 | ND | ND | Yes¶ | No |
| Golgin 97 | Yes | No | Yes¶ | No (×1.01) |
| Endocytic markers | ||||
| EEA1 | Yes | No | Yes | No (×0.83) |
| M6PR | Yes | No | Yes | No |
| Autophagic markers | ||||
| LC3 | Yes | No | Yes | No (×0.85) |
| p62/SQSTM1 | Yes | No | Yes | No |
| Other markers | ||||
| Ubiquitin | No∥ | No | Yes | No |
| ScMAS | No** | No | Yes | No |
| Heparan sulfate | No†† | No | ND | ND |
| GM3 ganglioside | ND | ND | No‡‡ | No |
| Alpha synuclein | Yes | No | ND | ND |
| Neurofilament | Yes | No | ND | ND |
ND, not done.
Analyses were performed on confocal brain sections.
Colocalizations observed on apotome sections of cultured neurons were confirmed on confocal sections. MPSIIIB to wild-type ratios of doubly positive vesicles are indicated when colocalization was observed.
Images were not conclusive on brain sections due to high-density punctuated staining.
Broader signal extending to cell processes in MPSIIIB.
Typical perinuclear Golgi staining, signal intensity and surface were not quantified.
Dispersed aggregates in MPSIIIB rostral cortex.
Clustered signals in neuronal soma in MPSIIIB rostral cortex lamina II/III.
Diffuse signal in neuronal soma in MPSIIIB rostral cortex.
More abundant signal in MPSIIIB.
Figure 3.
LAMP1-positive storage vesicles are not related to the endocytic pathway. A: Parasagittal rostral cortex sections of 8-month-old wild-type or MPSIIIB mice (cortex sections) and cultures of wild-type or MPSIIIB cortical neurons (cortical neuron cultures) were stained with anti-LAMP1 antibodies (in purple) in combination with anti-EEA1 (in green) or anti-M6PR (in green). Nuclei were counterstained in blue. Merged confocal images of cortical sections, or apotome images of cultured neurons, show that LAMP1 signals do not colocalize with endosomal vesicle markers. For single labeling, see Supplemental Figure 2 at http://ajp.amjpathol.org. Scale bars, 10 μm. B: Ten-day-old cortical neuron cultures established from wild-type or MPSIIIB mouse embryos were pulsed with biotinylated dextran for 5 minutes then fixed after 1 hour or 5 hours of chase. Biotinylated dextran was revealed with fluorescent streptavidin (in green) and cells were simultaneously immunolabeled with anti-LAMP1 antibodies (in purple). Nuclei were counterstained in blue. Apotome views show small doubly positive signals in soma and neurites (chase 1 hour, arrows). Note the absence of dextran signal in distended LAMP1-positive vesicles that are well visible in MPSIIIB neurites (chase 5 hours, arrowheads). Scale bars, 10 μm.
We examined dextran uptake, a compound internalized via nonspecific fluid-phase endocytosis and addressed to lysosomes. Cultured cortical neurons were briefly pulsed with biotinylated dextran, chased for various periods of time and stained for LAMP1. Dextran was internalized in small-size vesicles (<0.5 μm2, Figure 3B). Equivalent colocalization coefficients in small vesicles doubly stained for dextran and LAMP1 were scored in wild type (8.0 ± 1.0%) and MPSIIIB neurons (8.4 ± 1.8%) after a one-hour chase. Noticeably, examination after 5 hours did not detect biotinylated dextran in large-size (>1 μm2) LAMP1-positive storage vesicles, which were easily identified in neurites. This result suggested that large storage vesicles were not accessible to internalized dextran during the period of observation.
Storage Vesicles Are Not Related to the Macroautophagy Pathway
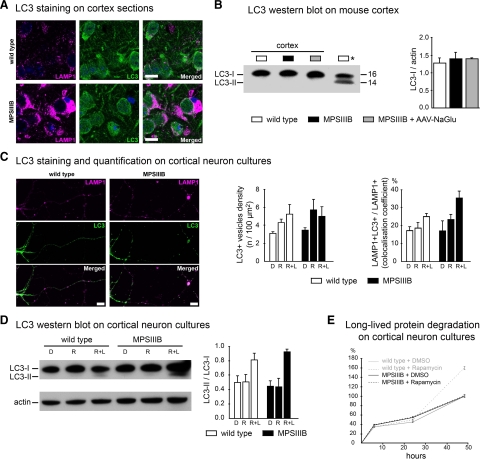
The detection of intracellular vesicles stained with anti-LC3 antibodies, a marker of autophagosomes, is an indicator of activated autophagy.36 We observed diffuse nonvesicular LC3 staining in wild-type and MPSIIIB mouse cortical sections (Figure 4A, Table 1). LAMP1-positive storage vesicles were negative for LC3. Activation of autophagy and translocation of LC3 to autophagosomes is associated with an increased proportion of the lipidated versus unlipidated forms of LC3.36 Western blots of cortical tissue extracts from wild-type or MPSIIIB mice did not detect LC3-II signal (Figure 4B). These results suggested that macroautophagy was not activated in the MPSIIIB mouse cortex.
Figure 4.
