Summary
The mitochondrial genome of Trypanosoma brucei, called kinetost DNA (kDNA), is a network of topologically interlocked DNA rings including several thousand minicircles and a few dozen maxicircles. kDNA synthesis involves release of minicircles from the network, replication of the free minicircles, and reattachment of the progeny. Here we report a new function of the mitochondrial topoisomerase II (TbTOP2mt). Although traditionally thought to reattach minicircle progeny to the network, here we show that it also mends holes in the network created by minicircle release. Network holes are not observed in wild type cells, implying that this mending reaction is normally efficient. However, RNAi of TbTOP2mt causes holes to persist and enlarge, leading to network fragmentation. Remarkably, these network fragments remain associated within the mitochondrion, and many appear to be appropriately packed at the local level, even as the overall kinetoplast organization is dramatically altered. The deficiency in mending holes is temporally the earliest observable defect in the complex TbTOP2mt RNAi phenotype.
Keywords: trypanosome, kDNA, mitochondrial DNA, replication
Introduction
Trypanosoma brucei, the African trypanosome, is a protozoan parasite that causes sleeping sickness. In addition to its significance as a pathogen, this organism has unusual biological features. A well-known example is its mitochondrial genome, called kinetoplast DNA (kDNA), which resides within the trypanosome’s single mitochondrion and which has a structure unique in nature [see (Liu et al., 2005; Lukes et al., 2002; Shlomai, 2004) for recent reviews on kDNA]. kDNA is a network of topologically interlocked DNA rings including a few dozen maxicircles (~23 kb) and several thousand minicircles (~1 kb). The maxicircles, like conventional mitochondrial DNAs, encode ribosomal RNAs and a handful of mitochondrial proteins such as subunits of respiratory complexes. Most maxicircle transcripts are edited by insertion or deletion of uridylate residues at specific internal sites, thus forming the open reading frame. Editing requires guide RNAs, mostly encoded by minicircles, to serve as templates for U-insertion or deletion [for a review see (Stuart et al., 2005)].
The organization of the kDNA network is remarkable. It is a planar structure whose topology resembles that of the chain mail of medieval armor. Previous studies on the kDNA of the related parasite Crithidia fasciculata have shown that each minicircle in the network has an average of 3 neighbors (Chen et al., 1995b). It is likely that T. brucei networks have a similar structure. We previously found that the number of neighbors, termed the minicircle valence, changes during the C. fasciculata cell cycle (Chen et al., 1995a), and we will return to this point in the Discussion. The kDNA network in vivo is condensed into a disk-shaped structure in the matrix of the cell’s single mitochondrion. The disk is physically attached to the basal body of the flagellum by a transmembrane filament system called the tripartite attachment complex (TAC) (Ogbadoyi et al., 2003).
The unusual network structure requires an unconventional replication mechanism [for reviews see (Liu et al., 2005; Shlomai, 2004)]. In the current model (we will focus here on replication of minicircles), covalently-closed minicircles are released from the network into a region between the kDNA disk and the mitochondrial membrane near the flagellar basal body. In this region, known as the kinetoflagellar zone, they encounter multiple replication proteins and initiate unidirectional replication as theta structures. The daughter minicircles are thought to segregate in the kinetoflagellar zone and then migrate to the antipodal sites, two assemblies of enzymes positioned on opposite sides of the kDNA disk. Some of these enzymes catalyze the final stages of minicircle replication, including primer removal and repair of most of the resulting gaps between Okazaki fragments. At least one gap remains that is thought to mark minicircles that have completed replication, thereby preventing a second round of DNA synthesis. Then the progeny minicircles are reattached to the network periphery adjacent to the antipodal sites. A topoisomerase II (TbTOP2mt), situated in the antipodal sites, has been shown to perform this function (Melendy et al., 1988; Wang and Englund, 2001; B. Liu, J.Wang, M.E. Lindsay, J. D. Griffith, P.T. Englund, unpublished data). As replication continues, the network elongates and when the minicircle copy number has doubled, the network splits in two and the remaining minicircle gaps are repaired. The TAC filament system mediates segregation of the progeny kDNA networks; the moving apart of the basal bodies pulls the kDNAs, via their connection by TAC, into the daughter cells.
Replication and segregation of a DNA network are immense topological challenges, probably requiring type II topoisomerase (topo II) activity at multiple steps in these pathways. Only one mitochondrial topo II, TbTOP2mt, is predicted from the genome sequence. This is surprising, given the fact that the mitochondrion contains six DNA polymerases (Klingbeil et al., 2002; Saxowsky et al., 2003) and six DNA helicases (B. Liu and P.T. Englund, unpublished data), all of which apparently have distinct functions in kDNA replication and maintenance. The gene for the mitochondrial topo II was first cloned from T. brucei (Strauss and Wang, 1990) and later shown to be homologous [68% identical at the protein level (Pasion et al., 1992)] to a protein purified from C. fasciculata (Melendy and Ray, 1989) that is localized predominantly in the antipodal sites (Melendy et al., 1988). Antibodies raised against the C. fasciculata protein as well as those raised against a recombinant fragment of the TbTOP2mt sequence (prepared by T. Kulikowicz and T. Shapiro, Johns Hopkins Medical School) both label the T. brucei antipodal sites as judged by immunofluorescence (Wang and Englund, 2001; Liu and Englund, 2007).
