Abstract
Histone deacetylase 6 (HDAC6) is structurally and functionally unique among the 11 human zinc-dependent histone deacetylases. Here we show that chemical inhibition with the HDAC6-selective inhibitor tubacin significantly enhances cell death induced by the topoisomerase II inhibitors etoposide and doxorubicin and the pan-HDAC inhibitor SAHA (vorinostat) in transformed cells (LNCaP, MCF-7), an effect not observed in normal cells (human foreskin fibroblast cells). The inactive analogue of tubacin, nil-tubacin, does not sensitize transformed cells to these anticancer agents. Further, we show that down-regulation of HDAC6 expression by shRNA in LNCaP cells enhances cell death induced by etoposide, doxorubicin, and SAHA. Tubacin in combination with SAHA or etoposide is more potent than either drug alone in activating the intrinsic apoptotic pathway in transformed cells, as evidenced by an increase in PARP cleavage and partial inhibition of this effect by the pan-caspase inhibitor Z-VAD-fmk. HDAC6 inhibition with tubacin induces the accumulation of γH2AX, an early marker of DNA double-strand breaks. Tubacin enhances DNA damage induced by etoposide or SAHA as indicated by increased accumulation of γH2AX and activation of the checkpoint kinase Chk2. Tubacin induces the expression of DDIT3 (CHOP/GADD153), a transcription factor up-regulated in response to cellular stress. DDIT3 induction is further increased when tubacin is combined with SAHA. These findings point to mechanisms by which HDAC6-selective inhibition can enhance the efficacy of certain anti-cancer agents in transformed cells.
Keywords: γH2AX, Chk2, DDIT3/CHOP/GADD153, apoptosis, histones
In humans, 11 zinc-dependent histone deacetylases (HDACs) have been identified and classified based on homology to yeast proteins: class I (HDACs 1, 2, 3, and 8), class IIa, (HDACs 4, 5, 7 and 9), class IIb (HDACs 6 and 10), and class IV (HDAC11). Class I HDACs are primarily nuclear proteins and display enzymatic activity toward histone substrates. Class IIa HDACs shuttle between the nucleus and cytoplasm. Class IIb HDACs are primarily cytoplasmic proteins and have nonhistone proteins as primary targets (1). Inhibiting HDAC enzymes has emerged as a promising approach for the treatment of cancers (2–5). Several different structural classes of HDAC inhibitors are currently in clinical development for the treatment of both hematologic and solid tumors. Two HDAC inhibitors, SAHA (vorinostat) and romidepsin (depsipeptide or FK228), have been approved by the US Food and Drug Administration for the treatment of cutaneous T cell lymphoma (6–8). Preclinical data with numerous cancer cell lines have shown synergistic and additive effects when combining HDAC inhibitors with various antitumor therapies (5). A number of combination therapies with HDAC inhibitors are being investigated in clinical trials for the treatment of neoplastic diseases (9). HDAC inhibitors currently in clinical development target several HDAC isoforms (10). The discovery of isoform-selective HDACi is important to elucidate the mechanism of action of specific HDAC enzymes and may offer a therapeutic advantage by minimizing toxicity.
This study focuses on the selective inhibition of HDAC6. HDAC6 is a structurally and functionally unique zinc-dependent HDAC. HDAC6 has two catalytic domains, a ubiquitin-binding zinc-finger domain, and a dynein-binding domain, and selectively deacetylates nonhistone proteins such as tubulin, HSP90, cortactin, and peroxiredoxins (1, 11–13). Selective inhibition of HDAC6 can affect a number of cellular pathways important in tumorigenesis. Hyperacetylation of HSP90 in response to HDAC6 inhibition reduces the chaperone association with its client proteins, resulting in polyubiquitination and proteasomal degradation of a number of HSP90 substrates (13). HDAC6 inhibition enhances α-tubulin acetylation, which stabilizes microtubules and is often associated with reduced cell movement (14). Through the ubiquitin-binding domain, HDAC6 has been shown to recruit polyubiquitinated proteins to dynein motors and to transport protein cargo to aggresomes (15). HDAC6 inhibition can abrogate HSP90 chaperone function when combined with the HSP90 inhibitor 17-AAG in human leukemia cells (16), augment the cytotoxic effects of paclitaxel (17), and enhance the cytotoxicity of the proteasome inhibitor bortezomib (18–21).