Macroautophagy is not altered in MPSIIIB mouse cortical neurons. A: Parasagittal sections of the rostral cortex of 8-month-old wild-type or MPSIIIB mice were immunolabeled for LAMP1 (in purple) and LC3 (in green). Nuclei were counterstained in blue. Confocal views show intense LAMP1 staining of MPSIIIB sections and similar diffuse LC3 staining pattern in wild-type and MPSIIIB. Scale bars, 10 μm. B: LC3-I to LC3-II conversion was analyzed by Western blots. Proteins were extracted from the rostral cortex of 8-month-old wild-type mice, untreated MPSIIIB mice, or MPSIIIB mice treated by intracerebral injection of AAV-NaGlu vector (MPSIIIB+AAV-NaGlu). The diagram on the right shows equivalent LC3-I signal intensities with respect to actin signals (means ± SEM from three mice). LC3-II was not detected in cortical tissue extracts in contrast to a positive control (Asterisk) provided by wild-type mouse cortical neuron cultures treated with rapamycin (250 nmol/L) and leupeptin (50 μmol/L) for 48 hours. C: Cultures of wild-type or MPSIIIB cortical neurons were treated at day 6 with vehicle (DMSO: D), or rapamycin (250 nmol/L, R), or rapamycin (250 nmol/L) and leupeptin (50 μmol/L, R + L). Neurons were fixed at day 8 and immunolabeled for LAMP1 (in purple) and LC3 (in green). Apotome views of neurons treated with rapamycin and leupeptin show LC3-positive vesicles in wild-type and MPSIIIB neurites (middle row). Scoring of LC3-positive vesicles along neurites in basal conditions and after drug treatment did not show significant difference between wild-type and MPSIIIB neurons (left diagram, means ± SEM from three independent cultures; P > 0.1, t-test). LC3 and LAMP1 signals did not colocalize more frequently in MPSIIIB than in wild-type cultures whatever the treatment (right diagram, means ± SEM from three independent cultures; P > 0.1, t-test). Scale bars, 10 μm. D: Proteins extracted at day 8 from neuron cultures maintained in basal conditions (DMSO: D), or incubated with rapamycin (R), or with rapamycin and leupeptin (R + L) for days 6 to 8 were analyzed by Western blots to examine LC3-I to LC3-II conversion. Signal quantification shows equivalent LC3-II/LC3-I ratios for each condition in wild-type and MPSIIIB neurons (means ± SEM from three independent cultures). E: The degradation of long-lived proteins was examined by metabolic labeling of cultured neurons with 3H-leucine. Ratios of 3H counts in culture supernatant (corresponding to the amount of degraded proteins) to cell pellet (corresponding to the number of labeled cells) were determined at day 8 in wild-type (gray lines) or MPSIIIB (black lines) neurons cultures, either in basal conditions (DMSO) or after treatment with rapamycin (250 nmol/L) for 48 hours (means ± SEM from three independent cultures, P > 0.1 for all time-points, Mann–Whitney test).
In cortical neuron cultures, LC3 showed both diffuse and punctate patterns in neurites (Figure 4C). The density of LC3-positive vesicles and the frequency of LAMP1 and LC3 colocalization (Figure 4C) were equivalent in wild-type and MPSIIIB neurites (Table 1). Western blots revealed comparable LC3-II/LC3-I ratios in cell protein extracts (Figure 4D). The turnovers of long-lived proteins were also comparable (Figure 4E). These results suggested that in basal condition activation of macroautophagy was equally low in MPSIIIB and wild-type neuron cultures.
Rapamycin is an inducer of autophagy, which stimulates autophagosome formation, as indicated by the translocation of LC3 to vesicles, and by the increased proportion of LC3-II. Rapamycin effects on autophagosome formation are more apparent in the presence of leupeptin, a drug that inhibits proteolysis after autophagosome fusion with lysosomes.36 In wild-type neuron cultures, rapamycin (250 nmol/L) slightly increased LC3 vesicle density in neurites (Figure 4C), did not modify LC3-II/LC3-I ratios, except when leupeptin (50 μmol/L) was added (Figure 4D), and increased long-lived protein proteolysis (Figure 4E). Rapamycin effects on MPSIIIB neurons were not uniform. Effects on LC3-positive vesicles and LC3-II/LC3-I ratios were similar to wild-type neurons (Figure 4, C and D), indicating that rapamycin stimulated autophagosome formation in MPSIIIB efficiently, as in wild-type neurons. However, the drug did not increase the proteolysis of long-lived proteins (Figure 4E), suggesting limited proteolytic capacities in MPSIIIB neuron lysosomes.
Accumulating Substances Are Not Associated with Storage Vesicles
Staining of cortical sections, or cortical neuron cultures for HS, ScMAS, ubiquitin, or GM3 gangliosides differed between wild-type and MPSIIIB mice (Table 1, see Supplemental Figure 3 at http://ajp.amjpathol.org). On brain sections, HS staining showed diffuse signal in MPSIIIB neuron soma, which was not observed in wild-type controls but which did not colocalize with LAMP1. As previously described,22 ScMAS formed aggregates in perinuclear areas of MPSIIIB neurons in cortical laminae II/III, which were not observed in wild-type cortex but which partially colocalized with LAMP1. ScMAS signal outside perinuclear aggregates was distinct from LAMP1. Ubiquitin formed aggregates throughout the MPSIIIB cortex, which were not stained with anti-LAMP1 antibodies. In cortical neuron cultures, ScMAS and ubiquitin staining was similar in wild-type and MPSIIIB neurons, despite more intense LAMP1 staining in MPSIIIB. Cultured MPSIIIB neurons showed more abundant and more intense GM3 ganglioside staining in vesicles located in cell soma, some of them being positive for LAMP1. In contrast, anti-GM3 antibodies did not stain LAMP1-positive storage vesicles in neurites.
Storage Vesicles Express Markers of the Early Secretory Pathway
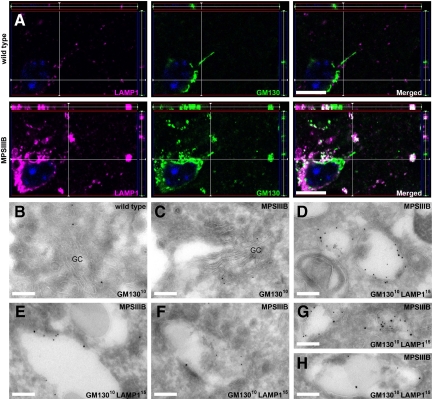
We next examined whether storage vesicles were related to the early steps of lysosome biogenesis. Mouse cortical sections were stained for sec23, a COPII coat component of vesicles exiting ER to reach the ER-to-Golgi intermediate compartment (ERGIC), ERGIC53, Golgi matrix protein 130 (GM130), p115, GRASP65, giantin and β-COP, all markers of ERGIC and Golgi complex,37 and Golgin97, a marker of the median and trans-Golgi network.38 With the exception of GM130, staining patterns were comparable in MPSIIIB and wild-type mouse brain and colocalization with LAMP1 was equally rare (Table 1). GM130 signal was more intense and broader in MPSIIIB rostral cortex (Figure 5A). Whereas GM130 and LAMP1 signals rarely colocalized in wild-type neuron soma or processes, colocalization was frequently observed in soma and processes of MPSIIIB neurons (Figure 5A). Immunoelectron microscopy confirmed juxtaposition of LAMP1 and GM130 signals in storage vesicle limiting membranes and showed that doubly stained vesicles were ninefold more frequent in MPSIIIB than in wild-type neurons (4.45 ± 0.69 vs. 0.53 ± 0.22 doubly stained vesicle per neuron profile, n = 20, P < 0.00001, t-test, Figure 5B-H). All examined MPSIIIB neuron profiles contained at least one doubly stained vesicle.
