Abstract
The CpG island methylator phenotype (CIMP-high, CIMP1) is a distinct phenotype associated with microsatellite instability (MSI) and BRAF mutation in colon cancer. Recent evidence suggests the presence of KRAS mutation-associated CIMP subtype (CIMP-low, CIMP2). We used cluster analysis, principal component analysis (PCA), and structural equation modeling (SEM), a novel strategy, to decipher the correlation structure of CpG island hypermethylation. Using a database of 861 colon and rectal cancers, DNA methylation at 16 CpG islands [CACNA1G, CDKN2A (p16/ink4a), CHFR, CRABP1, HIC1, IGF2, IGFBP3, MGMT, MINT-1, MINT-31, MLH1, NEUROG1, p14 (CDKN2A/arf), RUNX3, SOCS1, and WRN] was quantified by real-time PCR. Tumors were categorized into three groups: Group 1 with wild-type KRAS/BRAF (N = 440); Group 2 with mutant KRAS and wild-type BRAF (N = 308); and Group 3 with wild-type KRAS and mutant BRAF (N = 107). Tumors with mutant KRAS/BRAF (N = 6) were excluded. In unsupervised hierarchical clustering analysis, all but six markers (CACNA1G, IGF2, RUNX3, MGMT, MINT-1, and SOCS1) were differentially clustered with CIMP-high and CIMP-low according to KRAS and BRAF status. In SEM, the correlation structures between CIMP, locus-specific CpG island methylation, and MSI differed according to KRAS and BRAF status, which was consistent with PCA results. In conclusion, KRAS and BRAF mutations appear to differentially influence correlation structure of CpG island methylation. Our novel data suggest two distinct perturbations, resulting in differential locus-specific propensity of CpG methylation.
Epigenetic alterations are important mechanisms in human carcinogenesis. A number of tumor suppressor genes are aberrantly silenced by promoter CpG island methylation in colorectal cancer. A subset of colorectal cancers exhibit widespread promoter CpG island methylation, ie, the CpG island methylator phenotype (CIMP),1,2 which is a major cause of microsatellite instability (MSI) in sporadic colorectal cancer through epigenetic silencing of a mismatch repair gene MLH1.3,4,5 Colorectal cancers with high-level CIMP (ie, CIMP-high) show distinct characteristics including associations with older age, female sex, proximal tumor location, the presence of synchronous colorectal cancer,6 BRAF mutation, wild-type TP53, stable chromosomes, and inactive WNT/β-catenin CTNNB1,3,7,8,9,10,11,12 independent of MSI.4,5,13 Experimental and human correlative data suggest that DNMT3B (DNA methyltransferase 3B) may contribute to locus-specific CpG island methylation and CIMP-high in colorectal tumors.14,15,16 Therefore, the best preventive or therapeutic approaches for CIMP-high tumors will be very different from those for non–CIMP-high tumors.
In contrast to CIMP-high, the existence of CIMP-low as a distinct phenotype remains controversial. CIMP-low in colorectal cancer has been associated with KRAS mutation,11,17,18 low-level locus-specific methylation,19 and poor prognosis.20 CIMP-low and other proposed subtypes [“CIMP2”10 and “intermediate-methylation epigenotype (IME)”21] appear to share overlapping characteristics. Experimental evidence suggests contribution of KRAS or BRAF activation to locus-specific CpG island methylation,22,23 although there are conflicting data.24 However, the interrelationship between KRAS and BRAF mutations and various methylation markers have not been deciphered using a large number of tumors. Because KRAS and BRAF are commonly-mutated human oncogenes, it is of particular interest to understand how KRAS and BRAF mutations relate to locus-specific CpG island methylation in cancer cells, which may confer drug sensitivity or resistance.
In this study, we used a database of 861 colorectal cancers and biostatistical techniques including cluster analysis, principal component analysis (PCA) and structural equation modeling (SEM), the latter of which is a novel strategy to decipher the correlation structure of CpG island methylation in cancer. We have found that the correlation structure of locus-specific methylation varies according to KRAS and BRAF mutational status. The consistent data by cluster analysis, PCA and SEM increase confidence in our conclusions. Our novel data suggest a possible role of KRAS and BRAF mutation status in modifying propensity for CpG island methylation in a locus-specific manner during carcinogenic process.
Materials and Methods
Study Group
We used the databases of two large prospective cohort studies; the Nurses’ Health Study (NHS, N = 121,700 women followed since 1976),25 and the Health Professionals Follow-up Study (HPFS, N = 51,500 men followed since 1986).25 A subset of cohort participants developed colorectal cancer during prospective follow-up. Previous studies on the cohorts have described baseline characteristics of cohort participants and incident colorectal cancer cases and confirmed that our colorectal cancers were well representative as a population-based sample.25 We collected paraffin-embedded tissue blocks from hospitals where participants had undergone resections of primary colorectal cancers. Among our cohort studies, there was no significant difference in demographic features between cases with tissue available and those without available tissue.25 Based on availability of adequate tissue specimens and results (on CpG island methylation and KRAS and BRAF sequencing), a total of 861 colorectal cancer cases were included in this study. Histopathological features including tumor differentiation, mucinous features, and signet ring cells were examined by a pathologist (S.O.). Poor differentiation was defined as the presence of <50% glandular area. Considering the importance of MSI testing in screening for Lynch syndrome, we provide Supplemental Table 1 (available at http://ajp.amjpathol.org) to describe cases with MSI-high and unmethylated MLH1 in tumor. We have previously analyzed all of the 861 tumors for statuses of MSI, KRAS, BRAF, DNMT3B and the 16 CpG island methylation markers.4,15 However, our current study represents new analyses of principal component analysis (PCA) and structural equation modeling (SEM) on the tumor database. To our knowledge, an application of SEM to correlation structure analysis of CpG island methylation has not been done by any research group. Informed consent was obtained from all study subjects. Tissue collection and analyses were approved by the Harvard School of Public Health and Brigham and Women’s Hospital Institutional Review Boards.