In this study, we show that chemical inhibition of HDAC6 with a small-molecule inhibitor, tubacin (12), or genetic knockdown of HDAC6 in certain transformed cells, enhances cell death induced by topoisomerase II inhibitors etoposide or doxorubicin and the pan-HDAC inhibitor SAHA. Normal cells are resistant to cell death induced by the combination of tubacin plus etoposide, doxorubicin, or SAHA. Enhanced cell death in transformed cells is mediated in part via the intrinsic apoptotic pathway, as evidenced by enhanced PARP cleavage and partial inhibition of cell death by the pan-caspase inhibitor Z-VAD-fmk. Further, we found that HDAC6 inhibition with tubacin induces DNA damage and enhances DNA damage induced by etoposide or SAHA, as indicated by an increased accumulation of γH2AX, an early marker of DNA double-strand breaks (DSBs) and activation of the checkpoint protein Chk2. HDAC6 inhibition with tubacin induces the expression of cellular stress genes DDIT4 (RTP801/Dig2/REDD1) (22, 23) and DDIT3 (CHOP/GADD153) (24). The induction of DDIT3 is enhanced in transformed cells when tubacin is combined with SAHA.
These findings suggest that inhibition of HDAC6 can enhance the cytotoxic effects of DNA damaging agents in certain transformed cells at concentrations that do not affect normal cell viability and demonstrate mechanisms by which HDAC6-specific inhibition can enhance the efficiency of certain anticancer agents.
Results
Tubacin Enhances Transformed but Not Normal Cell Death Induced by Topoisomerase II Inhibitors and a Pan-HDAC Inhibitor.
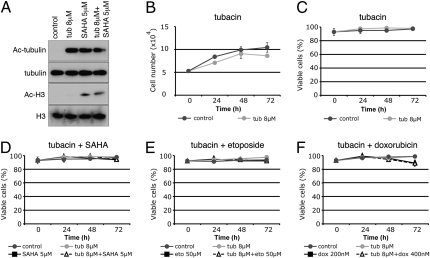
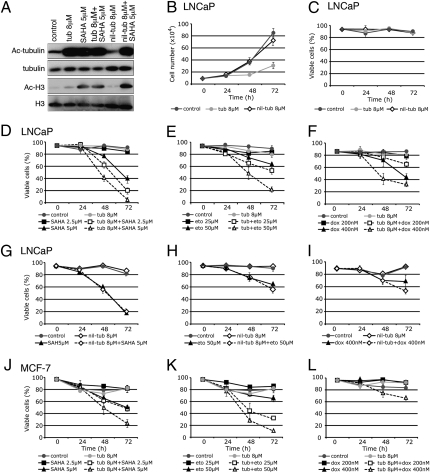
HDAC6 inhibition with tubacin results in the accumulation of acetylated α-tubulin, but not acetylated histones in normal human foreskin fibroblast (HFS) cells and transformed human prostate cancer (LNCaP) cells (Fig. 1A and Fig. 2A). Tubacin reduced the rate of growth of transformed and, to a lesser extent, normal cells, without loss of cell viability (Fig. 1 B and C and Fig. 2 B and C).
Fig. 1.
HFS cells are resistant to cell death induced by tubacin cultured in combination with SAHA, etoposide, or doxorubicin. (A) Western blot analysis probing with antibodies against acetylated α-tubulin and acetylated histone H3 in HFS cells cultured for 24 h with tubacin, SAHA, and tubacin plus SAHA. α-Tubulin and histone H3 are shown as loading controls. Cell growth (B) and viability (C) of HFS cells cultured with tubacin. Cell viability in HFS cells cultured with tubacin in combination with SAHA (D), etoposide (E), or doxorubicin (F). Error bars represent the SD of data points generated from experiments, which were all done in triplicate.
Fig. 2.
Tubacin enhances cell death induced by SAHA, etoposide, or doxorubicin in transformed LNCaP and MCF-7 cells. (A) Western blot analysis probing with antibodies against acetylated α-tubulin and acetylated histone H3 in LNCaP cells cultured for 24 h with nil-tubacin, tubacin, SAHA, tubacin plus SAHA, and nil-tubacin plus SAHA. α-Tubulin and histone H3 are shown as loading controls. Cell growth (B) and viability (C) of LNCaP cells cultured with tubacin or nil-tubacin. Cell viability of LNCaP cells cultured with tubacin in combination with (D) SAHA, (E) etoposide, or (F) doxorubicin. Cell viability of LNCaP cells cultured with nil-tubacin in combination with (G) SAHA, (H) etoposide, or (I) doxorubicin. Cell viability of MCF-7 cells cultured with tubacin in combination with (J) SAHA, (K) etoposide, or (L) doxorubicin. Error bars represent the SD of data points generated from experiments, which were all done in triplicate.
To assess whether specific inhibition of HDAC6 enhances cell death when combined with anticancer agents, cells were cultured with tubacin in combination with the topoisomerase II inhibitors etoposide or doxorubicin and the pan-HDAC inhibitor SAHA. In HFS cells, tubacin had no detectable effect on cell viability when combined with these anticancer agents (Fig. 1 D–F). In LNCaP cells, culture with 2.5 μM SAHA did not alter cell viability, whereas the combination of 2.5 μM SAHA plus 8 μM tubacin resulted in an 80% loss of cell viability after 72 h (Fig. 2D). Similarly, the combination of tubacin with 5 μM SAHA increased cell death compared with cultures with 5 μM SAHA alone (Fig. 2D). Tubacin also enhanced cell death induced by MS-275, which does not inhibit HDAC6 (10) (Fig. S1).