Figure 5.
LAMP1-positive storage vesicles express GM130 in the MPSIIIB mouse brain. A: Parasagittal rostral cortex sections of 8-month-old wild-type or MPSIIIB mice were immunolabeled for LAMP1 and GM130. Confocal immunofluorescence (LAMP1 in purple, GM130 in green, nuclei counterstained in blue) shows frequent colocalization of LAMP1 and GM130 signals (in white) in MPSIIIB cortical sections. Scale bars, 10 μm. B–H: Immunogold electron microscopy was performed on ultra-thin cryosections of the rostral cortex of 8-month-old wild-type (B) or MPSIIIB (C–H) mice. Sections were labeled with anti-GM130 antibodies revealed with 10-nm gold particles alone or in combination with anti-LAMP1 antibodies revealed with 15-nm gold particles. Golgi structures decorated with GM13010 nm particles are shown (B and C). Distended vesicles in which the limiting membrane is doubly labeled with LAMP115 nm and GM13010 nm were frequently observed in MPSIIIB neurons (D–H). GC, Golgi complex. Scale bars, 0.2 μm.
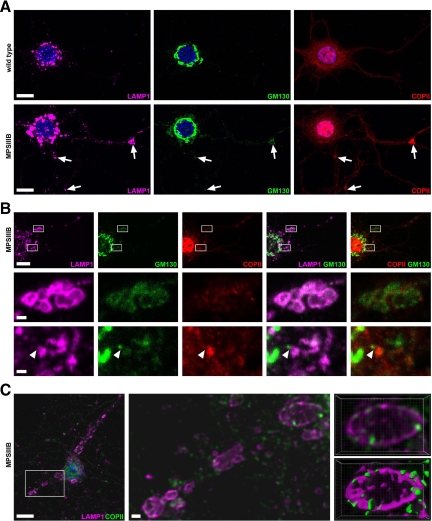
In addition to the markers examined in the brain, we stained cortical neuron cultures with antibodies against calnexin, a marker of ER, syntaxin 5 and Gs15, two IC and Golgi complex markers. With the exception of GM130, all markers showed comparable staining patterns in MPSIIIB and wild-type mouse neurons (Table 1). In wild-type neurons, GM130 signal most frequently surrounded the entire nucleus, sometime extending in neurites (79.3 ± 0.8% of neurons, n = 490). Less frequently, GM130 signal was polarized and exclusively juxtanuclear (20.7 ± 0.8%). This latter aspect was even less frequent in MPSIIIB neurons (11.0 ± 0.6%, n = 570, P < 0.001, χ2). Colocalization of LAMP1 and GM130 signals in the same vesicle was more frequently detected in MPSIIIB (15.2%, n = 828) than in wild-type neurites (4.9%, n = 570, Figure 6, A and B, P < 0.001, χ2). Genetic correction of deficient neurons with a lentivirus vector coding for the missing enzyme rescued the wild-type phenotype (18.3 ± 1.0% neurons with polarized and juxtanuclear GM130 signal; n = 327, P < 0.001, χ2; 2.0% of doubly-positive vesicles, n = 551). Although Sec23-COPII perinuclear staining pattern was comparable in MPSIIIB and wild-type neurons, it was more frequently associated with LAMP1 in MPSIIIB neurites (36%, n = 506, versus 23% of wild-type, n = 253, Figure 6, A and B). Deconvoluted confocal images revealed colocalization of LAMP1 and sec23-COPII in vesicle limiting membranes (Figure 6C). Triple staining for GM130, COPII, and LAMP1 detected the three signals in the same vesicles in MPSIIIB neurites but not in wild type neurites (Figure 6, A and B). Codetection of these three markers in limiting membranes suggested that storage vesicles accumulating in MPSIIIB neurons were related to pre-Golgi compartments, or Golgi complex.
Figure 6.
LAMP1-positive storage vesicles express GM130 and sec23COPII in MPSIIIB mouse neuron cultures. Wild-type or MPSIIIB cortical neurons cultures were fixed at day 11 and immunolabeled for LAMP1 (in purple), GM130 (in green), and sec23-COPII (in red in A and B, in green in C). Nuclei were counterstained in blue. A: Low-magnification confocal immunofluorescence views show vesicles triply positive for LAMP1, GM130, and COPII (arrows) in MPSIIIB neurites. Scale bars, 10 μm. B: Areas boxed in the low magnification view of a MPSIIIB neuron (upper row) were examined at high magnification. Middle row shows a cluster of distended vesicles costained with the three antibodies. Bottom row shows images consistent with the incorporation of a GM130-positive vesicle in a LAMP1-COPII-positive vesicular complex (arrowhead). Scale bars, 10 μm in upper row; 1 μm in middle and bottom rows. C: A low magnification confocal image of a MPSIIIB neuron costained with LAMP1 and COPII antibodies is shown on the left. A high-magnification deconvoluted image of the boxed area is shown in the middle. Contiguous staining for LAMP1 and COPII is visible in vesicle limiting membranes. Three-dimensional views of the vesicle in the upper right corner are shown in the right panels, without (top) or with (bottom) isosurface treatment. Scale bars, 10 μm in left image; 1 μm in middle image.