Two other topo II enzymes are predicted by the genome and are encoded by tandemly-linked genes named TbTOP2α and TbTOP2β (Kulikowicz and Shapiro, 2006). The predicted gene products are 74% identical, and they differ significantly only in the C-terminal region. RNAi of TbTOP2α causes a dramatic nuclear phenotype (e.g. failure to separate chromosomes during mitosis, leading to anucleate cells and cells with fragmented nuclei) consistent with the nuclear localization of this protein. There is as yet no evidence for expression of TbTOP2β protein. Although the localization and function of the putative TbTOP2β protein is unknown, its sequence clusters phylogenetically with other eukaryotic nuclear topo IIs. RNAi of TbTOP2α had no observable effect on kDNA, and RNAi of TbTOP2β did not produce any phenotype, so it is unlikely that either TbTOP2α or TbTOP2β plays a significant role in kDNA replication. If TbTOP2mt is the only mitochondrial topo II, we must consider the possibility that this enzyme has multiple functions in replication and segregation of kDNA. It is also possible that other type II topoisomerases, both nuclear and mitochondrial, have non-canonical sequences and therefore have yet to be identified.
In this work, we present compelling evidence for a new and unexpected function for TbTOP2mt. When a minicircle is individually released from the network for replication, several topological bonds must be broken nearly simultaneously, thus creating a small hole in the network. Since we never observe holes in wild type networks, these holes must be efficiently mended; that is, new topological bonds must form to fill the hole created by minicircle release. Now we have found that depletion of TbTOP2mt by RNAi causes these holes to persist, indicating that this enzyme is responsible for the mending reaction. Thus, we report for the first time that TbTOP2mt constantly remodels the network during replication to maintain a proper minicircle density and network structure.
Results
Thin-section EM of TbTOP2mt RNAi cells revealed an unusual kinetoplast structure
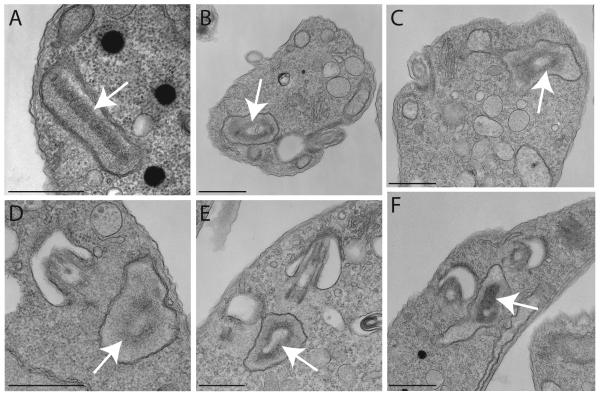
While investigating kinetoplast organization in trypanosomes with RNAi-mediated kDNA replication defects, we observed an unusual kinetoplast structure in cells in which TbTOP2mt had been knocked down by RNAi for 4 days. Instead of having a conventional disk-shaped kinetoplast, the RNAi cells often had kinetoplasts that appeared to have a hole in the middle (resembling a donut). In 26 randomly-chosen cells with a visible kinetoplast, 9 had holes (see examples in Fig. 1). We decided to investigate the origin and significance of these holes at the molecular level.
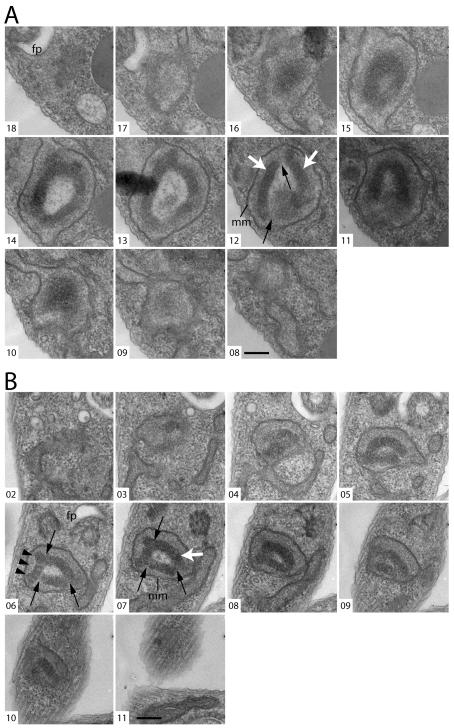
Figure 1. Thin-section EM of cells during TbTOP2mt RNAi reveals kinetoplasts with “holes”.
(A) A kinetoplast from an uninduced (day 0) cell. In this image, the kDNA has been sectioned parallel to the disk’s short axis. (B-F) Examples of kinetoplasts from cells after RNAi for 4 days showing a hole in the kinetoplast. White arrows indicate the kinetoplast. Scale bars for (A), (D), and (E), 600 nm. Scale bars for (B), (C), and (F), 800 nm.
RNAi of TbTOP2mt caused rapid shrinking of the kDNA network
kDNA minicircles undergo replication after being individually released from the network. This release, catalyzed by an unknown topoisomerase activity, must involve the nearly simultaneous breakage of several topological bonds. Thus, minicircle release would leave a small hole in the network. Subsequent release of other minicircles would form more holes, which would eventually merge to form larger holes. Inspection of many EMs of networks isolated from wild type cells has never revealed holes, implying the existence of an efficient mechanism to mend holes created by minicircle release. In contrast, EM of replicating networks from wild type C. fasciculata had revealed holes, implying that the mending activity in this parasite was less efficient (Perez-Morga and Englund, 1993a). We hypothesized that if holes in T. brucei networks are normally mended by TbTOP2mt, then holes might accumulate in networks from the RNAi cells. Our initial thought was that enlargement and merging of small holes might explain the kinetoplast holes shown in Fig. 1.