LNCaP cell death was markedly enhanced in cultures with tubacin and 25 μM or 50 μM etoposide compared with cultures with etoposide alone (Fig. 2E). Similarly, LNCaP cell death was enhanced in culture with tubacin plus doxorubicin compared with culture with doxorubicin alone (Fig. 2F). Tubacin also increased the sensitivity of MCF-7 human adenocarcinoma cells to SAHA-, etoposide-, and doxorubicin-induced cell death (Fig. 2 J–L). PC3 is a human prostate cancer cell line that is relatively resistant to SAHA induced cell death (25). PC3 cells cultured with a combination of SAHA plus tubacin resulted in a 25% decrease in cell viability only after 72 h in culture, whereas culture with SAHA alone did not induce any cell death (Fig. S2).
To evaluate whether the effect of tubacin in enhancing the cytotoxic effects of SAHA, etoposide, or doxorubicin was a result of selective inhibition of HDAC6, LNCaP cells were cultured with nil-tubacin, an analogue of tubacin that does not inhibit HDAC6 deacetylase activity as evidenced by lack of inducing acetylated tubulin (Fig. 2A) (12). Nil-tubacin did not increase cell death of LNCaP in combination culture with SAHA, etoposide, or doxorubicin (Fig. 2 G–I).
Down-Regulation of HDAC6 Expression in LNCaP Increases Sensitivity to Cell Death Induced by SAHA, Etoposide, or Doxorubicin.
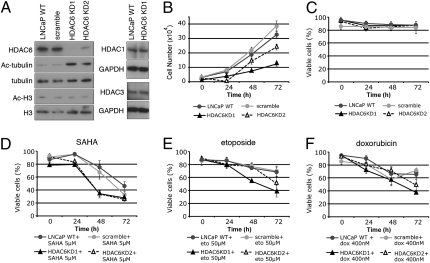
We next determined whether LNCaP cells in which HDAC6 expression was genetically suppressed were more sensitive than WT cells to SAHA-, etoposide-, or doxorubicin-induced cell death. Two sets of shRNA targeting different regions of HDAC6 mRNA were stably expressed in LNCaP cells, resulting in reduced HDAC6 levels and in the accumulation of acetylated tubulin (Fig. 3A). LNCaP cells in which HDAC6 levels were reduced did not accumulate acetylated histones, and the levels of HDAC1 and HDAC3 expression were not affected (Fig. 3A). Knockdown of HDAC6 resulted in a decrease in the rate of cell growth (Fig. 3B) and did not affect cell viability (Fig. 3C). HDAC6 knockdown of LNCaP cells cultured with SAHA for 48 h resulted in 70% cell death compared with approximately 30% cell death in WT and scramble shRNA transfected LNCaP cells (Fig. 3D). Increased sensitivity to etoposide- or doxorubicin-induced cell death was observed in LNCaP cells with reduced HDAC6 expression. Approximately 50% to 60% cell death occurred by 72 h in culture with etoposide or doxorubicin compared with approximately 30% cell death in WT and scramble shRNA transfected LNCaP cells (Fig. 3 E and F). Greater cell death was induced by doxorubicin and etoposide in HDAC6 KD1 than HDAC6 KD2 (Fig. 3 E and F), which may reflect more efficient down-regulation of HDAC6 in the HDAC6 KD1 cells (Fig. 3A).
Fig. 3.
Down-regulation of HDAC6 expression in LNCaP cells results in increased sensitivity to cell death induced by SAHA, doxorubicin, or etoposide. (A) Western blot analysis in LNCaP cells expressing shRNAs targeting two different sequences within HDAC6 (HDAC6 KD1 and HDAC6 KD2), WT LNCaP cells (LNCaP WT), and LNCaP cells expressing nontargeting scramble shRNA. Cell growth (B) and viability (C) in LNCaP WT, scramble, HDAC6 KD1, and HDAC6 KD2. Cell viability in LNCaP cells in which HDAC6 is down-regulated had increased sensitivity to SAHA- (D), doxorubicin- (E), and etoposide-induced (F) cell death. Error bars represent the SD of data points generated from experiments, which were all done in triplicate.
Taken together, these findings show that chemical inhibition of HDAC6 or genetically reduced HDAC6 expression increases the sensitivity of LNCaP cells to SAHA-, etoposide-, or doxorubicin-induced cell death.
Culture with Tubacin Plus SAHA or Etoposide Enhances Caspase-Dependent Apoptosis in LNCaP Cells.