Pre-Golgi Vesicular Trafficking Is Efficient
We examined ER to Golgi complex trafficking by treating cortical neuron cultures with brefeldin A (BFA). This selective inhibitor of the GTP-exchange factor for ADP-ribosylation factor 1 (Arf1) blocks COPI coat assembly and induces Golgi complex collapse.39,40 Twenty-five minutes after BFA treatment (1.25 μg/ml), giantin became localized to ER structures in both wild-type and MPSIIIB neurons, indicating efficient ER to Golgi complex transport. Golgi collapse was however slightly delayed in MPSIIIB neurons, indicating reduced sensitivity to BFA (Figure 7A). ER to Golgi complex trafficking was also investigated in neural cells by using ts045VSV-G-YFP, a well-known fluorescent protein misfolded and blocked in ER at nonpermissive temperature (40°C), which exits ER at permissive temperature (32°C) to successively reach COPII-coated vesicles, Golgi complex and the cell surface.41 Comparable evolution of ts045VSV-G-YFP staining patterns in wild type and MPSIIIB cells after 15, 17 and 60 minutes of temperature switch, indicated that VSV-G transport from ER to Golgi complex was not altered in MPSIIIB cells (see Supplemental Figure 4A at http://ajp.amjpathol.org). Equivalent LAMP1/ts045VSV-G-YFP co-localized area relative to cell surface after 15 minutes of temperature switch (3.4 ± 0.5% in wild-type cells versus 2.9 ± 0.4% in MPSIIIB cells, n = 15) indicated that the amount of newly synthetized LAMP1 protein exiting ER was not higher in MPSIIIB cells than in wild type cells (see Supplemental Figure 4B at http://ajp.amjpathol.org).
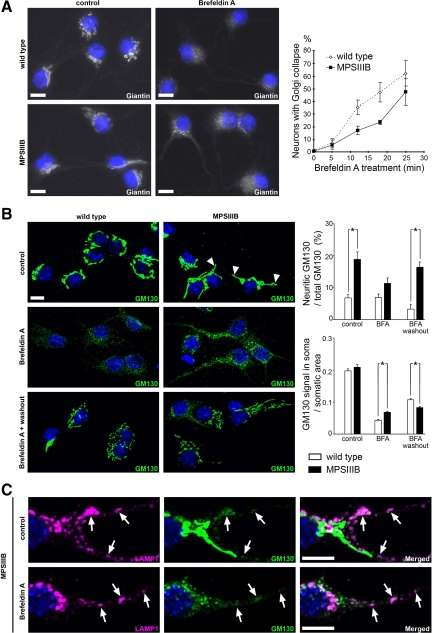
Figure 7.
MPSIIIB neurons are partially resistant to brefeldin A. Wild-type or MPSIIIB cortical neuron cultures were treated with brefeldin A (BFA, 1.25 μg/ml) at day 7, fixed, and immunolabeled for giantin (white in A), GM130 (green in B and C), or LAMP1 (purple in C). A: Dense perinuclear giantin staining of Golgi complex (left column) gives rise to diffuse signal spread over neuronal bodies and processes after 25 minutes of BFA treatment (middle column). Slower kinetics of disappearance of perinuclear giantin staining in MPSIIIB neurons, when compared with wild type, is shown in the right diagram (means ± SEM from signals quantified in at least 300 neurons per condition in three independent cultures, P > 0.1, t-test). Scale bars, 10 μm. B: In control conditions, MPSIIIB neurons show increased GM130 signal in proximal neurites (upper row, arrowheads, upper diagram on the right). After 25 minutes in the presence of BFA, GM130 staining was dispersed over the cell, but still more intense in MPSIIIB soma than in wild type (middle row, diagrams on the right). Reconstitution of Golgi structures was visible 4 hours after washout (lower row). GM130 signal in neurites was 5.2-fold more intense in MPSIIIB than in wild type (upper diagram on the right). Values are means ± SEM from signals quantified in at least 90 neurons per condition in two independent cultures. *P < 0.0001 (t-test). Scale bars, 10 μm. C: BFA did not affect LAMP1 staining pattern (left column), whereas perinuclear GM130 staining became dispersed (middle column). Vesicles doubly positive for LAMP1 and GM130 (right column), which are well visible in neurites, were not affected by BFA treatment (arrows). Scale bars, 10 μm.
Golgi Complex Alterations in MPSIIIB Neurons
When compared with wild-type, cultured MPSIIIB neurons showed increased GM130 signal, which predominated in neurites, and the proportion of neurons with GM130 staining in neurites was twice higher (Figure 7B). BFA treatment induces the redistribution of GM130 to punctate structures corresponding to dispersed Golgi matrix proteins.42 Although Golgi complex dispersion was observed in both MPSIIIB and wild-type BFA-treated neurons, remnant nondispersed GM130 signal was 1.6-fold more abundant in MPSIIIB (Figure 7B). Colabeling for GM130 and LAMP1 revealed that doubly stained vesicles in MPSIIIB neurites were resistant to BFA (Figure 7C), showing increased density on treatment (8.8 ± 1.3 per 100 μm2 in treated, versus 5.8 ± 0.8 in untreated MPSIIIB neurites). Four hours after BFA washout, quantification of GM130 staining signal in soma indicated equally efficient reconstitution of Golgi structures in wild type and MPSIIIB neurons (Figure 7B). Consistently with long-range trafficking from the soma,43 reconstitution was not yet visible 4 hours after washout in wild-type neurites. In contrast, GM130 staining was increased in MPSIIIB neurites. Signal was fourfold more intense than in wild-type neurites, and the proportion of neurons with GM130 signal in neurites was again twice higher.