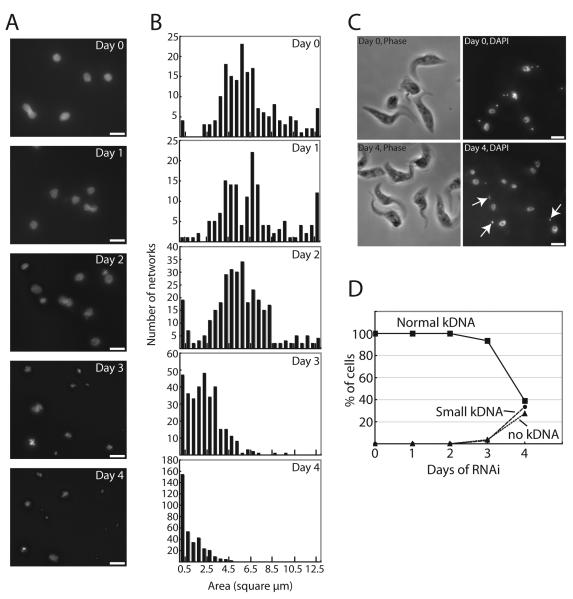
To test this possibility, we examined the structure of kDNA networks isolated from cells undergoing TbTOP2mt RNAi for 1 to 4 days. Our first approach was to stain the networks with DAPI and examine them by fluorescence microscopy. At this resolution we could never detect a large central hole in any of the networks (Fig. 2A). However, we did find a very dramatic shrinkage of the networks, especially at days 3 and 4. By day 4 there are virtually no normal-size networks (see images in Fig. 2A and surface area measurements in Fig. 2B). The small size of the networks, especially those at day 4, did not seem to fit with our thin section EMs in Fig. 1 and also with our images of DAPI-stained whole cells after 4 days of RNAi [about 40% of the latter had apparently normal-size kinetoplasts (Fig. 2C, D)]. However, our data on DAPI-stained cells did agree with published data (Wang and Englund, 2001). To resolve this discrepancy we next examined isolated kDNA networks by EM.
Figure 2. Networks become fragmented during TbTOP2mt RNAi.
(A) Networks were isolated from cells that had undergone RNAi for the indicated time (day 0 is the uninduced control), stained with DAPI, and visualized by fluorescence microscopy. Scale bar is 5 μm. (B) Histograms showing surface areas of DAPI-stained isolated networks measured by IPLab software. Note that scales on y-axis differ in different panels. Measurements of day 0 control networks agree with previous values (Zhao et al., 2008). (C) Images showing DAPI staining and phase imaging of cells on day 0 (uninduced control) and after four days of RNAi. Arrows indicate normal-sized kinetoplasts. Scale bar is 5 μm. (D) Graph showing kinetics of kDNA loss during TbTOP2mt RNAi.
Networks became loose during TbTOP2mt RNAi
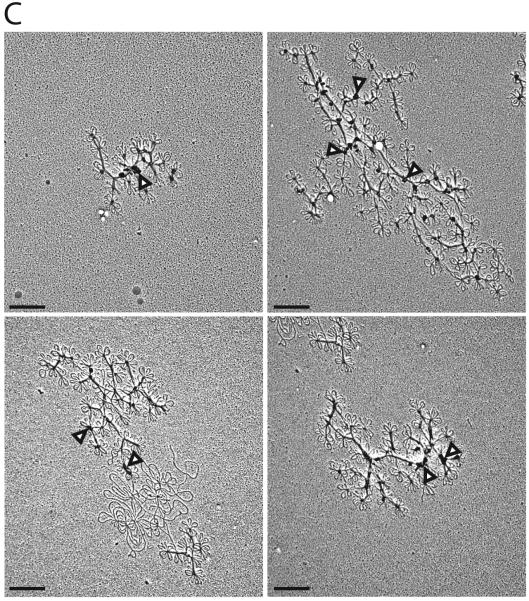
In previous studies, we found that TbTOP2mt RNAi causes networks to shrink and then disappear (Wang and Englund, 2001). However, we now were surprised to find by EM that networks from cells that had undergone RNAi for 1 or 2 days had actually grown in size (see examples of EMs in Fig. 3A and measurements of their surface areas in Fig. 3B). The fact that the network surface areas measured on the EMs was considerably larger (about 30 μm2) than those measured on fluorescence images (about 6.4 μm2) has been observed previously and has been attributed to differences in settling and adhesion to the polylysine-coated slide and to the EM grid (Guilbride and Englund, 1998). These differences may explain why the expansion in network surface area detected by EM (Fig. 3A, B) was not detectable by fluorescence microscopy (Fig. 2B).
Figure 3. Effect of time of TbTOP2mt RNAi on isolated kDNA networks.
(A) EMs of networks isolated from day 0 (uninduced), day 1, and day 2 of RNAi. Scale bar is 2 μm. Higher magnification images of the same networks are shown in Fig. 4A. (B) Network surface areas (9 networks per time point) were measured for days 0, 1, and 2. Areas were not measured for days 3 and 4 because the networks were too loose and they did not have a well-defined periphery (see examples of day 4 network fragments in panel C). One point per day is shown representing the average network area on each day. Bars show standard error. (C) EMs of networks isolated from cells after 4 days of RNAi. The “blobs” in these networks (arrowheads) were observed previously (Wang and Englund, 2001) and we are currently investigating their origin. Scale bar is 0.5 μm.
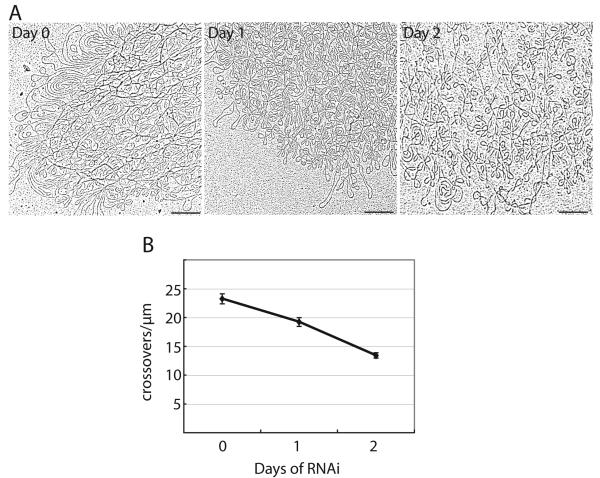
It seemed unlikely that this increase in surface area could be due to an increase in minicircle copy number, especially since we had previously shown by Southern blot that there was no change in abundance of total minicircles or maxicircles during the first 2 days of RNAi, although both declined thereafter (Wang and Englund, 2001). Indeed, close inspection of the day 1 and day 2 networks indicated that they had become looser, with many small holes evenly distributed throughout the network (Fig. 4A). We quantitated this apparent decrease in minicircle density by drawing 1 μm vertical lines at random positions on each network image and counting how many times each line was crossed by a DNA strand (Chen et al., 1995a). This analysis confirmed our observation that the TbTOP2mt RNAi networks were looser than wild-type (Fig. 4B).