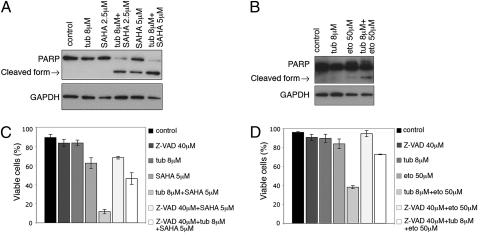
To investigate the pathway of cell death in LNCaP cells cultured with the combination of tubacin and SAHA or etoposide, the status of poly(ADP ribose) polymerase (PARP) and its proteolytic fragments were assayed. PARP is a 116-kDa nuclear protein that is specifically cleaved by caspase-3 into a 85-kDa fragment and serves as a marker of apoptosis (26). Cells cultured with 5 μM SAHA resulted in PARP cleavage (Fig. 4A). Culture with 2.5 μM SAHA (a concentration that does not induce LNCaP cell death; Fig. 2D) did not result in PARP cleavage (Fig. 4A). In cells cultured with tubacin and 2.5 μM or 5 μM SAHA, the level of full-length PARP decreased dramatically, with an increase in cleaved PARP (Fig. 4A). Similarly cells cultured with the combination of tubacin and etoposide induced PARP degradation (Fig. 4B). To further examine caspase-dependent activation in cells cultured with tubacin and SAHA or etoposide, the pan-caspase inhibitor Z-VAD-fmk was added to cultures 1 h prior to the addition of tubacin plus SAHA or tubacin plus etoposide. The addition of Z-VAD-fmk decreased cell death in LNCaP cells cultured with tubacin in combination with SAHA from 90% to 60% and in combination with etoposide from 65% to 25% (Fig. 4 C and D). These findings suggest that cell death induced by the combination of tubacin and SAHA or tubacin and etoposide is, in part, dependent on caspase activation.
Fig. 4.
Activation of the intrinsic apoptotic pathway is enhanced in transformed cells cultured with tubacin in combination with SAHA or etoposide. (A) Western blot analysis showing PARP degradation in LNCaP cells cultured with tubacin, SAHA, or simultaneous culture with tubacin and SAHA and (B) simultaneous culture of and tubacin and etoposide for 48 h. GAPDH is shown as a loading control. (C) Effect of the pan-caspase inhibitor Z-VAD-fmk on cell viability following a 48-h culture with tubacin, SAHA, or simultaneous addition of tubacin and SAHA and (D) 48-h culture with tubacin, etoposide, or simultaneous addition of tubacin and etoposide with and without Z-VAD-fmk. Error bars represent the SD of data points generated from experiments, which were all done in triplicate.
Tubacin Enhances the Accumulation of γH2AX and Phospho-Chk2 Induced by SAHA or Etoposide.
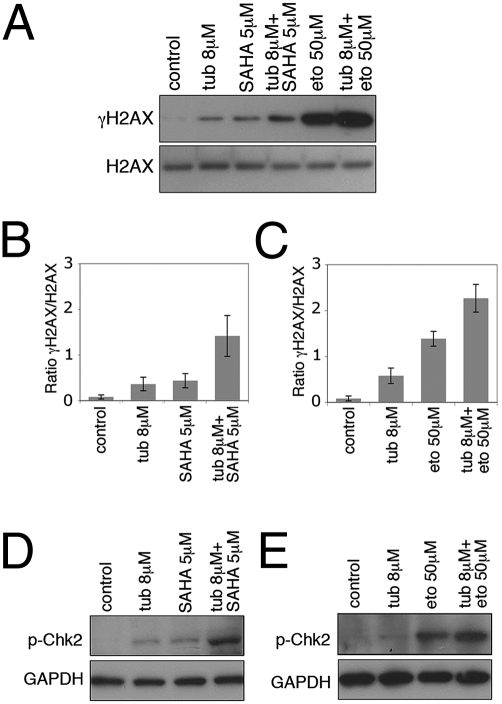
We examined whether selective inhibition of HDAC6 with tubacin activated a DNA damage response, which may account for tubacin-mediated cell cycle arrest. The accumulation of γH2AX (phosphorylation of histone H2AX), an early marker of DNA DSBs, increased in LNCaP cells cultured with tubacin, SAHA, or etoposide (Fig. 5A). The combination of tubacin with SAHA or with etoposide resulted in a more marked accumulation of γH2AX than in cells cultured with either compound alone (Fig. 5A). Quantitation of γH2AX levels showed an approximately sixfold increase in γH2AX when tubacin was combined with SAHA compared with SAHA alone (Fig. 5B) and a 1.5-fold increase when tubacin was combined with etoposide compared with etoposide alone (Fig. 5C).
Fig. 5.