DNMT3B Immunohistochemistry, KRAS and BRAF Sequencing, and Microsatellite Instability (MSI) Analysis
DNMT3B immunohistochemistry was performed as previously described.15 DNA was extracted from paraffin tissue, and PCR-Pyrosequencing targeted for KRAS codons 12 and 13,26 and BRAF codon 600 were performed.17 MSI status was determined using D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487.27 MSI-high was defined as the presence of instability in ≥30% of the markers, and MSI-low/microsatellite stability (MSS) as 0 to 29% unstable markers.
Real-Time PCR (MethyLight) for Quantitative DNA Methylation Analysis
Sodium bisulfite treatment on DNA and subsequent real-time PCR (MethyLight28) was validated as previously described.29 We quantified DNA methylation in 16 CpG islands,4 including the 5 CpG island methylator phenotype (CIMP)-specific promoters (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1)3 and 11 other CpG islands [CDKN2A (p16), CHFR, CRABP1, HIC1, IGFBP3, MGMT, MINT-1, MINT-31, MLH1, p14 (CDKN2A/ARF), and WRN].4,19 We used the database of 861 colorectal cancer cases with available molecular data, including methylation data on the 16 preselected CpG islands. COL2A1 was used to normalize for the amount of template bisulfite-converted DNA.29 Primers and probes were previously described.3,4 The PCR condition was initial denaturation at 95°C for 10 minutes followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. The percentage of methylated reference (PMR; ie, degree of methylation) at a specific locus was calculated by dividing the GENE:COL2A1 ratio of template amounts in a sample by the GENE:COL2A1 ratio of template amounts in SssI-treated human genomic DNA (presumably fully methylated) and multiplying this value by 100. While methylation positivity at each locus for CIMP classification was set as PMR ≥4 as previously validated,29 PMR value at each locus was used as a continuous variable to assess locus-specific methylation. CIMP-high was defined as methylation positivity at ≥6 of 8 methylation markers (CACNA1G, CRABP1, CDKN2A (p16), IGF2, MLH1, NEUROG1, RUNX3, and SOCS1), CIMP-low as 1 to 5 of the 8 markers, and CIMP-0 as 0/8 methylated markers.4
Statistical Analysis
We used the SAS system for Windows (version 9.1.3; SAS Institute, Cary, NC) for the all statistical analyses. The statistical significance level was set at P = 0.05. The χ2 test or the F-test was used to assess statistical significance of differences in the categorical variables or the continuous variables, respectively, between tumor groups. The correlations among the 16 CpG island methylation markers, CIMP (high versus low versus CIMP-0), and MSI score were studied by Spearman’s rank correlation, adjusted for age, sex, body mass index (BMI), family history of colorectal cancer (present versus absent), disease stage (ordinal; I, II, III and IV), tumor location (proximal versus distal), and DNMT3B. The Spearman’s rank correlation coefficient was designated as ρ. To estimate the correlation coefficient for either KRAS or BRAF mutation eliminating the effect of the other mutation, the correlation coefficients among the 16 CpG islands and KRAS (or BRAF) mutation were calculated after exclusion of BRAF (or KRAS) mutation.
To explore the latent correlation structure of CIMP and locus-specific CpG island methylation, we performed principal component analysis (PCA) and cluster analysis. We used the VARCLUS procedure of SAS (SAS Institute, Cary, NC) to cluster the 15 CpG island markers excluding MLH1 for each group of colorectal cancers. We excluded MLH1 from the cluster analysis because MLH1 methylation has been known to be in the intermediate path from CIMP-high to MSI-high.3,4,5 In contrast to standard clustering methods, which divide observations into clusters based on similar values, VARCLUS divides numeric variables based on similar correlation structure (ie, pairwise correlation values). This can be regarded as an analogy of the k-means method using correlation coefficients, and the resulting clusters are similar to those created by factor analysis with oblique rotation. However, the VARCLUS produces disjoint clusters as opposed to fuzzy clusters produced by factor analysis which have more straightforward biological interpretations. To assess the reliability of clustering results, we calculated approximated unbiased P value (AUp) via multiscale bootstrap resampling,30 which is commonly used in phylogenetic analysis. We generated 1000 bootstrap samples to estimate AUp. P value of a cluster is a value between 0 and 1, which indicates how strong the cluster is supported by data.
Structural equation modeling (SEM), a statistical technique for testing and estimating causal relationships,31 was used to assess the independent effects of the CIMP status on CpG island methylation and MSI status. SEM. has three advantages. First, it can simultaneously model both latent and observed variables. Second, it allows for effect decomposition; because the total causal effect is the sum of the values of all of the paths between two variables, an indirect effect is determined by subtracting the direct effect from the total. The third advantage is a path diagram, which graphically explains the hypothetical causality. Because we could not observe the true state of CIMP-high, we regarded methylation level of CpG island markers as a surrogate for the true state of CIMP-high. The correlations and path coefficients of MLH1, the other CpG island markers, and MSI were computed while controlling for possible confounding effects (sex, age, BMI, family history, tumor location, and DNMT3B status). We used DNMT3B as a potential confounder according to the aim of this study; our goal was to explore the relationships of KRAS and BRAF mutations with CIMP and CpG island methylation markers. The maximum likelihood method was used to estimate the parameters of SEM, and approximated t-tests were conducted to test individual path coefficients, following the t distribution with degrees of freedom of n − k − 1 (n: sample size; k: the parameters of the path model). We calculated asymptotic 95% confidence interval (CI) for the direct effects and bias-corrected and accelerated bootstrap CIs for the total and indirect effects based on 1000 bootstrap replicates. In addition, we performed multiple group analysis to test invariance of the structure across groups. Of the several fit indices available for the overall model test, we selected the goodness-of-fit index (GFI), the adjusted GFI (AGFI), and the root mean square error of approximation (RMSEA) as the fit index for the overall model test. The data probably do not fit the model if the AGFI is negative or much larger than 1 and RMSEA is 0.10. Good models have an RMSEA of 0.05 or less.