Golgi complex resident mannosidase II modifies saccharide chains born on most glycoproteins transiting through Golgi complex, conferring resistance to endoglycanase H digestion. As an exception to this rule, despite efficient addressing to lysosomes LAMP1 produced in cultured wild-type neurons conserved sensitivity to endoglycanase H (see Supplemental Figure 5A at http://ajp.amjpathol.org). Thus, LAMP1 digestion with endoglycanase H could not be used as an indicator of storage vesicle trafficking through Golgi complex. Because IDUA-GFP was addressed to these vesicles, we examined the susceptibility of endogenous IDUA to endoglycanase H. As well as in wild-type neurons, IDUA produced in MPSIIIB neurons was resistant to endoglycanase H, indicating that oligosaccharide chains generated on IDUA molecules present in the lumen of storage vesicles underwent trimming by Golgi mannosidase II (see Supplemental Figure 5B at http://ajp.amjpathol.org).
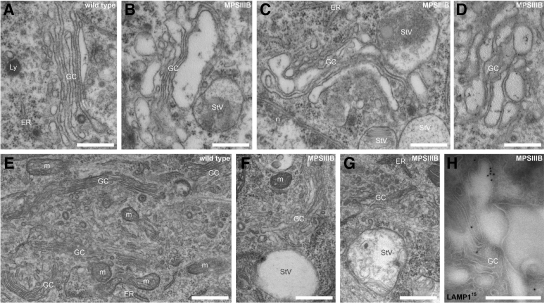
We assumed that observations made in MPSIIIB neurons could be related to Golgi complex alterations. This assumption was confirmed by ultrastructural studies in vivo and in vitro. Golgi complex profiles showed constant features in wild type neurons (n = 32). Ribbons typically consisted of 4 to 6 stacks of cisterna. Small vesicles (≤0.12 μm2), presumably representing fenestrated cisterna, were visible at one side. Thin saccules (≤0.15 μm wide), from which coated vesicles frequently budded, formed the other side (Figure 8, A–G). This typical asymmetrical architecture was not observed in MPSIIIB neurons (n = 49). Although Golgi ribbons were not more frequently fragmented, their architecture was disorganized to various extends (Figure 8). Milder modifications consisted in wider cisterna forming internal bulbs and giving rise to dilated saccules, especially at the rims (Figures 5C and 8, C and D). An increased amount of surrounding storage vesicles, the lumen of which was clear or crowded with internal membranes or other materials, was observed. Cisterna were sometime in continuity with LAMP1-positive storage vesicles (Figure 8H).
Figure 8.
Disorganisation of Golgi architecture in MPSIIIB neurons. Rostral cortex sections (A–D and H), or day 7 cortical neuron cultures (E–G) were processed for ultrastructural morphology (A–G), or immunogold labeling for LAMP1 on cryosections (H). Pictures show typical Golgi complex in wild-type (A and E) and MPSIIIB neuronal soma (B, C, D, F, and G). Cisterna distensions adjacent to storage vesicles are well visible in MPSIIIB neurons. A distended Cisterna decorated with LAMP1 antibodies emerging from a Golgi tubular structure, which is also decorated with anti-LAMP1 antibodies, is shown in H. ER, endoplasmic reticulum; GC, Golgi complex; Ly, lysosome; m, mitochondria; n, nucleus; StV, storage vesicle. Scale bars, 0.5 μm.
Discussion
Our studies of storage vesicles in MPSIIIB neurons showed that they were not associated with major engorgement or dysfunction in the endo-lysosomal or macroautophagy pathways. They did not exhibit markers of these compartments, and were not actively connected with these pathways. Alternatively, our results provide evidence for disease-related alterations involving Golgi complex, which give rise to abnormal vesicular structures related to storage vesicles.
Storage Vesicles Are Not Related to Post-Golgi Compartments
Consistent with previous descriptions,21,22 electron microscopy revealed abundant storage vesicles in MPSIIIB mouse rostral cortex neurons. Although to a lesser extent, they were also observed in pure MPSIIIB mouse cortical neuron cultures, indicating that their generation was cell autonomous and effective in short-term culture conditions. Storage vesicles displayed markers of lysosomes, including LAMP1 in the limiting membrane and the lysosomal hydrolase IDUA in the acidic lumen. They differed from lysosomes with respect to large size, heterogeneous content, and absence of movements in neurites. Disappearance after correction of the genetic defect indicated that storage vesicle accumulation was disease related. Storage vesicles clustering in neuronal processes induced distensions reminiscent of spheroid bodies,44,45 which formed bottlenecks affecting transport along neurites.
We did not find evidence for a strong association of LAMP1-positive storage vesicles with primary or secondary storage products. LAMP1 signal in neurites was most frequently not associated with HS, ScMAS, or ubiquitin in the MPSIIIB mouse rostral cortex, in contrast with previous observations in the soma of medial entorhinal or somatosensory cortical neurons.22 Increased LAMP1 signal in cultured MPSIIIB neurons was not associated with increased ScMAS and ubiquitin staining. Consistent with the notion that secondary stored products are distributed in separate structures,46 GM3 staining was frequently not associated with LAMP1, especially in MPSIIIB neurites. We concluded that primary and/or secondary storage products mostly accumulate in structures distinct from LAMP1-positive storage vesicles.
Fusion of lysosomes with late endosomes or amphisomes is the terminal step of endocytosis and macroautophagy, respectively. Previous studies in Nieman-Pick C disease47 and in the C. elegans model of mucolipidosis IV,48 two LSDs in which vesicular trafficking is affected in post-Golgi compartments, showed that material internalized by fluid phase endocytosis was addressed to storage lesions. In contrast, our results indicate that storage vesicles were not accessible to internalized cargo in MPSIIIB neurons. Dynamic exploration of macroautophagy did not reveal activation, or blockage in MPSIIIB neurons. Results from these dynamic assays combined to the absence of detection of markers of early endosomes, late endosome, or autophagosomes indicate that storage vesicles in MPSIIIB neurons are not connected to these pathways.