Tubacin enhances the accumulation of γH2AX and phospho-Chk2 induced by SAHA or etoposide. (A) Western blot analysis showing accumulation of γH2AX following a 24-h culture with tubacin, SAHA, etoposide, and the combinations of tubacin with SAHA or etoposide. H2AX is shown as a loading control. (B and C) Quantitation of γH2AX levels of Western blots cultured as described in A. Error bars represent the SD of data points generated from experiments which were all done in triplicates. (D and E) Western blot analysis of accumulation of phospho-Chk2 following a 24-h culture with tubacin, SAHA, etoposide, and the combinations of tubacin with SAHA or etoposide.
We assessed the activation of the checkpoint kinase Chk2 which is phosphorylated on Thr68 in response to DNA damage and has been implicated in both G1 and G2 checkpoint activation (27, 28). Culture with SAHA or etoposide alone resulted in the activation of Chk2, as shown by an increase of phospho-Chk2 (Fig. 5 D and E). The level of phospho-Chk2 was higher when tubacin was cultured in combination with SAHA or etoposide (Fig. 5 D and E). Thus, HDAC6 inhibition can potentiate the DNA damage and checkpoint response induced by SAHA or etoposide.
Tubacin Induces a G1 Arrest, Up-Regulates DDIT3 and DDIT4, and Down-Regulates DNA Replication Proteins.
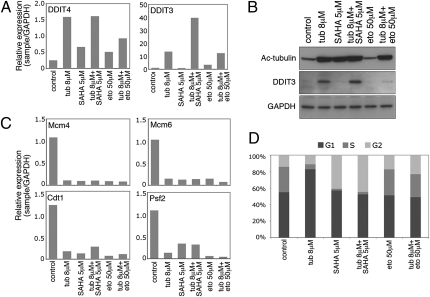
To further characterize the molecular pathways altered by tubacin, SAHA, and the combination of tubacin and SAHA, gene expression profiles were examined following culture of LNCaP cells for 2, 8, and 24 h. In SAHA cultured cells, approximately the same number of genes were up- and down-regulated at each time point (Table S1). In culture with tubacin alone, only one gene, DDIT4 (DNA-damage-inducible transcript 4), also known as RTP801/Dig2/REDD1, was up-regulated at least twofold at 2 h (Table S2). DDIT4 has been identified in mammalian cells as a gene induced in response to a variety of cellular stress, including agents that promote DNA damage and endoplasmic reticulum (ER) stress (22, 23). DDIT4 was induced to a similar level in LNCaP cells cultured for 24 h with tubacin plus SAHA (Table S2 and Fig. 6A).
Fig. 6.
Tubacin up-regulates DDIT3 and DDIT4, down-regulates replication proteins, and induces a G1 arrest. (A) Real-time PCR analysis on LNCaP cells cultured with tubacin, SAHA, etoposide, and the combinations of tubacin with SAHA or etoposide for 24 h. Primers used were against DDIT3 and DDIT4. (B) Western blot analysis probing with antibodies against acetylated α-tubulin and DDIT3. GAPDH is shown as a loading control. (C) Real-time PCR analysis on LNCaP cells cultured as described in A. Primers used were against Mcm4, Mcm6, Cdt1, and Psf2. (D) Cells cultured for 24 h as described in A were stained with propidium iodide and assessed by flow cytometry.
DDIT3 (DNA-damage-inducible transcript 3), also known as CHOP/GADD153, a transcription factor up-regulated in response to a variety of cellular stress, notably ER stress (24), was one of six genes up-regulated at least twofold at 8-h culture of LNCaP with tubacin (Table S2). Culture with SAHA alone did not induce DDIT3 at 8 h or 24 h (Table S2 and Fig. 6A). The combination of tubacin plus SAHA resulted in a 22-fold increase in DDIT3 gene expression compared with a sevenfold increase with tubacin alone at 24 h (Table S2). Increased expression of DDIT3 was confirmed on analysis at the protein level (Fig. 6B).
Microarray analysis of LNCaP cells cultured for 24 h with tubacin alone identified 72 genes down-regulated at least twofold (Table S3), of which approximately 40% were members of the cell cycle machinery (Table S3). Expression of several genes regulating G1/S transition and replication progression were down-regulated in culture with tubacin, SAHA, or etoposide alone and in combinations, including Mcm4, Mcm6, Cdt1, and Psf2 (Fig. 6C). At 24 h, there was an increase in cells arrested in G1 in LNCaP cells cultured with tubacin (Fig. 6D). SAHA induced an increase in cells arrested in G2, and there was an increase in cells in G2 in cultures with etoposide or etoposide plus tubacin (Fig. 6D).
Discussion
In this study, we show that selective chemical inhibition or genetic down-regulation of HDAC6 in transformed cells (LNCaP and MCF-7) results in increased sensitivity of these cells to the anticancer agents etoposide and doxorubicin or the pan-HDAC inhibitor SAHA. This effect is not observed in normal cells (i.e., HFS cells). These findings show that, at concentrations of SAHA, doxorubicin, or etoposide that are clinically attainable and tolerated (29–31), HDAC6-selective inhibition can enhance the therapeutic efficacy of these agents in certain transformed cells.