Figure 4. Effect of TbTOP2mt RNAi on minicircle density in kDNA networks.
(A) EMs of networks isolated from cells at day 0 and after 1 or 2 days of RNAi. These are higher magnification images of networks shown in Fig. 3A. Scale bar is 0.5 μm. (B) Network density was measured by drawing at least 7 random 1 μm vertical lines through each network and counting the number of times each line was crossed by a DNA strand (crossovers/μm) (Chen et al., 1995a). An average was calculated for each network, and those values averaged to give the single point per day shown on the graph. Error bars show standard error.
RNAi of other replication proteins did not loosen networks
To investigate whether RNAi of other kDNA replication proteins reduced minicircle density, we examined networks isolated from cells undergoing RNAi knockdown of p38 (Liu et al., 2006), ligase kα (Downey et al., 2005), and PIF1 helicase (B. Liu, J. Wang, M.E. Lindsay, J. Griffith, and P.T. Englund, unpublished data). Fig. 5A shows examples of networks from p38 RNAi cells at day 0 (uninduced for RNAi) and day 2. The latter is clearly smaller in size and appears to have a minicircle density comparable to that of the day 0 network. Fig. 5B presents quantitative data comparing the effect of RNAi of these three proteins with that of TbTOP2mt. In the early stages of RNAi (either 1 or 2 days after induction), there was a striking difference between TbTOP2mt and the other proteins. As already discussed, networks from TbTOP2mt RNAi cells increase in surface area and decline in minicircle density. In contrast, networks from the other three RNAi cell lines decreased in surface area and were unchanged in minicircle density. From this experiment we conclude that reduced minicircle density is not a general property of networks undergoing an RNAi-mediated block in kDNA replication. Instead it is a specific property of networks from TbTOP2mt RNAi cells.
Figure 5. Effect of RNAi on other kDNA replication proteins.
(A) EMs of networks undergoing RNAi for p38 at day 0 (prior to induction) and day 2. Scale bar is 1 μm. (B) Comparison of the effects of RNAi on TbTOP2mt and other kDNA replication proteins. Network surface areas (open squares) and minicircle density (closed diamonds) were measured as described in Methods. Nine or ten randomly-selected networks were included in each sample. TbTOP2mt data is same as shown in Figs. 3B and 4B.
Serial sectioning of kinetoplasts revealed that they are fragmented
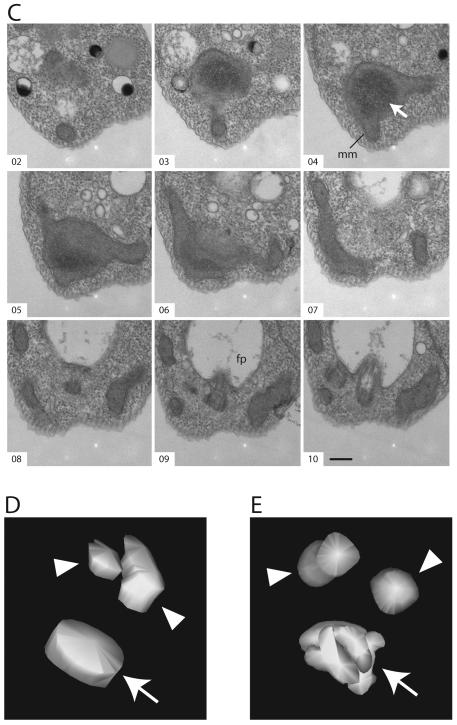
To investigate more rigorously the nature of the kinetoplast holes presented in Fig. 1, we examined serial thin sections through kinetoplasts in cells after four days of TbTOP2mt RNAi. Of 24 series [six containing the whole kinetoplast (8-15 sections) and 18 containing partial views of the kinetoplast (3-8 sections)], 16 (67%) revealed a hole. However, rather than a hole that extended completely through the kDNA disk, like the hole in a donut, we found that the hole was actually a hollow cavity within a vaguely sphere-shaped kinetoplast (Fig. 6A, B, E). The material within the cavities appears less electron dense than the kDNA or the surrounding mitochondrial matrix (Fig. 6A, B). Sections though the center of these kinetoplasts give the appearance of a ring structure, matching our initial observation. In non-induced cells, as in wild-type, the kDNA is a highly-organized electron dense disk-shaped structure positioned near the basal body [Fig. 6C, D, reviewed in (Klingbeil et al., 2001)].
Figure 6. Serial sections show fragmented kinetoplasts.
(A, B) Serial thin sections, viewed by TEM, through kinetoplasts after four days of RNAi. (C) Serial thin sections through a kinetoplast of an uninduced (day 0) cell. White arrows indicate kDNA. Black arrows indicate apparent discontinuities between kinetoplast fragments. Black arrowheads indicate a fragment where the striated structure is clearly visible. mm: mitochondrial membrane; fp: flagellar pocket. Scale bar is 250 nm. (D) Three-dimensional (3-D) reconstruction of a kinetoplast in an uninduced cell, based on the serial sections shown in C. (E) 3-D reconstruction of a kinetoplast after four days of RNAi, based on the serial sections shown in B. Arrowheads indicate the position of the basal bodies. Arrows indicate kDNA. Movies showing different views of the 3-D reconstructions based on panels (A), (B), and (C) can be found in Online Supplementary Material.
