Abstract
Plants respond to low levels of UV-B radiation with a coordinated photomorphogenic response that allows acclimation to this environmental stress factor. The key players in this UV-B response are COP1 (an E3 ubiquitin ligase), UVR8 (a β-propeller protein), and HY5 (a bZIP transcription factor). We have shown previously that an elevated UV-B–specific response is associated with dwarf growth, indicating the importance of balancing UV-B–specific signaling. Negative regulators of this pathway are not known, however. Here, we describe two highly related WD40-repeat proteins, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2, that interact directly with UVR8 as potent repressors of UV-B signaling. Both genes were transcriptionally activated by UV-B in a COP1-, UVR8-, and HY5-dependent manner. rup1 rup2 double mutants showed an enhanced response to UV-B and elevated UV-B tolerance after acclimation. Overexpression of RUP2 resulted in reduced UV-B–induced photomorphogenesis and impaired acclimation, leading to hypersensitivity to UV-B stress. These results are consistent with an important regulatory role for RUP1 and RUP2, which act downstream of UVR8–COP1 in a negative feedback loop impinging on UVR8 function, balancing UV-B defense measures and plant growth.
Keywords: abiotic stress, light signaling, photobiology, quercetin, sun simulator
UV-B (280–315 nm) radiation of wavelengths exceeding ∼295 nm as an integral part of the sunlight reaching the surface of the Earth induces a broad range of physiological responses. UV-B stress induces mostly unspecific damage responses in living organisms (1, 2); however, plants demonstrate UV-B–specific photoregulatory responses regulated by an as-yet molecularly unidentified UV-B receptor that is different from photoreceptors responding to the visible part of the light spectrum (3–5). This pathway is characterized molecularly by the involvement of the UVR8 (UV RESISTANCE LOCUS 8) protein, which was recently shown to enhance survival under simulated sunlight with realistic UV-B levels (6). In contrast, no difference in the performance of uvr8 mutants and WT was seen when the UV radiation was filtered out (6).
UVR8 is a β-propeller protein with a sequence similarity to the eukaryotic guanine nucleotide exchange factor RCC1 (7). Although UVR8 has little in vitro exchange activity, it interacts with histones and is associated with chromatin of the ELONGATED HYPOCOTYL 5 (HY5) promoter region (8). Moreover, UV-B radiation stimulates the rapid nuclear accumulation of UVR8, which is necessary but not sufficient for its function (9). Recent data show that UVR8 interacts with the multifunctional E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) in a UV-B–dependent manner (6).
The COP1 protein comprises an N-terminal RING-finger domain, a coiled-coil region, and a C-terminal WD40-repeat domain (10). The protein–protein interaction with UVR8 depends on the WD40-repeat domain in COP1 (6). Both COP1 and UVR8 then impinge on the transcriptional activation of the HY5 gene, which encodes a bZIP transcription factor with a central function in the UV-B signaling pathway (6, 8, 11, 12).
In addition to the transcriptional activation, COP1-mediated degradation of HY5 protein is inhibited under UV-B, probably due to the interaction of UVR8 with COP1 (6, 12). Despite the recent identification of important positive players and pathways, the “brakes” in UV-B–specific signaling are not well known. The recently described ROOT UVB SENSITIVE 1 (RUS1) protein seems to negatively regulate a postulated UV-B response pathway that is restricted to roots and thus differs from the COP1/UVR8 pathway (13). However, the UV-B–resistant but dwarfed phenotype of Arabidopsis lines overexpressing UVR8 clearly points to the need for tight control of the UV-B response in the latter pathway (6).
In response to visible light, the action of positive signaling factors downstream of the phytochrome (red/far-red) and cryptochrome (blue/UV-A) photoreceptors is counterbalanced by an important set of repressor proteins, including the four members of the SUPPRESSOR OF PHYA-105 (SPA) gene family and COP1, which interact and form complexes in vivo (14, 15). These proteins are repressors of light signaling in both dark-grown and light-grown seedlings, and their absence in mutant plants leads to marked dwarfism or seedling lethality (10, 15). In contrast, the COP1 protein positively regulates the UV-B–specific response independent of the SPA proteins (12).
Repressors of the COP1/UVR8-mediated UV-B–specific pathway were unknown until now. Here we describe two redundant UVR8-interacting WD40-repeat proteins, RUP1 and RUP2, that are important repressors of UV-B–induced photomorphogenesis and UV-B acclimation. These proteins play a crucial negative feedback regulatory role balancing UV-B–specific responses and ensuring normal plant growth.
Results
RUP1 and RUP2 Transcripts Are Rapidly and Transiently Induced by UV-B in a COP1-, UVR8-, and HY5-Dependent Manner.