The selective HDAC6 inhibitor tubacin (12) was found to cause accumulation of γH2AX, a marker of DNA DSBs. The combination of tubacin plus SAHA or etoposide markedly increased the accumulation of γH2AX and phospho-Chk2 in LNCaP cells. These findings suggest that HDAC6 inhibition increased etoposide or SAHA induced accumulation of DNA DSBs, which may explain, in part, the chemosensitizing effect of HDAC6 inhibition in transformed cells. Synergistic and additive tumor cell apoptosis has been observed when combining pan-HDAC inhibitors with cytotoxic therapies that induce DNA damage (32–34). Enhanced DNA damage observed in culture with combined inhibitors has been attributed to induction of histone hyperacetylation by the HDAC inhibitor, resulting in a more open chromatin structure, making DNA more susceptible to damage by various toxic agents (35). Pan-HDAC inhibitors such as SAHA can suppress DNA repair proteins in transformed cells, resulting in failure to repair DNA damage (36–38). HDAC6 inhibition does not cause accumulation of acetylated histones. Target proteins of HDAC6 include the chaperone protein HSP90 (13, 39). Acetylation of Hsp90 impairs its chaperone function and exposes its client proteins to degradation, which is associated with activation of the intrinsic apoptotic pathway. We found that tubacin markedly enhanced SAHA- or etoposide-induced transformed cell apoptosis, as evidenced by increased PARP cleavage and caspase-dependent cell death. Microarray analysis of LNCaP cells cultured with SAHA showed down-regulation of a number of genes involved in DNA damage and repair pathways (Table S4). This suggests that tubacin-induced accumulation of DNA breaks in LNCaP cells cultured with SAHA may result in part from an impaired capacity for DNA DSB repair. Several proteins involved in the DNA damage repair pathway are targets of lysine acetylation (40), and acetylation of DNA repair proteins has been shown to alter their activity (41, 42).
HDAC6 has a role in the removal of misfolded and damaged proteins through its ability to recruit polyubiquitinated proteins to dynein motors and transporting them to aggresomes (15, 43). Tubacin can inhibit the interaction of HDAC6 with dynein in multiple myeloma cells, resulting in the accumulation of ubiquitinated proteins (18). We found that tubacin induces the expression of DDIT3, a transcription factor that is up-regulated in response to ER stress (24). Furthermore, LNCaP cell culture with tubacin plus SAHA resulted in enhanced DDIT3 induction, which could contribute to the chemosensitizing effect of HDAC6 inhibition in combination with a pan-HDAC inhibitor in transformed cells. Culture with SAHA alone did not induce DDIT3 expression in LNCaP cells (Fig. 6 A and B). Tubacin and SAHA inhibit the catalytic activity of HDAC6, as evident through hyperacetylation of α-tubulin (Fig. 2A). The accumulation of DDIT3 in tubacin cultured cells may reflect tubacin inhibiting the interaction of HDAC6 with dynein, whereas the HDAC6-inhibitory effects of SAHA may be limited to the inhibition of HDAC6 catalytic activity.
The present findings suggest that combination therapy with a selective HDAC6 inhibitor and certain anticancer agents may be a strategy for therapy of sensitive tumors while sparing normal cells.
Materials and Methods
Cell Lines and Reagents.
HFS cells were obtained from the Yale Skin Diseases Research Center Core. LNCaP, MCF-7, and PC3 cell lines were obtained from the American Tissue Culture Collection and cultured according to directions of the supplier. Doxorubicin and etoposide were purchased from Sigma, Z-VAD-fmk from R&D Systems, and MS-275 from Calbiochem. Tubacin and nil-tubacin were provided by Stuart Schreiber, James Bradner, and Ralph Mazitschek (Harvard University, Cambridge, MA). In all studies, an equivalent amount of DMSO without the drug was added to the control culture medium.
Cell Growth and Viability.
To monitor cell growth and viability, cells were seeded in triplicate at 5 × 104 cells in 1 mL of medium in 24-well plates. The drugs were added at the indicated concentrations 24 h after seeding. Cells were harvested by trypsin digestion at 24 h, 48 h, and 72 h following drug additions. Cell number and viability were determined by trypan blue exclusion.
Western Blot Analysis.
Cells (1 × 106) were washed with PBS solution and lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 0.5 mM EDTA, 0.5% Nonidet P-40) containing protease inhibitors (Boehringer Mannheim). Antibodies used were HDAC6, HDAC1, HDAC3 (Santa Cruz Biotechnology), acetylated α-tubulin (Sigma), α-tubulin (Calbiochem), PARP (BD Pharmingen), γH2AX (Abcam), H2AX (Abcam), phospho-CHK2 (Cell Signaling), DDIT3 (Santa Cruz Biotechnology), GAPDH (Thermo Scientific), and Histone H3 (Active Motif). Quantitation of Western blots was performed using ImageJ software.