The kinetoplast in some of the series appeared to be composed of several fragments [Fig. 6A (section 12) and B (sections 6 and 7)]. To show the position of the kinetoplast fragments relative to each other and in relation to the basal body/flagellum complex, we traced the contours of these objects in the serial sections and built a three-dimensional model (Fig. 6E and Online Supplementary Material). In the model built from the kinetoplast in TbTOP2mt RNAi cells, the normal disk structure is severely altered, and the DNA fragments are seen tightly packed around one another. The position of the basal bodies relative to the kinetoplast appears unchanged.
The kDNA in non-induced or wild type cells has a characteristic striated structure when viewed in sections cut perpendicular to the plane of the disc (Fig. 1A), and appears punctate when sectioned parallel to the plane of the disc (Fig 6C, sections 3 and 4). This striated structure is thought to reflect the ordered arrangement of the minicircles, packed perpendicular to the plane of the condensed disk [reviewed in (Klingbeil et al., 2001)]. It was interesting that in the majority of the induced TbTOP2mt RNAi cells, each kDNA fragment is organized at the local level into a structure resembling a segment of a normal kDNA disk, both in thickness and in having the characteristic striations (for example see Fig 6B, section 6). These network fragments are detected only in the proper region of the mitochondrion, adjacent to the flagellar basal body and are not distributed throughout the mitochondrion.
Discussion
Previous studies indicated that TbTOP2mt is required for resolution of interlocked minicircle dimers (a replication intermediate), and also for reattachment of monomeric minicircle replication products to the periphery of the kDNA disk (Melendy et al., 1988; Wang and Englund, 2001; B. Liu, J. Wang, M.E. Lindsay, J. Griffith, and P.T. Englund, unpublished data). Other steps in kDNA replication that could require a topo II activity include minicircle release prior to replication and removal of positive supertwists introduced during unwinding of the minicircle or maxicircle parental strands. Finally, segregation of the fully-replicated sister networks could utilize a topo II.
Here we report a new function for TbTOP2mt in mending holes created by minicircle release. Our studies followed the unexpected finding that some kinetoplasts in TbTOP2mt RNAi cells have a large central hole (Fig. 1). Our initial thought was that release of minicircles for replication might be restricted to the network’s central region. Failure to mend the resulting holes in the network (because of knockdown of TbTOP2mt) could result in formation of the large kinetoplast holes visualized in Fig. 1. This hypothesis was based on our previous observations of C. fasciculata kDNA (Perez-Morga and Englund, 1993a). EMs of isolated replicating networks from wild type C. fasciculata cells revealed a central hole that we attributed to release of minicircles for replication. However, since these holes were less prominent in a subsequent study (Chen et al., 1995a), we assumed that the balance of minicircle release and mending of holes, which determines whether holes persist, is highly sensitive to experimental conditions. Since we never observed holes in T. brucei networks we assumed that holes must be mended efficiently. We will discuss in subsequent paragraphs that the anomalous kinetoplast structures in Fig. 1 are in fact due to a deficiency in hole mending but not in the way we originally assumed.
The evidence that TbTOP2mt functions to mend holes in the network is strong. EMs of networks isolated from cells during a course of TbTOP2mt RNAi revealed a striking loosening of the structure even after only one day of RNAi. Based on the uniform distribution of small holes throughout the network, minicircle release for replication must occur at random positions throughout the network and not be restricted to the central region. We found that as RNAi proceeds to days 3 and 4, the holes merge, and the networks get much looser and eventually are converted to small fragments (Fig. 2A, B; Fig. 3C). The reduction in network size is clearly demonstrated by surface area measurements of DAPI-stained isolated networks (Fig. 2A, B). After RNAi for 4 days, there are no normal-size networks, and since the isolation of kDNA used for these measurements included a centrifugation step, there likely was preferential loss of the smallest fragments. Thus, the average fragment size may be even smaller than indicated in the bar graphs in Fig. 2B. We previously published an EM of a network after 5 days of TbTOP2mt RNAi that was larger than the 4 day networks shown in Fig. 3C (Wang and Englund, 2001). However, in that study we did not evaluate the range of sizes in the population, which we now know is substantial (Fig. 2B).
There are three ways that network size could be reduced by RNAi of TbTOP2mt. One is shrinking caused by continued minicircle release without reattachment of the progeny, another is asymmetric division of the network (Wang et al., 2002), and a third is fragmentation due to the absence of hole mending. Although all these effects may contribute to the size reduction, there are three reasons that fragmentation must play a major role. First, we showed previously by Southern blot that 4 days of RNAi caused a 40% loss of both minicircles and maxicircles (Wang and Englund, 2001). Since the average network surface area was reduced from 6.4 ± 0.2 μm2 at day 0 to 1.0 ± 0.1 μm2 at day 4 (a reduction of 84%), there must also be significant fragmentation. Second, we found that in cells subjected to RNAi for 4 days, 40% had kinetoplasts that appeared normal in size (Fig 2C, D), but virtually no isolated networks were normal size; this comparison led to the conclusion that these kinetoplasts must be composed of multiple fragments. Third, our 3-dimensional reconstructions of kinetoplasts after 4 days of RNAi provided strong evidence that they are composed of multiple fragments (Fig. 6).
There are other important points about network loosening and mending of holes. Although network loosening is clearly a property of TbTOP2mt RNAi cells, the data in Fig. 5 demonstrate that loosening does not occur following knockdown of other kDNA replication proteins (p38, ligase kand PIF1 helicase), at least for the early stages of RNAi. At later stages there is some reduction of minicircle density (data not shown), and we will address this issue later in the Discussion. Our data also emphasize the importance of mending holes in the network. Without this activity, the networks undergo fragmentation and become disorganized within the mitochondrion, conditions that are obviously incompatible with propagation of the mitochondrial genome.