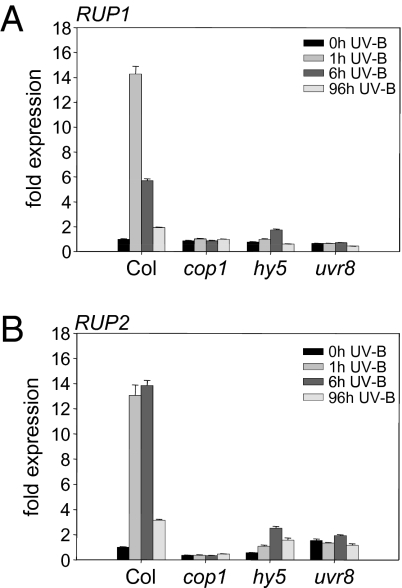
We previously analyzed specific responses to UV-B at the level of transcriptomic change (6, 11) and confirmed the transcriptional activation of several genes using the luciferase reporter (including At5g52250; see below) (16). We selected two genes induced early in response to narrowband UV-B irradiance encoding highly similar WD40-repeat proteins for detailed analysis. We named these genes REPRESSOR OF UV-B PHOTOMORPHOGENESIS (RUP) 1 and 2 (At5g52250 and At5g23730). Quantitative RT-PCR confirmed their early responsiveness to supplementary narrowband UV-B radiation (Fig. 1 A and B). Moreover, the UV-B–mediated up-regulation of both genes was found to depend on the presence of functional HY5, COP1, and UVR8 proteins (Fig. 1 A and B), showing these to be potential effectors of the main players.
Fig. 1.
RUP1 and RUP2 gene activation in response to UV-B depends on COP1, HY5, and UVR8. (A and B) Quantitative RT-PCR analysis of RUP1 (A) and RUP2 (B) gene activation in response to UV-B in cop1-4, hy5-215, and uvr8-6 mutants compared with WT Col. Four-day-old seedlings were irradiated with UV-B for the indicated times before harvesting. Representative data from three independent experiments are shown. Error bars represent SD of technical triplicates.
The RUP1 (385 aa) and RUP2 (368 aa) proteins are highly homologous, with 63% identity in an overlap of 349 amino acids (Fig. S1). Both proteins consist of seven WD40-repeats with apparently no additional domains. In transgenic Arabidopsis lines that constitutively express RUP1-YFP and RUP2-YFP under control of the CaMV 35S-promoter, both RUP-YFP fusion proteins localized to the nucleus and the cytoplasm (Fig. S2A). Their subcellular localization was similar in continuous darkness, white light, and white light supplemented with UV-B radiation (Fig. S2A). In agreement, RUP2-GFP protein expressed under its own promoter was detected in both cytosolic and nuclear fractions isolated from transgenic plants (Fig. S2B); however, the very low expression levels of RUP2-GFP in this line prevented microscopic analysis of its subcellular localization. Thus, RUP gene expression is induced by UV-B downstream of the UVR8-COP1 pathway, and the constitutively overexpressed RUP-YFP fusion proteins localize to both nucleus and cytoplasm, independent of the light conditions.
RUP Proteins Interact with UVR8.
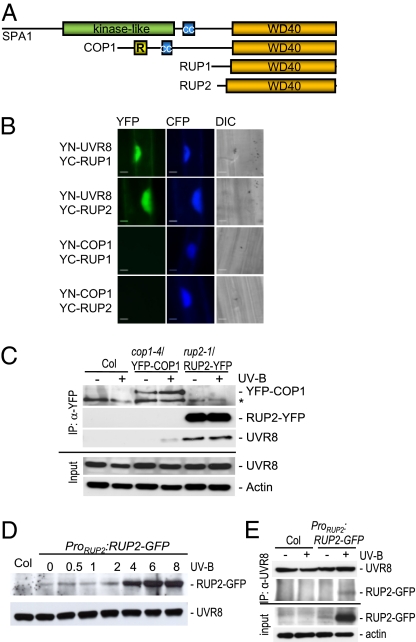
Interestingly, the closest relatives of the RUP proteins, based on sequence conservation of WD40-repeat domains, are the structurally related COP1 and SPA proteins (Fig. 2A and Fig. S3). The SPA proteins are repressors of photomorphogenesis with no role in UV-B signaling, whereas the COP1 protein represses photomorphogenesis but promotes UV-B–specific signaling (12). Our previous results demonstrated that the UV-B–dependent interaction of UVR8 with COP1 depends on the WD40 domain of COP1 (6). This prompted us to investigate whether RUP proteins also interact directly with UVR8, using the bimolecular fluorescent complementation (BiFC) assay (17) in transiently transformed mustard hypocotyl cells (6). Reconstitution of a functional YFP signal from the complementary “split YFP” parts attached to the UVR8 and either RUP1 or RUP2 proteins was clearly identified (Fig. 2B). No YFP signal was seen when YC-RUP1 and YC-RUP2 were used in combination with empty vector controls, YN-COP1, or YN-RUP2 and YN-RUP1, respectively, indicating that the interaction was specific (Fig. 2B and Fig. S4). In contrast to the UV-B–dependent interaction of UVR8 with COP1 (6), the interaction with RUP1 and RUP2 occurred to the same extent under conditions devoid of UV-B radiation (Fig. 2B).