Results
CpG Island Methylation, KRAS and BRAF Status in Colorectal Cancers
Using the database of 861 clinically and molecularly-annotated colorectal cancers in the two prospective cohort studies, we examined DNA methylation in the 5 CpG islands (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1) selected as CIMP (CpG island methylator phenotype) surrogate markers,3 and in 11 other CpG islands [CDKN2A (p16), CHFR, CRABP1, HIC1, IGFBP3, MGMT, MINT-1, MINT-31, MLH1, p14 (CDKN2A/ARF) and WRN]. We determined CIMP status using eight markers, including the five markers in the Weisenberger panel,3 CDKN2A, CRABP1, and MLH1, as previously validated.4 To evaluate the effect of KRAS and BRAF mutations on the correlation structure of the 16 individual CpG island methylation markers, we categorized tumors into four groups (Table 1): Group 1 with both wild-type KRAS and BRAF; Group 2 with mutant KRAS and wild-type BRAF; Group 3 with wild-type KRAS and mutant BRAF; and Group 4 with both mutant KRAS and BRAF. Because the sample size of Group 4 was very small (N = 6), we made comparisons between Groups 1, 2, and 3 (N = 855), excluding Group 4 for further analyses.
Table 1.
Clinical, Pathological, and Molecular Characteristics of Colorectal Cancer Cases According to KRAS and BRAF Mutational Status
| Clinical, pathological, or molecular feature | All cases | Group 1 (Wild-type KRAS/BRAF) | Group 2 (Mutant KRAS, wild-type BRAF) | Group 3 (Wild-type KRAS, mutant BRAF) | Group 4 (Mutant KRAS/BRAF) | P value |
|---|---|---|---|---|---|---|
| Total N | 861 | 440 | 308 | 107 | 6 | |
| Sex | <0.0001 | |||||
| Male, n (%) | 385 (45) | 202 (43) | 158 (51) | 24 (22) | 1 (17) | |
| Female, n (%) | 476 (55) | 238 (54) | 150 (49) | 83 (78) | 5 (83) | |
| Mean age ± SD, years | 66.6 ± 8.4 | 66.0 ± 8.6 | 67.1 ± 8.3 | 67.6 ± 7.3 | 64.9 ± 6.5 | 0.075 |
| Mean body mass index ± SD | 26.6 ± 4.7 | 26.6 ± 4.6 | 26.4 ± 4.6 | 26.5 ± 5.4 | 29.2 ± 4.1 | 0.94 |
| Family history of colorectal cancer | 0.98 | |||||
| Absent, n (%) | 649 (75) | 331 (75) | 232 (75) | 81 (75) | 5 (83) | |
| Present, n (%) | 212 (25) | 109 (25) | 76 (25) | 26 (25) | 1 (17) | |
| Smoking status | 0.70 | |||||
| Never, n (%) | 342 (40) | 182 (42) | 113 (38) | 45 (42) | 2 (33) | |
| Current or past, n (%) | 506 (60) | 254 (58) | 186 (62) | 62 (58) | 4 (67) | |
| Tumor location | <0.0001 | |||||
| Proximal colon, n (%) | 365 (44) | 137 (33) | 138 (47) | 87 (83) | 3 (50) | |
| Distal colon, n (%) | 270 (33) | 168 (40) | 88 (30) | 13 (13) | 1 (17) | |
| Rectum, n (%) | 189 (23) | 116 (27) | 67 (23) | 4 (3.8) | 2 (33) | |
| Tumor stage | 0.0007 | |||||
| I, n (%) | 194 (26) | 112 (29) | 65 (24) | 16 (16) | 1 (17) | |
| II, n (%) | 247 (32) | 128 (33) | 75 (27) | 40 (39) | 4 (67) | |
| III, n (%) | 222 (29) | 107 (28) | 91 (33) | 23 (23) | 1 (17) | |
| IV, n (%) | 101 (13) | 36 (9.4) | 42 (15) | 23 (23) | 0 | |
| Tumor grade | <0.0001 | |||||
| Low, n (%) | 766 (91) | 399 (92) | 275 (95) | 78 (73) | 4 (67) | |
| High, n (%) | 78 (9) | 33 (8) | 14 (5) | 29 (27) | 2 (33) | |
| CIMP status* | <0.0001 | |||||
| CIMP-0, n (%) | 400 (46) | 255 (58) | 136 (44) | 7 (7) | 2 (33) | |
| CIMP-low, n (%) | 333 (39) | 150 (34) | 156 (51) | 25 (23) | 2 (33) | |
| CIMP-high, n (%) | 128 (15) | 35 (8.0) | 16 (5) | 75 (70) | 2 (33) | |
| MSI status | <0.0001 | |||||
| MSI-low/MSS, n (%) | 727 (85) | 385 (89) | 285 (93) | 54 (50) | 3 (50) | |
| MSI-high, n (%) | 124 (15) | 48 (11) | 20 (6.6) | 53 (50) | 3 (50) | |
| DNMT3B expression | 0.0005 | |||||
| (−), n (%) | 645 (85) | 327 (87) | 225 (86) | 69 (71) | 3 (60) | |
| (+), n (%) | 115(15) | 49 (13) | 36 (14) | 29 (28) | 2 (40) |
The percentage number (%) indicates the proportion of cases with a specific clinical, pathological, or molecular feature within each group. Groups 1–3 were used in further analyses; Group 4 was excluded from further analysis.