The 3-dimensional reconstructions of the kinetoplast structure demonstrate, remarkably, that the network fragments appear to remain associated in the proper region of the mitochondrion, near the flagellar basal body, and do not diffuse to other parts of the mitochondrial matrix. It is most likely that these fragments remain associated because of their continued linkage to TAC. In some cases, fragments may also be held together by proteins (other than those associated with TAC) or DNA links (perhaps a single minicircle or maxicircle) which are broken during isolation.
The images in Fig. 1, suggesting a large hole in the kinetoplast, turned out to be misleading. After analysis of serial sections, we realized that fragments of kDNA were arranged in the cell such that they enclosed a hollow cavity. In some sections through these structures, the kinetoplast looked like a donut as we had observed in Fig. 1. It is not clear why a fragmented network would adopt such an organization. If the network fragments are less restricted spatially than a normal kDNA disk, they may assume abnormal arrangements. Alternatively, if the mitochondrial space containing the kDNA is restricted to a defined size and the network fragments, plus the space between them, actually takes up more space than a normal disk, the fragments may be forced to fold over on themselves. Finally, when the duplicated basal bodies begin to separate, they apply tension on the replicated sister kinetoplasts through their connection via TAC. In the case of a fragmented kinetoplast, the TAC filaments may be unable to attach equally to all of the fragments. Only the strongly-linked fragments would be held in close proximity to the basal bodies; weakly-linked segments could assume folded-over conformations.
It is striking that some kinetoplast fragments appear to have a normal organization at the local level. Like a conventional kDNA disk, they appear striated and have a thickness that is about half the circumference of a minicircle. In wild type kinetoplasts, these characteristics are thought to reflect stretched out minicircles lined up side by side and interlocked with their neighbors (Klingbeil et al., 2001). We speculate that kDNA binding proteins package the DNA normally at the local level even when the network is fragmented and overall kinetoplast organization is lost. There are several candidates for such proteins. One group of kinetoplast-associated proteins are highly basic histone H1-like proteins thought to contribute to proper packaging of kDNA (Lukes et al., 2001; Xu et al., 1996). Another is a highly-basic kinetoplast-associated protein, named p19, currently being studied in our lab (Z. Zhao, J. Wang, and P.T. Englund, unpublished data).
TbTOP2mt localizes predominantly to the antipodal sites (Liu and Englund, 2007; Melendy et al., 1988; Wang and Englund, 2001). However, to mend holes some of this enzyme must be positioned throughout the kinetoplast disk. Perhaps less TbTOP2mt is needed for hole mending than for minicircle reattachment, and previous localization experiments could only detect areas where TbTOP2mt is the most abundant. In addition, in C. fasciculata it has been shown that the localization of this enzyme is dynamically regulated during the cell cycle, and can at times be found in areas other than the antipodal sites, including in the kDNA disk (Johnson and Englund, 1998). Within the kDNA disk TbTOP2mt may not only mend holes but also control network topology in other ways. For example, we previously reported that a topo II activity in C. fasciculata must control network topology during the cell cycle (Chen et al., 1995a). We demonstrated that the number of neighbors for each minicircle, called the minicircle valence, varies in a cell cycle-dependent manner (Chen et al., 1995a). In a pre-replication network, each minicircle has a valence of 3 (Chen et al., 1995b). As replication proceeds and the minicircle copy number increases, the space occupied by the network apparently does not increase (we had proposed that the kinetoplast is prevented from expanding by the mitochondrial membrane). Thus, the growing network becomes more densely packed with minicircles and the valence rises to six. Changes in valence require topoisomerase action. Only when replication is complete is there an expansion of the space occupied by the kinetoplast. The network again remodels, reducing the valence to 3 and doubling the network’s surface area. Then the network divides in two, forming two progeny identical to the parent. Similar variations in minicircle density were reported in one EM study of T. brucei networks at different stages of replication (although there were no quantitative measurements) (Hoeijmakers and Weijers, 1980). However, such variation was not detected in another study (Ferguson et al., 1994). If remodeling reactions occur during T. brucei kDNA replication, a topo II must be distributed throughout the kinetoplast disk. Earlier in the Discussion we mentioned that in the later stages of RNAi of p38 and PIF1 helicase, we did detect some reduction in minicircle density (data not shown). One explanation for this effect could be that the shrinking network occupied a space appropriate for a normal size network. Network remodeling would therefore cause expansion of the network to fill the available space with a corresponding reduction in minicircle density. Since both reactions require changes in topology of double stranded, covalently-closed DNA circles within the kDNA disk, reactions that must be performed by a topo II, we speculate that mending of holes and network remodeling are both catalyzed by TbTOP2mt. We further speculate that the kDNA network in situ is a dynamic structure with topological bonds continuously undergoing breakage and rejoining, reactions almost certainly catalyzed by TbTOP2mt.
One surprising feature of the TbTOP2mt RNAi phenotype is that knockdown does not appear to affect minicircle release. We found previously that covalently-closed monomeric minicircles are present during the first four days of RNAi, indicating that minicircle release continues after knockdown of TbTOP2mt (Wang and Englund, 2001). This conclusion is consistent with our current data, where the appearance of holes and fragmentation of the network during RNAi indicate continued minicircle release. It is possible that a novel decatenating activity is responsible for release as a traditional topo II may be unable to catalyze, in a concerted fashion, the near simultaneous breakage of three topological bonds in order to release a single minicircle. Finally, TbTOP2mt may indeed be involved in minicircle release, either alone or complexed with other proteins, but it must function at a very low concentration that is present even after RNAi knockdown.
The consequences of TbTOP2mt knockdown in the cell appear multifaceted and complex, and it is clear that we have only just begun to understand how this remarkable enzyme functions in kDNA replication. TbTOP2mt certainly has multiple functions. How these different functions are regulated, either spatially, temporally, through post-translational modifications or protein-protein interactions, will be a fascinating subject for future study.