Fig. 2.
The RUP proteins interact directly with the UVR8 protein. (A) Schematic comparison of the protein domain structures of the three groups of WD40-repeat–containing repressors of photomorphogenesis (see also Fig. S3). (B) BiFC visualization of YC-RUP1 and YC-RUP2 interaction with YN-UVR8, but not with YN-COP1. A Pro35S:CFP control plasmid was always cobombarded to identify transformed cells before the analysis of YFP fluorescence. Specific CFP and YFP filter sets were used for microscopic analysis. Differential interference contrast images are shown. (Scale bar: 10 μm.) (C) Coimmunoprecipitation of endogenous UVR8 with RUP2-YFP. Coimmunoprecipitation of proteins using anti-YFP antibodies in extracts from rup2-1/Pro35S:RUP2-YFP transgenic seedlings. Four-day-old seedlings were irradiated with UV-B for 4 h (+UV-B) or mock-treated under a cutoff filtering out UV-B (−UV-B). An asterisk indicates a nonspecific cross-reacting band. (D) UV-B–responsive accumulation of RUP2-GFP protein expressed under its own promoter. Total protein was isolated from 4-d-old rup1 rup2/ProRUP2:RUP2-GFP transgenic seedlings that were irradiated with UV-B for the indicated times before harvesting. The protein gel blot was sequentially probed with anti-GFP and anti-UVR8 antibodies. (E) Coimmunoprecipitation of RUP2-GFP expressed under its own promoter with endogenous UVR8. Coimmunoprecipitation of proteins using anti-UVR8 antibodies in extracts from rup1 rup2/ProRUP2:RUP2-GFP transgenic seedlings and nontransgenic Col controls. Four-day-old seedlings were irradiated with UV-B for 6 h (+UV-B) or mock-treated under a cutoff filtering out UV-B (−UV-B).
To further investigate RUP-UVR8 interaction in planta, we performed coimmunoprecipitation experiments using transgenic lines constitutively expressing YFP-tagged RUP2 in rup2-1 mutants. In agreement with the BiFC data, endogenous UVR8 protein was coimmunoprecipitated with RUP2-YFP from rup2-1/Pro35S:RUP2-YFP, independent of UV-B radiation (Fig. 2C). In contrast, no coimmunoprecipitation of UVR8 was found for control plants (Col), and YFP-COP1 coimmunoprecipitated UVR8 only under supplemental UV-B (Fig. 2C), as described previously (6). In agreement with the transcriptional activation of the RUP2 gene by UV-B (Fig. 1B), a clear increase in RUP2-GFP protein level was detected in response to supplemental UV-B when RUP2-GFP was expressed under its own promoter (ProRUP2:RUP2-GFP; Fig. 2D). Accordingly, in this line, UVR8 coimmunoprecipitated RUP2-GFP only under supplemental UV-B when the latter was expressed (Fig. 2E). Thus, the RUP proteins are UV-B–induced UVR8-interacting proteins that likely play a role in UV-B signaling.
rup1 rup2 Double Mutants Are Hypersensitive to Photomorphogenic UV-B Radiation.
To analyze the involvement of the two RUP proteins in UVR8-mediated UV-B signaling, we isolated homozygous knockout mutants for the RUP1 and RUP2 genes. RUP1 and RUP2 are encoded by intronless genes, and we identified T-DNA insertional lines with impaired functionality of both proteins (Fig. S5A). Indeed, in both mutants, the T-DNA insertion led to the absence of the respective mRNAs, as determined by RNA gel blot analysis (Fig. S5B). Because RUP1 and RUP2 probably are functionally redundant, as suggested by their high sequence conservation, we also generated a double-null mutant. As expected, both transcripts were missing from the rup1 rup2 mutant (Fig. S5B).