Histone Extraction.
Cells (1 × 106) were washed with PBS solution and lysed in lysis buffer (10 mM Tris-HCl, pH 6.5, 25 mM KCl, 10 mM MgCl2, 1% Triton X-100, 8.6% sucrose) containing protease inhibitors. Samples were centrifuged at 600 × g for 5 min at 4 °C and the pellets were resuspended in TE buffer (10 mM Tris-HCl, pH 7.4, 13 mM EDTA) and centrifuged at 600 × g for 5 min at 4 °C. The pellets were resuspended in 0.2 M H2SO4 followed by incubation on ice for 1 h and vortexed every 15 min for 10 s during the incubation. Following centrifugation at 10,000 × g at 4 °C for 10 min, supernatants were incubated with cold acetone for at least 1 h. The samples were centrifuged at 10,000 × g for 10 min at 4 °C and the pellets dried and resuspended in distilled water.
RNA Interference.
shRNA lentiviral particles targeting HDAC6 mRNA, HDAC6KD1 (HDAC6 knockdown 1) at 1.7 × 107 TU/mL and HDAC6KD2 (HDAC6 knockdown 2) at 1.9 × 107 TU/mL, and nontargeting “scramble” shRNA control particles (SHC002V) at 1.1 × 107 TU/mL were purchased from Sigma-Aldrich and transfected according to the manufacturer's instructions using polybrene (Millipore). The 21-nt sequence corresponding to HDAC6 mRNA for HDAC6KD1 is 5′-CATCCCATCCTGAATATCCTT-3′ and that for HDAC6KD2 is 5′-GCACAGTCTTATGGATGGCTA-3′. For each shRNA, 5 × 105 cells were infected at a multiplicity of infection of 2.
Microarray Analysis.
Alterations in gene expression were evaluated by microarray using the Illumina human cDNA array containing cDNA probes representing the whole genome. LNCaP cells (1 × 106) were seeded in 10-cm–diameter cell culture dishes and incubated for 24 h before culture with DMSO (control), 8 μM tubacin, 5 μM SAHA, or 8 μM tubacin and 5 μM SAHA for 2 h, 8 h, and 24 h. Triplicate samples were prepared for each drug treatment at each time point. Poly(A)+ mRNA was isolated from cells using TRIzol reagent according to the manufacturer's protocol (Invitrogen). The data were analyzed using the Bioconductor packages for the R statistical system. The output from Beadstudio was processed using the LUMI package. The normalization method used was quantile and the signal levels were log (base 2) transformed. To determine genes that are differentially expressed between the various sample types, the LIMMA package was used.
Quantitative Real-Time PCR.
One milligram of total RNA was reverse-transcribed using the Thermoscript RT-PCR system (Invitrogen) at 52 °C for 1 h. Resultant cDNA (20 ng) was used in a quantitative PCR with a 7500 Real-Time PCR System (Applied Biosystems) using predesigned primers for DDIT3, DDIT4, Mcm4, Mcm6, Cdt1, Psf2, and GAPDH (Applied Biosystems). Amplification was carried for 40 cycles (95 °C for 15 s, 60 °C for 1 min). To calculate the efficiency of the PCR and to assess the sensitivity of each assay, we performed a six-point standard curve (5, 1.7, 0.56, 0.19, 0.062, and 0.021 ng). Triplicate CT values were averaged, and amounts of target were interpolated from the standard curves and normalized to GAPDH.
Cell Cycle Analysis.
Cells (106) were harvested at 24-h culture with the indicated drugs, washed with PBS solution, and fixed in methanol. Cells were then resuspended in a buffer containing 50 mg/mL propidium iodide and 100 mg/mL RNase A. Samples were analyzed using a Becton Dickinson FACSCalibur flow cytometer. Data were collected for 10,000 events and analyzed using FlowJo software.
Supplementary Material
Acknowledgments
We thank Stuart Schreiber, James Bradner, and Ralph Mazitschek (Harvard University, Cambridge, MA) for the gift of tubacin and nil-tubacin; Nick Socci for his assistance with microarray analysis; the Memorial Sloan-Kettering Cancer Center genomics core facility for microarray and quantitative real-time PCR; and Becky Liu for technical assistance. This study was supported in part by National Institutes of Health Grant P30CA08748-44, CapCure Foundation, the David Koch Foundation, and the Experimental Therapeutics Center at Memorial Sloan-Kettering Cancer Center.
Footnotes
Conflict of interest statement: Memorial Sloan-Kettering Cancer Center and Columbia jointly hold patents on suberoylanilide hydroxamic acid (SAHA, Vorinostat) and related compounds that were exclusively licensed in 2001 to ATON Pharma, a biotechnology start-up that was wholly acquired by Merck, Inc., in April 2004.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013754107/-/DCSupplemental.