Experimental Procedures
Trypanosome strains and culture
Procyclic form T. brucei [cell line 29-13, provided by Dr. George Cross, Rockefeller University (Wirtz et al., 1999)] was maintained at 28 °C in SDM-79 medium (Brun et al., 1979) supplemented with 10% fetal bovine serum (FBS) and appropriate antibiotics. RNAi cell lines contained either a stem-loop vector for TbTOP2mt (Wang and Englund, 2001), or pZJM (Wang et al., 2000) constructs for p38 (Liu et al., 2006) and PIF1 (unpublished). For RNAi of ligase kα, a 554 bp PCR product (nucleotides 555 to 1109 of the coding sequence) was cloned into pZJM. The cell line used for RNAi is strain 29-13 (Wirtz et al., 1999), which was maintained in the presence of G418 (15 μg/ml) and hygromycin (50 μg/ml); the RNAi vector was maintained by phleomycin (2.5 μg/ml). RNAi was induced by tetracycline (1 μg/ml) or doxycycline (1 μg/ml) with comparable results.
DAPI staining of cells and isolated kDNA
For DAPI (4′, 6-diamidino-2-phenylindole) staining of intact cells, trypanosomes undergoing RNAi were adhered to 0.1% poly-L-lysine coated slides and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (5 min, room temperature). Slides were washed (at least 2 times, 5 min) in PBS, and then stained with DAPI (2 μg/ml in PBS). Slides were mounted in Vectashield (Vector Laboratories) and examined by fluorescence microscopy using a Zeiss Axioscop microscope equipped with a 100X phase 3 Plan-Neofluar objective (Zeiss, 1.3 numerical aperture) and a Retiga Exi CCD camera (QImaging). Images were acquired using IPLab software (Scanalytics) and were cropped and adjusted uniformly for brightness and contrast and using IPLab and Adobe Photoshop. For each sample, at least 120 cells were categorized as to whether they had normal, small, or no visible kDNA.
kDNA networks [isolated according to (Perez-Morga and Englund, 1993b)] were allowed to settle on slides coated with 0.01% poly-L-lysine (Sigma) for 30 min. Following a brief wash with PBS, they were stained with DAPI (2 μg/ml, 2 min) and mounted in Vectashield for fluorescence microscopy. Images were captured and adjusted as described above except that a Zeiss 63X phase 3 Plan-Neofluar objective (numerical aperture 1.25) was used. Network areas were measured using IPLab software (Zhao et al., 2008).
Electron microscopy of isolated kDNA networks
kDNA networks were isolated by centrifugation (Perez-Morga and Englund, 1993b), spread on nitrocellulose-coated EM grids and metal-shadowed (Perez-Morga and Englund, 1993c; Zhao et al., 2008). Images were captured on a Hitachi 7600 transmission electron microscope equipped with a DVC 1412M-FW digital camera and AMT Image Capture Engine software (5.4.2.307). Brightness and contrast of images were adjusted uniformly using Adobe Photoshop.
Surface areas of isolated networks on electron micrographs were measured using a polar planimeter (Keuffel and Esser). To estimate minicircle density in isolated networks, 1 μm vertical lines (7 to 20 per network in electron micrographs, with more lines used for larger networks) were drawn at random positions in each network using Adobe Illustrator. We then counted how many times each line was crossed by a DNA strand (Chen et al., 1995a). This analysis was performed on the same networks used for area measurements.
Thin section EM, serial sectioning and 3D reconstruction analysis
Uninduced control cells and cells induced for RNAi for four days were fixed in 2.5% glutaraldehyde, post-fixed in osmium tetroxide, and embedded in Epon as described (Baines and Gull, 2008). Thin sections were examined by an FEI Tecnai 12 transmission electron microscope. The same blocks were also used to cut serial sections (thickness ~80-100 nm). Images were captured on a Gatan Ultrascan 1000 CCD camera and DigitalMicrograph software. Image galleries were assembled using Image J (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997-2007) and brightness and contrast of images were adjusted uniformly using Adobe Photoshop. 3-D reconstructions were made with IMOD [http://bio3d.colorado.edu/imod/index.html; (Kremer et al., 1996)], using the program Midas for manual alignment of images and 3dmod for modeling of the kDNA and the basal bodies. Snapshots created in 3dmod were imported into ImageJ to make the movies.
Supplementary Material
Acknowledgements
We thank Gokben Yildirir for valuable technical support, Mike Shaw for help with electron microscopy, and our lab colleagues for valuable discussion. Thanks also to Arnab Roy Chowdhury and Jane Scocca for commenting on the manuscript. This work was funded by the National Institutes of Health (grant AI058613 to P.T.E.) and The Wellcome Trust and EP Abraham Trust (to K.G.). K.G. is a Wellcome Trust Principal Research Fellow.
List of abbreviations
- (kDNA)
kinetoplast DNA
- (topo II)
topoisomerase II
- (TAC)
tripartitate attachment complex
Footnotes
Supplementary Material Videos showing rotation of 3-D reconstructions of serial section EMs are provided online as supplemental material. Included are a wild type kinetoplast (serial sections shown in Fig. 6C and 3-D reconstruction in Fig. 6D) and a kinetoplast from a cell after 4 days of TbTOP2mt RNAi (serial sections shown in Fig. 6B and 3-D reconstruction in Fig. 6E).