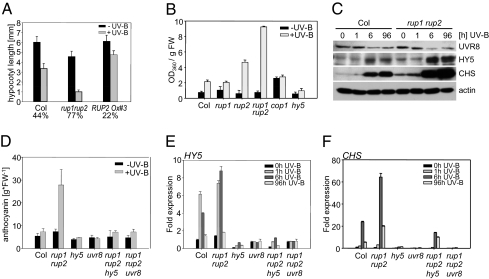
Interestingly, rup1 rup2 double mutants showed a strong hypersensitivity to supplementary narrowband UV-B radiation at the level of hypocotyl growth inhibition (Fig. 3A), and flavonoid (Fig. 3B) and anthocyanin accumulation (Fig. 3D and Fig. S5C). The rup1 single mutant was similar to WT, whereas the rup2 single mutant already showed weak UV-B hypersensitivity (Fig. 3B). The UV-B hypersensitivity of the rup1 rup2 double mutant also was reflected in the molecular analysis, which showed more strongly induced UV-B–responsive HY5 and CHS gene expression than in the WT (Fig. S5 D and E). The gene expression data also were reflected in the differences in HY5 and CHS protein levels under UV-B (Fig. 3C). There was no major difference in UVR8 protein level between the rup1 rup2 double mutant and WT, indicating that rup1 rup2 hypersensitivity is not due to elevated UVR8 levels (Fig. 3C).
Fig. 3.
RUP1 and RUP2 are repressors of UV-B–induced photomorphogenesis mediated by UVR8 and HY5. (A) Hypocotyl length of 4-d-old seedlings grown with or without supplemental UV-B. Numbers below the bars show the relative hypocotyl growth inhibition by UV-B as a percentage. Error bars represent SD (n = 30). (B) UV-B–induced flavonoid accumulation is enhanced in rup2 mutants and especially in rup1 rup2 double mutants. Error bars represent SD (n = 3). (C) Immunoblot analysis of UVR8, HY5, CHS, and actin (loading control) protein levels in 4-d-old seedlings irradiated with UV-B for the indicated times before harvesting. (D) Anthocyanin measurements of 4-d-old seedlings grown with or without supplemental UV-B showing that UV-B hypersensitivity of rup1 rup2 double mutants depends on functional UVR8 and HY5 proteins. Error bars represent SD (n = 3). (E and F) Quantitative RT-PCR analysis of HY5 (E) and CHS (F) gene activation in response to UV-B in rup1 rup2 hy5 and rup1 rup2 uvr8 triple mutants compared with WT Col, rup1 rup2, hy5-215, and uvr8-6. Four-day-old seedlings were irradiated with UV-B for the indicated times before harvesting. Error bars represent the SD of technical triplicates.
It should be noted that the RUP1 and RUP2 genes are also transcriptionally activated by red, far-red, and blue light, indicating a more general role in light responses (Fig. S6A). However, our initial analysis found no involvement of RUP1 and RUP2 in the response to these wavelengths at the level of hypocotyl growth inhibition and expression of the HY5 and CHS marker genes (Fig. S6 B–F). Thus, we conclude that RUP1 and RUP2 proteins have a major negative regulatory function in the UV-B photoregulatory response of Arabidopsis.
UV-B Hypersensitivity of rup1 rup2 Depends on Functional UVR8 and HY5 Proteins.
To examine whether the UV-B hypersensitivity of the rup1 rup2 mutant requires HY5 and UVR8 proteins, we generated rup1 rup2 hy5 and rup1 rup2 uvr8 triple mutants and analyzed their UV-B responses. Measurement of UV-B–induced anthocyanin accumulation as well as HY5 and CHS gene expression showed that both uvr8 and hy5 suppress the rup1 rup2 mutant phenotype and thus are epistatic to rup1 rup2 (Fig. 3 D–F). Thus, both UVR8 and HY5 proteins are required for UV-B hypersensitivity in rup1 rup2 double mutants.
RUP2 Overexpression Results in Blockage of UV-B–Specific Signaling.
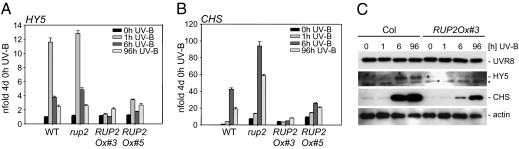
Because the genetic loss-of-function data strongly indicated a redundant function for RUP1 and RUP2 as crucial repressors of UV-B–induced photomorphogenesis, we generated CaMV 35S promoter–driven RUP2 overexpression lines. RUP2 overexpression resulted in a strong block of UV-B–induced expression of the HY5 and CHS marker genes (Fig. 4 A and B), strongly supporting RUP2's role as a repressor of UV-B signaling. Protein gel blot analysis confirmed the repressive effect on CHS and HY5 at the protein level and demonstrated that the hyposensitivity is not due to down-regulation of UVR8 protein in the RUP2 overexpression lines (Fig. 4C).