CIMP-high was defined as ≥6/8 methylated markers using the 8-marker CIMP panel [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1].
CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stability; SD, standard deviation.
Correlations between the 16 CpG Island Markers and KRAS and BRAF Mutations
We assessed the relationship between KRAS/BRAF mutations and each of the 16 CpG island methylation markers after adjusting for clinical features (Table 2; for unadjusted data, see Supplemental Table 2 at http://ajp.amjpathol.org). Except for MGMT, all of the other 15 CpG island markers positively correlated with BRAF mutation (adjusted ρ, 0.15 to 0.59; P < 0.001). Only WRN showed a modest positive correlation with KRAS mutation (adjusted ρ, 0.17; P < 0.0001). Three other markers (IGFBP3, MINT-1, and p14) were positively but weakly associated with KRAS mutation (adjusted ρ, 0.10 to 0.11; P < 0.034), while MLH1 was negatively correlated with KRAS mutation (adjusted ρ, −0.14; P = 0.0012). These results implied that not all methylation markers exhibited similar correlations with BRAF or KRAS mutation, and methylation patterns appeared to be locus-specific.
Table 2.
Correlation between Each of the 16 CpG Island Methylation Markers and KRAS or BRAF Mutation
| CpG island methylation marker |
KRAS mutation
|
BRAF mutation
|
||
|---|---|---|---|---|
| ρ* | P value | ρ* | P value | |
| CACNA1G | 0.0094 | 0.83 | 0.40 | <0.0001 |
| CDKN2A (p16) | −0.012 | 0.77 | 0.40 | <0.0001 |
| CHFR | −0.028 | 0.52 | 0.30 | <0.0001 |
| CRABP1 | 0.0096 | 0.83 | 0.40 | <0.0001 |
| HIC1 | −0.022 | 0.61 | 0.14 | 0.0042 |
| IGF2 | −0.0076 | 0.86 | 0.43 | <0.0001 |
| IGFBP3 | 0.097 | 0.026 | 0.19 | 0.0001 |
| MGMT | 0.059 | 0.18 | 0.0046 | 0.93 |
| MINT-1 | 0.10 | 0.017 | 0.26 | <0.0001 |
| MINT-31 | 0.014 | 0.75 | 0.34 | <0.0001 |
| MLH1 | −0.14 | 0.0012 | 0.38 | <0.0001 |
| NEUROG1 | 0.066 | 0.13 | 0.47 | <0.0001 |
| p14 (CDKN2A/ARF) | 0.11 | 0.015 | 0.28 | <0.0001 |
| RUNX3 | 0.0046 | 0.92 | 0.59 | <0.0001 |
| SOCS1 | −0.092 | 0.035 | 0.21 | <0.0001 |
| WRN | 0.17 | <0.0001 | 0.37 | <0.0001 |
A ρ value represents Spearman’s partial correlation coefficient adjusted for age, sex, BMI, smoking status, tumor stage, and location.
CIMP-High Cluster and CIMP-Low Cluster According to KRAS and BRAF Mutation Status
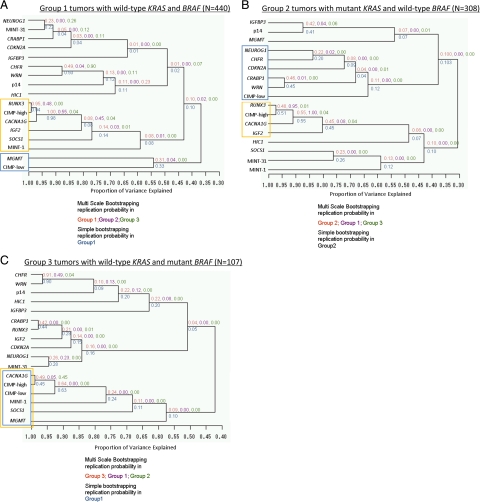
To examine differences in the correlation structures between groups, next we performed a clustering analysis of methylation markers in each tumor group (Figure 1, A–C). Table 3 shows which markers were clustered with CIMP-high or CIMP-low status in each tumor group. Replication rates were consistently higher within each group than across groups. This implied that these clusters were specific to KRAS and BRAF mutational status and that the specific correlation structure of the methylation markers was highly variable according to KRAS and BRAF status. Only CACNA1G clustered as a CIMP-high surrogate marker in all three groups (AUp ≥0.5), and it clustered with RUNX3 reproducibly in BRAF-wild-type tumors (Group 1, AUp = 1.0; Group 2, AUp = 0.55). CHFR and WRN clustered with high reproducibility only in the BRAF-mutant group (Group 3, AUp = 0.91). These results remained essentially unchanged when we analyzed methylation marker variables as bivariate at various cut-off points (data not shown), confirming the robustness of our findings.
Figure 1.
Unsupervised hierarchical clustering analysis in each tumor group. The correlation structure of the 15 CpG island methylation markers varies between the three tumor groups (A–C). A CIMP-high or CIMP-low cluster is indicated by an orange or blue circle, respectively. Group 1 (A) shows two latent clusters, CIMP-high and CIMP-low. Group 2 (B) shows two latent clusters, CIMP-high and CIMP-low. Note a difference in clustering compared to A. In Group 3 (C), CIMP-high and CIMP-low essentially colocalize. Red values are AU (approximately unbiased) P values estimated with the bootstrap samples from the samples with which the original cluster was generated, and purple and green values are AU P values estimated with the bootstrap samples from the other two groups. Blue values are bootstrap probability values with the 1000 bootstrap samples. P value of a cluster is a value between 0 and 1, which indicates how strong the cluster is supported by data.
Table 3.