References
- Baines A, Gull K. WCB is a C2 domain protein defining the plasma membrane - sub-pellicular microtubule corset of kinetoplastid parasites. Protist. 2008;159:115–125. doi: 10.1016/j.protis.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Brun R, Jenni L, Tanner M, Schonenberger M, Schell KF. Cultivation of vertebrate infective forms derived from metacyclic forms of pleomorphic Trypanosoma brucei stocks. Short communication. Acta Trop. 1979;36:387–390. [PubMed] [Google Scholar]
- Chen J, Englund PT, Cozzarelli NR. Changes in network topology during the replication of kinetoplast DNA. EMBO J. 1995a;14:6339–6347. doi: 10.1002/j.1460-2075.1995.tb00325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rauch CA, White JH, Englund PT, Cozzarelli NR. The topology of the kinetoplast DNA network. Cell. 1995b;80:61–69. doi: 10.1016/0092-8674(95)90451-4. [DOI] [PubMed] [Google Scholar]
- Downey N, Hines JC, Sinha KM, Ray DS. Mitochondrial DNA ligases of Trypanosoma brucei. Eukaryot Cell. 2005;4:765–774. doi: 10.1128/EC.4.4.765-774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson ML, Torri AF, Perez-Morga D, Ward DC, Englund PT. Kinetoplast DNA replication: mechanistic differences between Trypanosoma brucei and Crithidia fasciculata. J Cell Biol. 1994;126:631–639. doi: 10.1083/jcb.126.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbride DL, Englund PT. The replication mechanism of kinetoplast DNA networks in several trypanosomatid species. J Cell Sci. 1998;111(Pt 6):675–679. doi: 10.1242/jcs.111.6.675. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH, Weijers PJ. The segregation of kinetoplast DNA networks in Trypanosoma brucei. Plasmid. 1980;4:97–116. doi: 10.1016/0147-619x(80)90086-4. [DOI] [PubMed] [Google Scholar]
- Johnson CE, Englund PT. Changes in organization of Crithidia fasciculata kinetoplast DNA replication proteins during the cell cycle. J Cell Biol. 1998;143:911–919. doi: 10.1083/jcb.143.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingbeil MM, Drew ME, Liu Y, Morris JC, Motyka SA, Saxowsky TT, Wang Z, Englund PT. Unlocking the secrets of trypanosome kinetoplast DNA network replication. Protist. 2001;152:255–262. doi: 10.1078/1434-4610-00066. [DOI] [PubMed] [Google Scholar]
- Klingbeil MM, Motyka SA, Englund PT. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol Cell. 2002;10:175–186. doi: 10.1016/s1097-2765(02)00571-3. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kulikowicz T, Shapiro TA. Distinct genes encode type II Topoisomerases for the nucleus and mitochondrion in the protozoan parasite Trypanosoma brucei. J Biol Chem. 2006;281:3048–3056. doi: 10.1074/jbc.M505977200. [DOI] [PubMed] [Google Scholar]
- Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Liu B, Molina H, Kalume D, Pandey A, Griffith JD, Englund PT. Role of p38 in replication of Trypanosoma brucei kinetoplast DNA. Mol Cell Biol. 2006;26:5382–5393. doi: 10.1128/MCB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Englund PT. The rotational dynamics of kinetoplast DNA replication. Mol Microbiol. 2007;64:676–690. doi: 10.1111/j.1365-2958.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- Lukes J, Hines JC, Evans CJ, Avliyakulov NK, Prabhu VP, Chen J, Ray DS. Disruption of the Crithidia fasciculata KAP1 gene results in structural rearrangement of the kinetoplast disc. Mol Biochem Parasitol. 2001;117:179–186. doi: 10.1016/s0166-6851(01)00348-6. [DOI] [PubMed] [Google Scholar]
- Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, Englund PT. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendy T, Sheline C, Ray DS. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell. 1988;55:1083–1088. doi: 10.1016/0092-8674(88)90252-8. [DOI] [PubMed] [Google Scholar]
- Melendy T, Ray DS. Novobiocin affinity purification of a mitochondrial type II topoisomerase from the trypanosomatid Crithidia fasciculata. J Biol Chem. 1989;264:1870–1876. [PubMed] [Google Scholar]
- Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasion SG, Hines JC, Aebersold R, Ray DS. Molecular cloning and expression of the gene encoding the kinetoplast-associated type II DNA topoisomerase of Crithidia fasciculata. Mol Biochem Parasitol. 1992;50:57–67. doi: 10.1016/0166-6851(92)90244-e. [DOI] [PubMed] [Google Scholar]
- Perez-Morga D, Englund PT. The structure of replicating kinetoplast DNA networks. J Cell Biol. 1993a;123:1069–1079. doi: 10.1083/jcb.123.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Morga DL, Englund PT. The attachment of minicircles to kinetoplast DNA networks during replication. Cell. 1993b;74:703–711. doi: 10.1016/0092-8674(93)90517-t. [DOI] [PubMed] [Google Scholar]
- Perez-Morga DL, Englund PT. Microtechnique for electron microscopy of DNA. Nucleic Acids Res. 1993c;21:1327–1328. doi: 10.1093/nar/21.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxowsky TT, Choudhary G, Klingbeil MM, Englund PT. Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J Biol Chem. 2003;278:49095–49101. doi: 10.1074/jbc.M308565200. [DOI] [PubMed] [Google Scholar]
- Shlomai J. The structure and replication of kinetoplast DNA. Curr Mol Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- Strauss PR, Wang JC. The TOP2 gene of Trypanosoma brucei: a single-copy gene that shares extensive homology with other TOP2 genes encoding eukaryotic DNA topoisomerase II. Mol Biochem Parasitol. 1990;38:141–150. doi: 10.1016/0166-6851(90)90214-7. [DOI] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Englund PT. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–4683. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Drew ME, Morris JC, Englund PT. Asymmetrical division of the kinetoplast DNA network of the trypanosome. EMBO J. 2002;21:4998–5005. doi: 10.1093/emboj/cdf482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Xu CW, Hines JC, Engel ML, Russell DG, Ray DS. Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol Cell Biol. 1996;16:564–576. doi: 10.1128/mcb.16.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Lindsay ME, Chowdhury A. Roy, Robinson DR, Englund PT. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 2008;27:143–154. doi: 10.1038/sj.emboj.7601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.