Fig. 4.
Overexpression of RUP2 represses the UV-B response. (A and B) Quantitative RT-PCR analysis of HY5 (A) and CHS (B) gene activation in response to UV-B in two independent RUP2 overexpression lines compared with WT Col and the rup2-1 single mutant. Error bars represent the SD of technical triplicates. (C) Immunoblot analysis of UVR8, HY5, CHS, and actin (loading control) protein levels. In A–C, 4-d-old seedlings were irradiated with UV-B for the indicated times before harvesting. RUP2Ox#3/#5=Pro35S:RUP2 in rup2-1, lines 3 and 5.
rup1 rup2 Acclimatizes Better to UV-B than WT, and RUP2 Overexpression Lines Are Impaired in UV-B Tolerance.
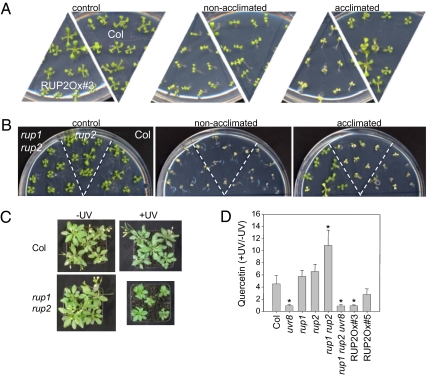
Taken together, our data indicate an important role of the RUP proteins as repressors of the UVR8/COP1-mediated UV-B photomorphogenic pathway. We previously showed that UV-B acclimation is absent in uvr8 mutants and enhanced in UVR8 overexpression lines (6). Because our data indicated similar phenotypes for RUP2 overexpression and the uvr8 mutation and for UVR8 overexpression and the rup1 rup2 double mutation, we directly tested the importance of RUP proteins for UV-B acclimation by combining weak, narrowband UV-B exposure with subsequent broadband UV-B stress. As described previously (6), exposure of WT seedlings for 7 d to narrowband UV-B that activated photomorphogenic responses resulted in enhanced tolerance to a subsequent broadband UV-B stress treatment (Fig. 5A). This acclimation effect was absent in RUP2 overexpression lines and enhanced in rup1 rup2 double mutants (Fig. 5 A and B).
Fig. 5.
The RUP proteins negatively regulate UV-B acclimation and tolerance. (A) Arabidopsis seedlings were grown for 7 d under white light (control and nonacclimated) or white light supplemented with narrowband UV-B (acclimated). Seedlings were then irradiated for 1.5 h (nonacclimated and acclimated) with broadband UV-B under a WG305 cutoff filter, or subjected to a 1.5-h mock treatment (control) under a WG345 filter (−UV-B). Treated seedlings were further grown for 7 d under standard conditions without UV-B before being photographed. (B) Identical treatment as that for A, except that the seedlings were exposed to broadband UV-B under a WG305 cutoff filter for 2 h (nonacclimated and acclimated). (C) Some 25-d-old rup1 rup2 and WT Col plants grown in sunlight simulators under realistic conditions (+UV) or with the UV portion specifically filtered out (−UV). (D) Quercetin accumulation (+UV/−UV) in 27-d-old plants grown under sun simulator conditions. Values are mean ± SD (n = 3). An asterisk indicates statistically significant differences from Col (P < 0.05, unpaired Student's t test).
To examine the importance of the RUP proteins under more realistic conditions, we grew plants in sun simulators with a natural spectral balance throughout the UV-to-infrared spectrum (18). Under these realistic conditions, rup1 rup2 mutant plants were clearly tolerant to UV-B radiation but were dwarfed and dark green (Fig. 5C), very similar to the UVR8 overexpressor phenotype described previously (6). The UV-B response mediated by the COP1/UVR8 pathway is associated with the accumulation of flavonol glycosides; thus, we quantified the relative levels of the flavonol quercetin under the sun simulator growth conditions. In WT Col, we found an ∼5-fold UV-B–mediated increase in quercetin level, similar to that in rup1 and rup2 single mutants (Fig. 5D). In contrast, the UV-B response at the level of quercetin accumulation was increased by ∼11-fold in the rup1 rup2 double mutant (Fig. 5D). It also should be noted that although the RUP2 overexpression lines demonstrated no dramatic difference from WT in overall growth phenotype, they did exhibit a reduced UV-B response in terms of quercetin accumulation (Fig. 5D). Thus, we conclude that the RUP proteins influence the important balance between two connected products of UV-B–specific signaling, namely growth inhibition and the mounting of UV-B defense measures.