References
- 1.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: Overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 2.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: Current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280:201–210. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Mehnert JM, Kelly WK. Histone deacetylase inhibitors: Biology and mechanism of action. Cancer J. 2007;13:23–29. doi: 10.1097/PPO.0b013e31803c72ba. [DOI] [PubMed] [Google Scholar]
- 4.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namdar M, Marks PA. In: in Histone Deacetylase Inhibitors in Medicine. El-Osta A, Karagiannis TC, editors. Signpost: Research; 2010. pp. 9–39. [Google Scholar]
- 6.Duvic M, et al. Evaluation of the long-term tolerability and clinical benefit of vorinostat in patients with advanced cutaneous T-cell lymphoma. Clin Lymphoma Myeloma. 2009;9:412–416. doi: 10.3816/CLM.2009.n.082. [DOI] [PubMed] [Google Scholar]
- 7.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 8.Campas-Moya C. Romidepsin for the treatment of cutaneous T-cell lymphoma. Drugs Today (Barc) 2009;45:787–795. doi: 10.1358/dot.2009.45.11.1437052. [DOI] [PubMed] [Google Scholar]
- 9.Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: Current status and overview of recent clinical trials. Drugs. 2009;69:1911–1934. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Bradner JE, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parmigiani RB, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci USA. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bali P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 14.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 16.Rao R, et al. HDAC6 inhibition enhances 17-AAG—mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–1893. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 17.Marcus AI, et al. The synergistic combination of the farnesyl transferase inhibitor lonafarnib and paclitaxel enhances tubulin acetylation and requires a functional tubulin deacetylase. Cancer Res. 2005;65:3883–3893. doi: 10.1158/0008-5472.CAN-04-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hideshima T, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci USA. 2005;102:8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bazzaro M, et al. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clin Cancer Res. 2008;14:7340–7347. doi: 10.1158/1078-0432.CCR-08-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawada J, Zou P, Mazitschek R, Bradner JE, Cohen JI. Tubacin kills Epstein-Barr virus (EBV)-Burkitt lymphoma cells by inducing reactive oxygen species and EBV lymphoblastoid cells by inducing apoptosis. J Biol Chem. 2009;284:17102–17109. doi: 10.1074/jbc.M809090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Gonzalez A, et al. Role of the aggresome pathway in cancer: targeting histone deacetylase 6-dependent protein degradation. Cancer Res. 2008;68:2557–2560. doi: 10.1158/0008-5472.CAN-07-5989. [DOI] [PubMed] [Google Scholar]
- 22.Lin L, Qian Y, Shi X, Chen Y. Induction of a cell stress response gene RTP801 by DNA damaging agent methyl methanesulfonate through CCAAT/enhancer binding protein. Biochemistry. 2005;44:3909–3914. doi: 10.1021/bi047574r. [DOI] [PubMed] [Google Scholar]
- 23.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinszner H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Ngo L, Perez G, Dokmanovic M, Marks PA. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proc Natl Acad Sci USA. 2006;103:15540–15545. doi: 10.1073/pnas.0607518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullen P. PARP cleavage as a means of assessing apoptosis. Methods Mol Med. 2004;88:171–181. doi: 10.1385/1-59259-406-9:171. [DOI] [PubMed] [Google Scholar]
- 27.Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 28.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 29.Rahman A, Carmichael D, Harris M, Roh JK. Comparative pharmacokinetics of free doxorubicin and doxorubicin entrapped in cardiolipin liposomes. Cancer Res. 1986;46:2295–2299. [PubMed] [Google Scholar]
- 30.Hande KR, et al. Pharmacokinetics of high-dose etoposide (VP-16-213) administered to cancer patients. Cancer Res. 1984;44:379–382. [PubMed] [Google Scholar]
- 31.Kelly WK, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23:3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karagiannis TC, El-Osta A. Modulation of cellular radiation responses by histone deacetylase inhibitors. Oncogene. 2006;25:3885–3893. doi: 10.1038/sj.onc.1209417. [DOI] [PubMed] [Google Scholar]
- 33.Kim MS, et al. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 34.Marchion DC, et al. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92:223–237. doi: 10.1002/jcb.20045. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-López W, Folle GA, Obe G, Jeppesen P. Chromosome regions enriched in hyperacetylated histone H4 are preferred sites for endonuclease- and radiation-induced breakpoints. Chromosome Res. 2001;9:69–75. doi: 10.1023/a:1026747801728. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Attenuated DNA damage repair by trichostatin A through BRCA1 suppression. Radiat Res. 2007;168:115–124. doi: 10.1667/RR0811.1. [DOI] [PubMed] [Google Scholar]
- 37.Adimoolam S, et al. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc Natl Acad Sci USA. 2007;104:19482–19487. doi: 10.1073/pnas.0707828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munshi A, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 39.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 41.Chen CS, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67:5318–5327. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 42.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyault C, et al. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.