Storage Vesicles Are Related to Pre-Golgi or Golgi Compartments
Vesicles transporting LAMP1 and bearing the COPII coatomer and/or the tethering protein GM130 are produced at ER exit sites in normal neurons and further delivered to Golgi complex. We actually detected doubly positive LAMP1/GM130, or LAMP1/COPII vesicles in wild-type neurons. However, doubly stained vesicles were ninefold more abundant in MPSIIIB than in wild-type neurons, and triply stained vesicles were exclusively detected in MPSIIIB neurons. Examination of LAMP1-positive vesicle exit from ER using ts045VSV-G indicated that they were not produced more actively in MPSIIIB neurons.
Several results demonstrate that at least a fraction of LAMP1-GM130 doubly positive vesicles was unique to MPSIIIB neurons, and related to storage vesicles. i) GM130 signal was increased in MPSIIIB neurons, and part of the additional signal corresponded to tubulo-vesicular structures, sometime expanding in neurites to form enlarged vesicles. These vesicles were positive for LAMP1, resistant to BFA treatment and cleared after genetic correction. ii) Proteins participating in the formation of GM130 complexes, like p115, syntaxin 5, or GRASP65, were not associated with LAMP1 more frequently in MPSIIIB than in wild-type neurons, indicating that they were not assembled with GM130 molecules present on LAMP1vesicles. iii) Electron microscopy revealed alterations of Golgi architecture. These alterations were reminiscent, albeit not identical, to the phenotype induced by the silencing GM130.49 They were very similar to the alterations induced by the silencing of cargo receptors, which also induces partial resistance of cis-Golgi to BFA.50 Consistent with physical connections with Golgi cisterna, the glycoprotein IDUA contained in storage vesicles was resistant to endoglycanase H digestion. Normal kinetics of ts045VSV-G trafficking, and almost normal response to BFA of LAMP1-negative Golgi complex structures suggested that traffic in pre-Golgi compartments and Arf1-mediated control were essentially preserved.39,40 Taken together, these results favor the hypotheses that storage vesicles in MPSIIIB neurons were related to defects affecting Golgi complex.
Golgi Complex Disorganization May Result from Disabled GM130 Tethering Complexes
GM130 is part of a tethering complex that functions in membrane transport and in cis-terna stacking at the Golgi complex level.51,52 It mediates homotypic tethering and coalescence of tubulovesicular carriers of the intermediate compartment, facilitating their incorporation into stacks,53 and of cis-Golgi cisterna, ensuring proper architectural organization of Golgi ribbon.49 Because Golgi complex organization was altered in MPSIIIB neurons, we assume that GM130 control was at least in part defective. Defects may result from the generation of structures that contain GM130 but lack the other components of the tethering complex, which are necessary for function, like GRAPS65, p115 or syntaxin 5. The COPII protein sec23 can be associated with this complex through its interaction with the cargo transporter p24.54 The presence of other proteins known to directly interact with GM130, like Rab1,55 YSK1,56 or ZFPL1,57 was not examined. While total GM130 amounts were not increased in MPSIIIB neurons, as shown by Western blot, lack of proper interaction with molecular partners may affect GM130 complex-mediated vesicle tethering and fusion, leading to the generation a dead end compartment unable to interact and/or fuse with adjacent compartments, and which progressively accumulates. Material contained in these vesicles certainly participates in storage. We ignore at this stage how the abnormal production of HS oligosaccharides, which is the initial pathogenic event in MPSIIIB, could induce the formation of defective tethering-fusion complexes. One hypothesis is that high sugar concentration in vesicular lumen alters interactions of glycoprotein cargos with their transport receptors. Defective interaction may affect proper assembly of coat proteins and/or tethering-fusion complexes.37
Disorganisation of Golgi Compartments May Account for Neuropathology Development
Our observations documented the impact of storage vesicles on trafficking along neurites. The formation of vesicle clusters likely participates in neurite dystrophy and spheroid body formation, a pathology previously described in LSDs and in other neurodegenerative disorders.45,58 Alteration of axono-dendritic transport has deleterious consequences on neuron function and survival.
Modification of glycosphingolipid metabolism is likely related to neuropathology in LSDs.59 Mechanisms responsible for the secondary accumulation of GM2 and GM3 gangliosides are not understood in MPSIIIB. Because glycosphingolipid synthesis depends on proper trafficking through Golgi stacks,60 our observation of Golgi complex disorganization in MPSIIIB could be relevant to this phenomenon.
In addition to its role on Golgi ribbon formation, GM130 plays cardinal roles in cell division, migration, and polarization that are possibly altered in MPSIIIB cells. It regulates mitotic spindle formation61 and centrosome morphology and function.62 Through the recruitment of the γ-tubulin-interacting protein AKAP450 on the Golgi membranes, it controls microtubule nucleation at the Golgi complex,63 and determines cell migration and cargo transport to the periphery.56 Exploration of these functions in MPSIIIB and other LSDs might shed light on the cascades of events leading to neurodegeneration.
Acknowledgments
We thank Stéphane Blanchard for his help in the preparation of lentivirus vectors. We thank Pr. Elizabeth Neufeld for the gift of the MPSIIIB mouse strain, Dr. David Palmer for anti-ScMAS antibodies, Dr. Chiara Zurzolo for ts045VSV-G-YFP plasmid DNA. We are grateful to Emmanuelle Perret and Pascal Roux at the Imagopole/Plateforme d’Imagerie Dynamique de l’Institut Pasteur for their help with confocal microscopy and image processing, and to Anne Nosjean for her help in immunohistochemistry and macroautophagy studies.
Footnotes
Address reprint requests to Jean Michel Heard, M.D., Ph.D., Unité Rétrovirus et Transfert Génétique, INSERM U622, Department of Neuroscience, Institut Pasteur 28 rue du Dr Roux, 75015 Paris, France. E-mail: jmheard@pasteur.fr.