Discussion
The survival of sessile plants in sunlight is ensured by UV-protective responses that are largely regulated by the UV-B–specific UVR8–COP1 pathway. Activation of these responses must be well balanced; a reduced response results in UV-B damage and cellular death (as in the uvr8 mutant), whereas an exaggerated response results in impaired growth and dwarfism (as produced by UVR8 overexpression) (6). Here we show that the Arabidopsis RUP1 and RUP2 proteins are crucial repressors of UV-B–induced photomorphogenesis that result in an adequate and balanced UVR8/COP1-mediated UV-B response.
Previous work has shown that specific perception of UV-B radiation by a postulated UV-B receptor results in rapid UVR8–COP1 interaction (6). This allows the COP1/UVR8-mediated activation of numerous genes, including HY5, which confers UV acclimation and protection (6, 8, 12), and RUP1 and RUP2, which provide negative feedback regulation through direct interaction with UVR8 (Fig. 2 D and E; see also the model shown in Fig. S7). The latter two genes encode β-propeller proteins belonging to the very diverse superfamily of WD40-repeat regulatory proteins, which comprises 237 potential proteins in Arabidopsis containing four or more copies of the WD40 motif (19). The common defining feature of these proteins is an ≈40-aa stretch typically ending in Trp-Asp (WD), but there is only limited amino acid sequence conservation otherwise. In many instances, repeated WD40 motifs act as sites for protein–protein interaction, and many proteins containing WD40 repeats are known to serve as platforms for the assembly of protein complexes (19). The two RUP proteins show significant sequence conservation in their seven WD40-repeat domains with the COP1 and SPA1–SPA4 proteins, including a conserved 16-aa DWD (DDB1-binding WD40) motif (20, 21). Members of the DWD motif–containing subset of WD40 proteins were shown to act as substrate receptors for DDB1-CUL4-ROC1–based E3 ubiquitin ligases (22). Such a function remains to be described for the RUP proteins, however. Nevertheless, a phylogenetic analysis based on the WD40-repeat region indicated that the most closely related sequences to RUP1 and RUP2 encoded in the Arabidopsis genome are the SPA proteins and COP1, with which they share about 33% and 37% identity in the WD40 domain, respectively. COP1 and SPA protein family members function as repressors under visible light devoid of UV-B (10, 15), a light environment prevalent in laboratory experiments, which neglects the influence of the UV-B radiation intrinsic to sunlight. A detailed understanding of the regulatory role of UV-B is needed to understand the control by light of plant growth and development, however. Recent experiments using sun simulator conditions to analyze the performance of the uvr8 mutant and UVR8 overexpression lines demonstrated the importance of the UV-B–specific photoregulatory pathway (6). The related phenotypes of RUP2 overexpression and rup1 rup2 double mutants described here demonstrate the importance of these UV-B–specific repressors under UV-B radiation, similar to the COP1 and SPA proteins in conditions devoid of UV-B. Indeed, the phenotype of the rup1 rup2 double mutant under supplemental UV-B radiation (e.g., short hypocotyl, dwarfism, high anthocyanin) is very reminiscent of the cop1 and combinatorial spa mutants grown under white light without UV-B.
The SPA–COP1 E3 ligase complexes are a point of convergence downstream of multiple light signals and constitute a central repressor of photomorphogenesis that is inactivated by visible light in an as-yet unknown molecular manner (14). Interestingly, the coiled-coil domain and the WD40-repeat domain of SPA1 are sufficient for its function, and the kinase-related domain apparently is not required (23, 24). Arabidopsis COP1 interacts with the four members of the SPA protein family (SPA1–SPA4) with their coiled-coil domains (25). In contrast, RUP proteins have no coiled-coil domain. In agreement with this, we found no interaction with COP1. However, similar to the activation of RUP1 and RUP2 by UV-B, the levels of SPA1, SPA3, and SPA4 transcripts were increased by red, far-red, and blue light, consistent with a negative feedback role in light-grown seedlings (23).
In contrast to RUP1 and RUP2, expression of the other main factors (COP1 and UVR8) responsible for the UV-B response in Arabidopsis is constitutive and not regulated by UV-B. Nevertheless, a change in the abundance of interacting proteins, such as the RUP1 and RUP2 (or SPA) proteins, in response to an exogenous signal could alter the activity and/or specificity of the complex as a whole. Thus, the regulation of RUP1 and RUP2 expression seems crucial to the adjustment of plant growth and development to changes in the light environment. Our data suggest that a rapid increase in RUP1 and RUP2 abundance is necessary to prevent overstimulation when seedlings are exposed to UV-B.