Results of the Cluster Analysis of the 15 CpG Island Methylation Markers (H, Clustered with CIMP-High Status; L, Clustered with CIMP-Low Status)
| CpG island methylation marker | Group 1 (Wild-type KRAS/BRAF) | Group 2 (Mutant KRAS, wild-type BRAF) | Group 3 (Wild-type KRAS, mutant BRAF) |
|---|---|---|---|
| CACNA1G | H | H | H, L |
| CDKN2A (p16) | L | ||
| CHFR | L | ||
| CRABP1 | L | ||
| HIC1 | H | ||
| IGF2 | H | H | |
| IGFBP3 | |||
| MGMT | L | H, L | |
| MINT-1 | H | H | H, L |
| MINT-31 | H | ||
| NEUROG1 | L | ||
| p14 (CDKN2A/ARF) | |||
| RUNX3 | H | H | |
| SOCS1 | H | H | H, L |
| WRN | L |
In this clustering analysis, splitting criterion was specified as the eigenvalues of all clusters being greater than one. Blank indicates no clustering with CIMP-high or CIMP-low.
CIMP, CpG island methylator phenotype.
As an additional analysis of methylation marker correlation structure, we also performed principal component analysis (PCA) using the 15 CpG island methylation markers (excluding MLH1, described above) in each tumor group according to KRAS and BRAF mutational status (see Supplemental Figure 1 at http://ajp.amjpathol.org). In all groups, at least 12 of 15 principal components were needed to account for 90% of the total variability, and the eigenvalues of the first four (in Group 2 and Group 3) or three (in Group 1) principal components were greater than 1.0. This supports our conclusion that the correlation structure of specific methylation markers varies within each tumor group, while the global structures of the three tumor groups differ significantly according to KRAS and BRAF status.
Causal Modeling of CIMP and Locus-Specific CpG Island Methylation by Structural Equation Modeling (SEM)
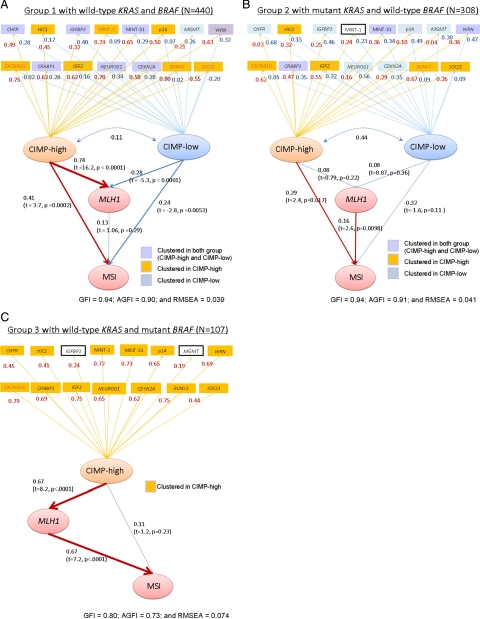
Based on the results of the previous studies3,4,5,10,11,17,18 and our clustering analysis, we constructed causal models as represented in Figure 2, A–C. Two latent variables (“CIMP-high” and “CIMP-low,” or one latent variable “CIMP-high” in Group 3) were adopted to explain the relationship between CIMP, MLH1, and MSI. These causal models were also based on the facts that KRAS and BRAF mutations, and CpG island methylation occur early in colorectal carcinogenic process whereas MSI occurs relatively late,32,33,34,35 and the assumption that the latent “CIMP-high” or “CIMP-low” status in colonic cells influences methylation in locus-specific CpG islands including MLH1. Grouping tumors by KRAS and BRAF mutational status seemed reasonable, given a recent study that has shown clinically significant level of CpG island methylation appears to be later than BRAF mutation events.36
Figure 2.
Path diagrams among CIMP status, MSI, MLH1, and the other 15 CpG island markers in structural equation modeling (SEM) analysis. Path coefficients with t and P values are shown in parentheses. A, B, and C demonstrate differences in correlation structures of CIMP status, locus-specific CpG island methylation, and microsatellite instability (MSI) across the tumor groups. In Group 1 (A), the direct effect of CIMP-high on MLH1 (0.74) is much larger than that of CIMP-high on MSI (0.41). In Group 2 (B), CIMP-high is not significantly associated with MLH1, but directly associated with MSI (0.29; P = 0.017). In Group 3 (C), the direct effect of CIMP-high on MLH1 is substantial (0.67), whereas that of CIMP-high on MSI is not significant. MSI, microsatellite instability.
Figure 2, A–C shows only 19 or 18 variables of interest so that the results are easier to read; other clinical and pathological variables (sex, age, family history of colorectal cancer, stage, tumor location, and DNMT3B status) were also included in the models to adjust path coefficients. In each tumor group, we recategorized the 15 CpG islands to a latent class (CIMP-high or CIMP-low) according to the size of the path coefficient between CIMP status and each of the 15 markers, which corresponded to the factor loading. There was only one latent CIMP class in Group 3, in agreement with the results by clustering and PCA. Our models exhibited a good fit for tumor Group 1 and Group 2 (GFI = 0.94, AGFI = 0.90, 0.91 and RMSE = 0.04); for Group 3, the fit indices showed lower values (GFI = 0.8, AGFI = 0.73 and RMSE = 0.07) due to the smaller number of model parameters. RMSE for Group 3 was relatively larger than that for other groups; however, it was shown that the model would fit the data modestly. We observed that this categorization of the 15 CpG islands in each tumor group almost matched with the results by the cluster analysis. This implied that the indicator of CIMP status defined by the observed 8 CIMP markers would reflect the latent CIMP status. The χ2 difference tests for the multiple group analysis indicated that there was a significant difference in the factor loadings between the tumor groups (P < 0.000001).