Supported by Agence Nationale de la Recherche (grant ANR-08-MNP-023) and the Conny-Maeva Foundation.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Walkley SU. Pathogenic cascades in lysosomal disease-Why so complex? J Inherit Metab Dis. 2009;32:181–189. doi: 10.1007/s10545-008-1040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 2002;21:1289–1300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- Pacheco CD, Kunkel R, Lieberman AP. Autophagy in Niemann–Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- Takamura A, Higaki K, Kajimaki K, Otsuka S, Ninomiya H, Matsuda J, Ohno K, Suzuki Y, Nanba E. Enhanced autophagy and mitochondrial aberrations in murine G(M1)-gangliosidosis. Biochem Biophys Res Commun. 2008;367:616–622. doi: 10.1016/j.bbrc.2007.12.187. [DOI] [PubMed] [Google Scholar]
- Woloszynek JC, Coleman T, Semenkovich CF, Sands MS. Lysosomal dysfunction results in altered energy balance. J Biol Chem. 2007;282:35765–35771. doi: 10.1074/jbc.M705124200. [DOI] [PubMed] [Google Scholar]
- Raben N, Baum R, Schreiner C, Takikita S, Mizushima N, Ralston E, Plotz P. When more is less: excess and deficiency of autophagy coexist in skeletal muscle in Pompe disease. Autophagy. 2009;5:111–113. doi: 10.4161/auto.5.1.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- Winslow AR, Rubinsztein DC. Autophagy in neurodegeneration and development. Biochim Biophys Acta. 2008;1782:723–729. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Schiavo G. Liaisons dangereuses: autophagy, neuronal survival and neurodegeneration. Curr Opin Neurobiol. 2008;18:504–515. doi: 10.1016/j.conb.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Pelled D, Lloyd-Evans E, Riebeling C, Jeyakumar M, Platt FM, Futerman AH. Inhibition of calcium uptake via the sarco/endoplasmic reticulum Ca2+-ATPase in a mouse model of Sandhoff disease and prevention by treatment with N-butyldeoxynojirimycin. J Biol Chem. 2003;278:29496–29501. doi: 10.1074/jbc.M302964200. [DOI] [PubMed] [Google Scholar]
- Lloyd-Evans E, Pelled D, Riebeling C, Bodennec J, de-Morgan A, Waller H, Schiffmann R, Futerman AH. Glucosylceramide and glucosylsphingosine modulate calcium mobilization from brain microsomes via different mechanisms. J Biol Chem. 2003;278:23594–23599. doi: 10.1074/jbc.M300212200. [DOI] [PubMed] [Google Scholar]
- Ginzburg L, Futerman AH. Defective calcium homeostasis in the cerebellum in a mouse model of Niemann-Pick A disease. J Neurochem. 2005;95:1619–1628. doi: 10.1111/j.1471-4159.2005.03534.x. [DOI] [PubMed] [Google Scholar]
- Bodennec J, Pelled D, Riebeling C, Trajkovic S, Futerman AH. Phosphatidylcholine synthesis is elevated in neuronal models of Gaucher disease due to direct activation of CTP:phosphocholine cytidylyltransferase by glucosylceramide. FASEB J. 2002;16:1814–1816. doi: 10.1096/fj.02-0149fje. [DOI] [PubMed] [Google Scholar]
- Walkley SU. Secondary accumulation of gangliosides in lysosomal storage disorders. Semin Cell Dev Biol. 2004;15:433–444. doi: 10.1016/j.semcdb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Tessitore A, del PMM, Sano R, Ma Y, Mann L, Ingrassia A, Laywell ED, Steindler DA, Hendershot LM, d'Azzo A. GM1-ganglioside-mediated activation of the unfolded protein response causes neuronal death in a neurodegenerative gangliosidosis. Mol Cell. 2004;15:753–766. doi: 10.1016/j.molcel.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Wei H, Kim SJ, Zhang Z, Tsai PC, Wisniewski KE, Mukherjee AB. ER and oxidative stresses are common mediators of apoptosis in both neurodegenerative and non-neurodegenerative lysosomal storage disorders and are alleviated by chemical chaperones. Hum Mol Genet. 2008;17:469–477. doi: 10.1093/hmg/ddm324. [DOI] [PubMed] [Google Scholar]
- Sano R, Annunziata I, Patterson A, Moshiach S, Gomero E, Opferman J, Forte M, d'Azzo A. GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca(2+)-dependent mitochondrial apoptosis. Mol Cell. 2009;36:500–511. doi: 10.1016/j.molcel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Yu WH, Rozengurt N, Zhao HZ, Lyons KM, Anagnostaras S, Fanselow MS, Suzuki K, Vanier MT, Neufeld EF. Mouse model of Sanfilippo syndrome type B produced by targeted disruption of the gene encoding alpha-N-acetylglucosaminidase. Proc Natl Acad Sci USA. 1999;96:14505–14510. doi: 10.1073/pnas.96.25.14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazantsev S, Yu WH, Zhao HZ, Neufeld EF, Ohmi K. Lysosomal accumulation of SCMAS (subunit c of mitochondrial ATP synthase) in neurons of the mouse model of mucopolysaccharidosis III B. Mol Genet Metab. 2007;90:393–401. doi: 10.1016/j.ymgme.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmi K, Kudo LC, Ryazantsev S, Zhao HZ, Karsten SL, Neufeld EF. Sanfilippo syndrome type B, a lysosomal storage disease, is also a tauopathy. Proc Natl Acad Sci USA. 2009;106:8332–8337. doi: 10.1073/pnas.0903223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausseil J, Desmaris N, Bigou S, Attali R, Corbineau S, Vitry S, Parent M, Cheillan D, Fuller M, Maire I, Vanier M, Heard JM. Early neurodegeneration progresses independently of microglial activation by heparan sulfate in the brain of mucopolysaccharidosis IIIB mice. PloSONE. 2008;3:e2296. doi: 10.1371/journal.pone.0002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitry S, Ausseil J, Hocquemiller M, NBigou S, Coura R, Heard JM. Synaptophysin expression defect in cortical neurons of the mouse model of mucopolysaccharidosis type IIIB. Mol Cell Neurosci. 2009;41:8–18. doi: 10.1016/j.mcn.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Neufeld EF, Muenzer J. Scriver ALB CR, Sly W S, Valle D, editors. New York: McGraw-Hill,; The mucopolysaccharidoses. 2001:p. pp. 3421–3452. [Google Scholar]