Interestingly, RNAi of a gene encoding a RUP-related protein in tomato, LeCOP1LIKE, results in field-grown plants with exaggerated photomorphogenesis, dark-green leaves, and elevated fruit carotenoid levels (26). This finding led to the conclusion that LeCOP1LIKE is involved in light signal transduction and functions as a negative regulator of fruit pigmentation. However, it should be noted that the closest homolog of LeCOP1LIKE in Arabidopsis is RUP1, and thus the enhanced photomorphogenesis described for the LeCOP1LIKE-RNAi lines under natural conditions in the field might be linked instead to its potential function as a repressor of UV-B signaling in tomato. Thus, manipulating UV-B signaling might provide another way to modify the nutrient quality of plants (27).
In summary, we have shown that the UV-B–specific response impinging on plant growth is precisely balanced by the UV-B–activated and UVR8-interacting RUP1 and RUP2 proteins. RUP1 and RUP2 are early responsive genes that function as negative regulators of the UV-B response in Arabidopsis through direct interaction with the UVR8 protein. This negative feedback loop prevents an exaggerated photomorphogenic UV-B response that would strongly affect plant growth and development.
Materials and Methods
Plant Material and UV-B Irradiation.
cop1-4, hy5-215, uvr8-6, rup1-1 (SALK_060638), and rup2-1 (SALK_108846) are in the Columbia ecotype (Col) (6, 28–30). Plants were grown and irradiated exactly as described previously (6, 12). The condition of the treatments in the sun simulator was a 14-h day period with mean photosynthetically active radiation (400–700 nm) of 730 μmol m−2 s−1 and 12 h of UV-B irradiance with a biologically effective dose of 500 mW m−2 [weighted by the generalized plant action spectrum (31), normalized at 300 nm]. Controls were grown excluding the entire UV radiation spectrum. Spectroradiometric measurements were performed using a double-monochromator system (model DTM-300; Bentham) and are shown in Fig. S8. The temperature was maintained at 23 °C during the day and 18 °C at night. The relative humidity was kept constant at 60%.
RNA Extraction and Analysis by Real-Time PCR.
Arabidopsis RNA was isolated with the Plant RNeasy Kit (Qiagen) according to the manufacturer's instructions. Quantitative real-time PCR was performed in a 96-well format using the 7300 Real-Time PCR System and TaqMan probes (Applied Biosystems). cDNA was synthesized from 50 ng of RNA with random hexamers using the TaqMan Reverse-Transcription Reagent Kit (Applied Biosystems). Quantitative PCR reactions were performed using the ABsolute QPCR Rox Mix Kit (ABgene), following the manufacturer's instructions. The gene-specific probes and primers were as follows: RUP1 (At5g52250), probe 6-FAM-CGCATCCACCGGATCAGACGCT-TAMRA with RUP1_for (5′-TCTCTTTCCGCCGTTGTTTC-3′) and RUP1-rev (5′-CCGGTAGGGTCGAACTCGAT-3′); RUP2 (At5g23730), probe 6-FAM-TCGCTACCGCCGGGATTTCAAGA-TAMRA with RUP2_for (5′-TGAATTCGATCCCACTGATAACA-3′) and RUP2-rev (5′-AGGGAGGCCGTAAAAACGA-3′); and CHS (At5G13930) and HY5 (At5g11260) as described previously (6). cDNA concentrations were normalized to a standard of 18S rRNA transcript levels using the Eukaryotic 18S rRNA Kit (Applied Biosystems). Expression was determined in triplicate.
Immunoprecipitation Assays and Protein Gel Blot Analysis.
Immunoprecipitation of YFP-COP1 and RUP2-YFP using monoclonal anti-GFP antibodies (Invitrogen) and protein A-agarose (Roche Applied Science) was performed as described previously (6). For protein gel blot analysis, total cellular proteins (10 μg) or immunoprecipitates were separated by electrophoresis in 10% SDS-polyacrylamide gels and electrophoretically transferred to PVDF membranes according to the manufacturer's instructions (Bio-Rad). Polyclonal anti-UVR8 (6), anti-HY5 (12), anti-actin (Sigma-Aldrich), anti-CHS (Santa Cruz Biotechnology), and monoclonal anti-GFP (BAbCO) were used as primary antibodies, with HRP-conjugated protein A (Pierce) or anti-rabbit, anti-goat, and anti-mouse immunoglobulins (DAKO) used as secondary antibodies. Signal detection was performed using the ECL Plus Western Detection Kit (GE Healthcare).
Extraction and Measurement of Flavonoids and Anthocyanins.
Anthocyanins were extracted and quantified as described previously (32). Flavonoid measurements were analyzed as described previously (12). Experiments were carried out at least in triplicate. Analysis of the soluble phenolic compound quercetin from plants grown in the sun simulator was performed by reversed-phase-HPLC as described previously (33).