This SEM analysis revealed an interesting potential pathogenetic relationship. Specifically, in BRAF-wild-type tumors (Group 1 and Group 2), there appeared to be a significant relationship (P < 0.02) between CIMP-high and MSI which was not mediated through MLH1 methylation. Besides MSI caused by MLH1 methylation in BRAF-mutated CIMP-high tumors (Figure 2C), MSI has been known to be caused by different mechanisms in different tumors. Hereditary nonpolyposis colorectal cancer (HNPCC) due to a germline mutation plus a second hit in a mismatch repair gene has been implicated in approximately 2% of colorectal cancers in the general population.37 Certainly, HNPCC could not explain the majority of colorectal cancers in Group 1 and Group 2. Additional studies are necessary to elucidate mechanisms of the potential link between CIMP-high and MSI in BRAF-wild-type tumors.
In agreement with the results of the cluster analysis and PCA, correlation structures of CIMP status and locus-specific CpG island methylation by SEM differed between Group 1, Group 2 and Group 3. Thus, colorectal preneoplastic or neoplastic cells might have had differential propensity of CpG island methylation in a locus-specific manner, according to KRAS and BRAF mutational status.
Discussion
This study represents the first large-scale investigation that used causal modeling to assess the complex interrelationship between KRAS and BRAF mutations, the CpG island methylator phenotype (CIMP) and locus-specific CpG island methylation. In particular, an application of structure equation modeling (SEM) to correlation structure analysis of CpG island methylation is quite novel. We examined the database of 855 colorectal cancers by cluster analysis, principal component analysis (PCA), and SEM, a combination of techniques that has not been used in any of the previous studies on CIMP.38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66 Our sample size was large enough to perform SEM analysis using a number of variables, while adjusting for potential confounding by clinical and pathological variables including DNMT3B expression. The 16 methylation markers included the five markers (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1) that were selected from 195 CpG islands3 and validated as the CIMP-high surrogate markers.4
We have shown that the interrelationship between CIMP and locus-specific methylation at the 16 CpG islands substantially differ according to background of KRAS and BRAF mutational status. Our data suggest a possible role of KRAS and BRAF mutations in differentially modifying a propensity of CpG island methylation in a locus-specific manner. Although unlikely given the high statistical significance, there is a possibility that the observed differential associations might have occurred by chance. Because our current analysis is exploratory by nature, a validation by independent dataset is necessary.
In SEM, we assumed certain causal relations. It has been well-known that MLH1 methylation causes most sporadic MSI-high colorectal cancers.3,4,5 In addition, we could assume that the “latent CIMP-high or CIMP-low status” might cause methylation in locus-specific CpG islands including MLH1. MSI has been known to occur relatively late in carcinogenic process, in contrast to KRAS and BRAF mutations and CpG island methylation; all of the latter changes could be seen in early adenomas and polyps.32,33,34,35 It is also reasonable to classify tumors by KRAS and BRAF mutational status before clustering, PCA and SEM analyses. A recent study has shown clinically significant level of CpG island methylation appears to be later than BRAF mutation events.36 Thus, in carcinogenic process, KRAS and BRAF mutations appear to occur early, followed by CpG island methylation, and then followed by MSI. Thus, our temporal assumptions and models appear to be appropriate.
A comprehensive examination of genetic and epigenetic alterations is important in cancer research. The relations between CIMP, MSI, and mutations in KRAS and BRAF have been evaluated in a number of previous studies.3,4,5,10,11,18,38,39 Thus far, cluster analyses have been used to evaluate CpG island methylation markers,3,4,10,21,54,65,66 and multivariate logistic regression analyses have been used to assess the independent relationship of CIMP with each of clinical, pathological, and other molecular features, using a large number of tumors.4,5 Nonetheless, no previous study has used PCA or SEM, and our current study is the first to demonstrate differential correlation structures of locus-specific CpG island methylation according to KRAS and BRAF mutational status.
Whether there is a causal relationship between mutations in KRAS and BRAF and CIMP remains enigmatic and has been a subject of active investigations. In particular, the relationship between BRAF mutation and CIMP-high,3,4,5,10,11 and that between KRAS mutation and CIMP-low11,17,18 (CIMP210 or IME21) have recently drawn much attention. It is speculated that KRAS and BRAF mutations may contribute to CpG island methylation in colorectal cancer.39 Although literature data are currently scant, there has been experimental evidence that suggests contribution of KRAS or BRAF mutation to locus-specific CpG island methylation.22,23 In one study using colon epithelial cell line NCM460, a transfection of mutant BRAF appears to induce MLH1 methylation,23 and in another study using NIH 3T3 cells, transformation by activated RAS caused DNA methylation at specific CpG islands.22 Consistent with these previous findings,22,23 we have shown that the correlation structures of CIMP status and CpG island methylation markers differ substantially by KRAS and BRAF mutational status. As noted above, the specific differences could represent a chance variation, but the overall correlation structure is reproducibly modified as KRAS/BRAF status varies. It is thus plausible that KRAS and BRAF mutational status differentially influence the propensity of CpG island methylation in a locus-specific manner.
The relationship between WRN methylation and KRAS mutation warrants discussion. It has been shown that KRAS mutation transgenic mouse develop senescent lung adenomas which display classic senescence markers including up-regulation of CDKN2A (p16), and that progression to adenocarcinoma appears to require premalignant cells to bypass senescence.67 WRN helicase is important in maintenance of proper telomere function, and epigenetic silencing of WRN may be one of mechanisms to bypass senescence in colorectal neoplasia.
In summary, in our large database of colorectal cancers, the correlation structures of the 16 methylation markers and CIMP status differed significantly according to KRAS and BRAF mutational status. Therefore, our data support a possible role of KRAS and BRAF status in differentially modifying cellular propensity for locus-specific CpG island methylation. Further studies are necessary to elucidate a mechanistic link between KRAS or BRAF mutation, CIMP, and locus-specific CpG island methylation in colorectal cancer.