- Hadfield MG, Ghatak NR, Nakoneczna I, Lippman HR, Myer EC, Constantopoulos G, Bradley RM. Pathologic findings in mucopolysaccharidosis type IIIB (Sanfilippo’s sydnrome B). Arch Neurol. 1980;37:645–650. doi: 10.1001/archneur.1980.00500590069012. [DOI] [PubMed] [Google Scholar]
- Tamagawa K, Morimatsu Y, Fujisawa K, Hara A, Taketomi T. Neuropathological study and chemico-pathological correlation in sibling cases of Sanfilippo syndrome type B. Brain Dev. 1985;7:599–609. doi: 10.1016/s0387-7604(85)80008-5. [DOI] [PubMed] [Google Scholar]
- Cressant A, Desmaris N, Verot L, Brejot T, Froissart R, Vanier MT, Maire I, Heard JM. Improved behavior and neuropathology in the mouse model of Sanfilippo type IIIB disease after adeno-associated virus-mediated gene transfer in the striatum. J Neurosci. 2004;24:10229–10239. doi: 10.1523/JNEUROSCI.3558-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DN, Bayliss SL, Westlake VJ. Batten disease and the ATP synthase subunit c turnover pathway: raising antibodies to subunit c. Am J Med Genet. 1995;57:260–265. doi: 10.1002/ajmg.1320570230. [DOI] [PubMed] [Google Scholar]
- Chen F, Vitry S, Hocquemiller M, Desmaris N, Ausseil J, Heard JM. alpha-l-Iduronidase transport in neurites. Mol Genet Metab. 2006;87:349–358. doi: 10.1016/j.ymgme.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Franceschini I, Vitry S, Padilla F, Casanova P, Tham TN, Fukuda M, Rougon G, Durbec P, Dubois-Dalcq M. Migrating and myelinating potential of neural precursors engineered to overexpress PSA-NCAM. Mol Cell Neurosci. 2004;27:151–162. doi: 10.1016/j.mcn.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells [see comments]. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Heldermon CD, Hennig AK, Ohlemiller KK, Ogilvie JM, Herzog ED, Breidenbach A, Vogler C, Wozniak DF, Sands MS. Development of sensory, motor and behavioral deficits in the murine model of Sanfilippo syndrome type B. PLoS ONE. 2007;2:e772. doi: 10.1371/journal.pone.0000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquemiller M, Vitry S, Bigou S, Bruyere J, Ausseil J, Heard JM. GAP43 overexpression and enhanced neurite outgrowth in mucopolysaccharidosis type IIIB cortical neuron cultures. J Neurosci Res. 2010;88:202–213. doi: 10.1002/jnr.22190. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, Stephens DJ. Assembly, organization, and function of the COPII coat. Histochem Cell Biol. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Tai G, Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol Biol Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Hirschberg K, Lippincott-Schwartz J. Secretory pathway kinetics and in vivo analysis of protein traffic from the Golgi complex to the cell surface. FASEB J. 1999;13 Suppl 2:S251–S256. doi: 10.1096/fasebj.13.9002.s251. [DOI] [PubMed] [Google Scholar]
- Puri S, Linstedt AD. Capacity of the golgi apparatus for biogenesis from the endoplasmic reticulum. Mol Biol Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- Walkley SU, Baker HJ, Rattazzi MC, Haskins ME, Wu JY. Neuroaxonal dystrophy in neuronal storage disorders: evidence for major GABAergic neuron involvement. J Neurol Sci. 1991;104:1–8. doi: 10.1016/0022-510x(91)90208-o. [DOI] [PubMed] [Google Scholar]
- McGlynn R, Dobrenis K, Walkley SU. Differential subcellular localization of cholesterol, gangliosides, and glycosaminoglycans in murine models of mucopolysaccharide storage disorders. J Comp Neurol. 2004;480:415–426. doi: 10.1002/cne.20355. [DOI] [PubMed] [Google Scholar]
- Lachmann RH, te Vruchte D, Lloyd-Evans E, Reinkensmeier G, Sillence DJ, Fernandez-Guillen L, Dwek RA, Butters TD, Cox TM, Platt FM. Treatment with miglustat reverses the lipid-trafficking defect in Niemann-Pick disease type C. Neurobiol Dis. 2004;16:654–658. doi: 10.1016/j.nbd.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, Hauri HP. The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol Biol Cell. 2008;19:1976–1990. doi: 10.1091/mbc.E07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez C, Garcia-Mata R, Hauri HP, Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J Biol Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- Seemann J, Jokitalo EJ, Warren G. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol Biol Cell. 2000;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A, Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating JR, Martens GJ. The p24 family and selective transport processes at the ER-Golgi interface. Biol Cell. 2009;101:495–509. doi: 10.1042/BC20080233. [DOI] [PubMed] [Google Scholar]
- Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis–Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, Gettemans J, Barr FA. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CF, Ghanekar Y, Frost L, Diao A, Morrison D, McKenzie E, Lowe M. ZFPL1, a novel ring finger protein required for cis-Golgi integrity and efficient ER-to-Golgi transport. EMBO J. 2008;27:934–947. doi: 10.1038/emboj.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace EA, Rabiner CA, Busciglio J. Characterization of neuronal dystrophy induced by fibrillar amyloid beta: implications for Alzheimer’s disease. Neuroscience. 2002;114:265–273. doi: 10.1016/s0306-4522(02)00241-5. [DOI] [PubMed] [Google Scholar]
- Walkley SU, Vanier MT. Secondary lipid accumulation in lysosomal disease. Biochim Biophys Acta. 2009;1793:726–736. doi: 10.1016/j.bbamcr.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL. Mechanisms of transport through the Golgi complex. J Cell Sci. 2009;122:443–452. doi: 10.1242/jcs.032581. [DOI] [PubMed] [Google Scholar]
- Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Sutterlin C. The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]