Bimolecular Fluorescence Complementation.
The RUP1 and RUP2 coding sequences were transferred into Gateway-compatible BiFC binary vectors, pE-SPYNE-GW and pE-SPYCE-GW (kindly provided by W. Dröge-Laser, University of Göttingen). Transient transformation of mustard seedlings using the Biolistic PDS-1000/He System (Bio-Rad) and BiFC assays were carried out as described previously (34).
Epifluorescence and Light Microscopy.
For epifluorescence and light microscopy, the seedlings were transferred to glass slides and examined with a Zeiss Axioskop II microscope. Excitation and detection of YFP and CFP were performed with standard YFP and CFP filter sets, respectively (AHF analysentechnik). Documentation of representative cells was done by photography using a Zeiss Axiocam digital camera system.
Supplementary Material
Acknowledgments
We thank Patrick King for editing the manuscript, Luca Rizzini (University of Freiburg, Germany) for providing an RUP2 entry clone, and Markus Funk and Susanne Stich for providing excellent technical assistance. We also thank the Nottingham Arabidopsis Stock Centre (University of Nottingham, Loughborough, UK) for providing the SALK mutant lines. This work was supported by the Emmy Noether Programme of the Deutsche Forschungsgemeinschaft (UL341/1-1 to R.U.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914532107/-/DCSupplemental.
References
- 1.Ulm R. Molecular genetics of genotoxic stress signalling in plants. Topics Curr Genet. 2003;4:217–240. [Google Scholar]
- 2.Herrlich P, Karin M, Weiss C. Supreme EnLIGHTenment: Damage recognition and signaling in the mammalian UV response. Mol Cell. 2008;29:279–290. doi: 10.1016/j.molcel.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulm R, Nagy F. Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol. 2005;8:477–482. doi: 10.1016/j.pbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 5.Brosche M, Strid A. Molecular events following perception of ultraviolet-B radiation by plants. Physiol Plant. 2003;117:1–10. [Google Scholar]
- 6.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown BA, et al. A UV-B–specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiserli E, Jenkins GI. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B–specific signaling component UVR8 and activates its function in the nucleus. Plant Cell. 2007;19:2662–2673. doi: 10.1105/tpc.107.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi C, Deng XW. COP1: From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong H, et al. Role of root UV-B sensing in Arabidopsis early seedling development. Proc Natl Acad Sci USA. 2008;105:21039–21044. doi: 10.1073/pnas.0809942106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20:2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell. 2004;16:2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safrany J, et al. Identification of a novel cis-regulatory element for UV-B–induced transcription in Arabidopsis. Plant J. 2008;54:402–414. doi: 10.1111/j.1365-313X.2008.03435.x. [DOI] [PubMed] [Google Scholar]
- 17.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiel S, et al. A phytotron for plant stress research: How far can artificial lighting compare to natural sunlight? J Plant Physiol. 1996;148:456–463. [Google Scholar]
- 19.van Nocker S, Ludwig P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics. 2003;4:50. doi: 10.1186/1471-2164-4-50. Available at http://www.biomedcentral.com/1471-2164/4/50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson S, Xiong Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fittinghoff K, et al. Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 2006;47:577–590. doi: 10.1111/j.1365-313X.2006.02812.x. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Wang H. The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 2006;47:564–576. doi: 10.1111/j.1365-313X.2006.02811.x. [DOI] [PubMed] [Google Scholar]
- 25.Hoecker U, Quail PH. The phytochrome A–specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem. 2001;276:38173–38178. doi: 10.1074/jbc.M103140200. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA. 2004;101:9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen MAK, Hectors K, O'Brien NM, Guisez Y, Potters G. Plant stress and human health: Do human consumers benefit from UV-B–acclimated crops? Plant Sci. 2008;175:449–458. [Google Scholar]
- 28.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 29.McNellis TW, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldwell MM. Solar UV irradiation and the growth and development of higher plants. In: Giese AC, editor. Photophysiology. Vol. 6. New York: Academic; 1971. pp. 131–177. [Google Scholar]
- 32.Noh B, Spalding EP. Anion channels and the stimulation of anthocyanin accumulation by blue light in Arabidopsis seedlings. Plant Physiol. 1998;116:503–509. doi: 10.1104/pp.116.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turunen M, et al. The effects of UV exclusion on the soluble phenolics of young Scots pine seedlings in the subarctic. Environ Pollut. 1999;106:219–228. doi: 10.1016/s0269-7491(99)00070-6. [DOI] [PubMed] [Google Scholar]
- 34.Stolpe T, et al. In planta analysis of protein–protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma. 2005;226:137–146. doi: 10.1007/s00709-005-0122-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.