Acknowledgments
We deeply thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires; hospitals and pathology departments throughout the United States for generously providing us with tumor tissue materials; Frank Speizer, Walter Willett, Susan Hankinson, Meir Stampfer, and many other staff members who implemented and have maintained the cohort studies.
Footnotes
Address reprint requests to Shuji Ogino, M.D., Ph.D., M.S., (Epidemiology), Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital, Harvard Medical School, 44 Binney St., Room JF-215C, Boston, MA 02115. E-mail: shuji_ogino@dfci.harvard.edu.
Supported by U.S. National Institutes of Health [P01 CA87969 (S.H.), P01 CA55075 (W.W.), P50 CA127003 (C.S.F.), K07 CA122826 (S.O.), R01 CA151993 (S.O.)]; the Bennett Family Fund for Targeted Therapies Research; and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. K.N. was supported by a fellowship grant from the Japan Society for the Promotion of Science. Y.B. was supported by a fellowship grant from the Uehara Memorial Foundation.
N.T., C.H., K.N., and Y.B. contributed equally to this study.
Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268:177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, Giovannucci EL, Fuchs CS, Ogino S. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz W, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Nosho K, Kure S, Irahara N, Shima K, Baba Y, Spiegelman D, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009;137:1609–1620, e1603. doi: 10.1053/j.gastro.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetta B, Grieu F, Li W, Ruszkiewicz A, Caruso M, Moore J, Watanabe G, Kawakami K. APC gene methylation is inversely correlated with features of the CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2006;119:2272–2278. doi: 10.1002/ijc.22237. [DOI] [PubMed] [Google Scholar]
- Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, Hamilton SR, Issa JP. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, Chapusot C, Piard F. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Slattery ML, Sweeney C, Herrick J, Wolff RK, Albertsen H. APC mutations and other genetic and epigenetic changes in colon cancer. Mol Cancer Res. 2007;5:165–170. doi: 10.1158/1541-7786.MCR-06-0398. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Meyerhardt JA, Fuchs CS, Ogino S. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–577. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, Meissner A, Jaenisch R. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosho K, Shima K, Irahara N, Kure S, Baba Y, Kirkner GJ, Chen L, Gokhale S, Hazra A, Spiegelman D, Giovannucci EL, Jaenisch R, Fuchs CS, Ogino S. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski P, Myszka A, Ramsey D, Misiak B, Gil J, Laczmanska I, Grzebieniak Z, Sebzda T, Smigiel R, Stembalska A, Sasiadek MM. Polymorphisms in methyl-group metabolism genes and risk of sporadic colorectal cancer with relation to the CpG island methylator phenotype. Cancer Epidemiol. 2010;34:338–344. doi: 10.1016/j.canep.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchow Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Ohnishi M, Nosho K, Suemoto Y, Kirkner GJ, Meyerhardt JA, Fuchs CS, Ogino S. CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci. Mod Pathol. 2008;21:245–255. doi: 10.1038/modpathol.3800982. [DOI] [PubMed] [Google Scholar]
- Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklof V, Rutegard J, Oberg A, Van Guelpen BR. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res. 2010;16:1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, Aburatani H, Kaneda A. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Moyer MP, Jass JR. Role of BRAF-V600E in the serrated pathway of colorectal tumourigenesis. J Pathol. 2007;212:124–133. doi: 10.1002/path.2160. [DOI] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, Fuchs CS. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. Unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat. 2004;32:2616–2641. [Google Scholar]
- Joreskog KG. Structural analysis of covariance and correlation matrices. Psychometrika. 1978;43:443–477. [Google Scholar]
- Rashid A, Shen L, Morris JS, Issa J-PJ, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA. Comparison of microsatellite instability. CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- Velho S, Moutinho C, Cirnes L, Albuquerque C, Hamelin R, Schmitt F, Carneiro F, Oliveira C, Seruca R. BRAF. KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis? BMC Cancer. 2008;8:255. doi: 10.1186/1471-2407-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Kakar S, Cun L, Deng G, Kim YS. Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer. 2008;123:2587–2593. doi: 10.1002/ijc.23840. [DOI] [PubMed] [Google Scholar]
- Vaughn CP, Wilson AR, Samowitz WS. Quantitative evaluation of CpG island methylation in hyperplastic polyps. Mod Pathol. 2010;23:151–156. doi: 10.1038/modpathol.2009.150. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A. The incidence of Lynch syndrome. Fam Cancer. 2005;4:233–237. doi: 10.1007/s10689-004-5811-3. [DOI] [PubMed] [Google Scholar]
- Sanchez JA, Krumroy L, Plummer S, Aung P, Merkulova A, Skacel M, DeJulius KL, Manilich E, Church JM, Casey G, Kalady MF. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg. 2009;96:1196–1204. doi: 10.1002/bjs.6683. [DOI] [PubMed] [Google Scholar]
- Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134:1950–1960, 1960 e1951. doi: 10.1053/j.gastro.2008.02.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- Ward RL, Williams R, Law M, Hawkins NJ. The CpG island methylator phenotype is not associated with a personal or family history of cancer. Cancer Res. 2004;64:7618–7621. doi: 10.1158/0008-5472.CAN-03-3978. [DOI] [PubMed] [Google Scholar]
- Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- Frazier ML, Xi L, Zong J, Viscofsky N, Rashid A, Wu EF, Lynch PM, Amos CI, Issa JP. Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer. Cancer Res. 2003;63:4805–4808. [PubMed] [Google Scholar]
- de Vogel S, Wouters KA, Gottschalk RW, van Schooten FJ, de Goeij AF, de Bruine AP, Goldbohm RA, van den Brandt PA, Weijenberg MP, van Engeland M. Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3086–3096. doi: 10.1158/1055-9965.EPI-09-0289. [DOI] [PubMed] [Google Scholar]
- Hughes LA, van den Brandt PA, de Bruine AP, Wouters KA, Hulsmans S, Spiertz A, Goldbohm RA, de Goeij AF, Herman JG, Weijenberg MP, van Engeland M. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS One. 2009;4:e7951. doi: 10.1371/journal.pone.0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehall VL, Wynter CV, Walsh MD, Simms LA, Purdie D, Pandeya N, Young J, Meltzer SJ, Leggett BA, Jass JR. Morphological and molecular heterogeneity within nonmicrosatellite instability-high colorectal cancer. Cancer Res. 2002;62:6011–6014. [PubMed] [Google Scholar]
- Kim JC, Choi JS, Roh SA, Cho DH, Kim TW, Kim YS. Promoter methylation of specific genes is associated with the phenotype and progression of colorectal adenocarcinomas. Ann Surg Oncol. 2010;17:1767–1776. doi: 10.1245/s10434-009-0901-y. [DOI] [PubMed] [Google Scholar]
- Kambara T, Simms LA, Whitehall VLJ, Spring KJ, Wynter CVA, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, Jass JR, Tanaka N, Matsubara N. Colorectal cancer with mutation in BRAF. KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- Cheng YW, Pincas H, Bacolod MD, Schemmann G, Giardina SF, Huang J, Barral S, Idrees K, Khan SA, Zeng Z, Rosenberg S, Notterman DA, Ott J, Paty P, Barany F. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan AS, Chan TL, Wu TT, Rashid A, Hamanaka Y, Hinoda Y, Shannon RL, Wang X, Morris J, Issa JP, Yuen ST, Leung SY, Hamilton SR. Epigenetic-genetic interactions in the APC/WNT. RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res. 2008;14:2560–2569. doi: 10.1158/1078-0432.CCR-07-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi K, Shen L, Jelinek J, Watanabe Y, Ahmed S, Kaneko K, Kogo M, Takano T, Imawari M, Hamilton SR, Issa JP. Concordant DNA methylation in synchronous colorectal carcinomas. Cancer Prev Res (Phila Pa) 2009;2:814–822. doi: 10.1158/1940-6207.CAPR-09-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arain MA, Sawhney M, Sheikh S, Anway R, Thyagarajan B, Bond JH, Shaukat A. CIMP status of interval colon cancers: another piece to the puzzle. Am J Gastroenterol. 2010;105:1189–1195. doi: 10.1038/ajg.2009.699. [DOI] [PubMed] [Google Scholar]
- Gonzalo V, Lozano JJ, Munoz J, Balaguer F, Pellise M, Rodriguez de Miguel C, Andreu M, Jover R, Llor X, Giraldez MD, Ocana T, Serradesanferm A, Alonso-Espinaco V, Jimeno M, Cuatrecasas M, Sendino O, Castellvi-Bel S, Castells A. Aberrant gene promoter methylation associated with sporadic multiple colorectal cancer. PLoS One. 2010;5:e8777. doi: 10.1371/journal.pone.0008777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English DR, Young JP, Simpson JA, Jenkins MA, Southey MC, Walsh MD, Buchanan DD, Barker MA, Haydon AM, Royce SG, Roberts A, Parry S, Hopper JL, Jass JJ, Giles GG. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev. 2008;17:1774–1780. doi: 10.1158/1055-9965.EPI-08-0091. [DOI] [PubMed] [Google Scholar]
- Van Guelpen B, Dahlin AM, Hultdin J, Eklof V, Johansson I, Henriksson ML, Cullman I, Hallmans G, Palmqvist R. One-carbon metabolism and CpG island methylator phenotype status in incident colorectal cancer: a nested case-referent study. Cancer Causes Control. 2010;21:557–566. doi: 10.1007/s10552-009-9484-y. [DOI] [PubMed] [Google Scholar]
- Deng G, Nguyen A, Tanaka H, Matsuzaki K, Bell I, Mehta KR, Terdiman JP, Waldman FM, Kakar S, Gum J, Crawley S, Sleisenger MH, Kim YS. Regional hypermethylation and global hypomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. Int J Cancer. 2006;118:2999–3005. doi: 10.1002/ijc.21740. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Curtin K, Sweeney C, Levin TR, Potter J, Wolff RK, Albertsen H, Samowitz WS. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Albertsen H, Sweeney C, Herrick J, Caan BJ, Anderson KE, Wolff RK, Slattery ML. Association of smoking. CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006;98:1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Igarashi S, Nojima M, Maruyama R, Yamamoto E, Kai M, Akashi H, Watanabe Y, Yamamoto H, Sasaki Y, Itoh F, Imai K, Sugai T, Shen L, Issa JP, Shinomura Y, Tokino T, Toyota M. IGFBP7 is a p53 responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis. 2010;31:342–349. doi: 10.1093/carcin/bgp179. [DOI] [PubMed] [Google Scholar]
- Goel A, Li MS, Nagasaka T, Shin SK, Fuerst F, Ricciardiello L, Wasserman L, Boland CR. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130:1950–1961. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- Ferracin M, Gafa R, Miotto E, Veronese A, Pultrone C, Sabbioni S, Lanza G, Negrini M. The methylator phenotype in microsatellite stable colorectal cancers is characterized by a distinct gene expression profile. J Pathol. 2008;214:594–602. doi: 10.1002/path.2318. [DOI] [PubMed] [Google Scholar]
- Ang PW, Loh M, Liem N, Lim PL, Grieu F, Vaithilingam A, Platell C, Yong WP, Iacopetta B, Soong R. Comprehensive profiling of DNA methylation in colorectal cancer reveals subgroups with distinct clinicopathological and molecular features. BMC Cancer. 2010;10:227. doi: 10.1186/1471-2407-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]