Abstract
Inositol phosphate (IP) kinases constitute an emerging class of cellular kinases linked to multiple cellular activities. Here, we report a previously uncharacterized cellular function in Hedgehog (Hh) signaling for the IP kinase designated inositol hexakisphosphate kinase-2 (IP6K2) that produces diphosphoryl inositol phosphates (PP-IPs). In zebrafish embryos, IP6K2 activity was required for normal development of craniofacial structures, somites, and neural crest cells. ip6k2 depletion in both zebrafish and mammalian cells also inhibited Hh target gene expression. Inhibiting IP6 kinase activity using N(2)-(m-(trifluoromethy)lbenzyl) N(6)-(p-nitrobenzyl)purine (TNP) resulted in altered Hh signal transduction. In zebrafish, restoring IP6K2 levels with exogenous ip6k2 mRNA reversed the effects of IP6K2 depletion. Furthermore, overexpression of ip6k2 in mammalian cells enhanced the Hh pathway response, suggesting IP6K2 is a positive regulator of Hh signaling. Perturbations from IP6K2 depletion or TNP were reversed by overexpressing smoM2, gli1, or ip6k2. Moreover, the inhibitory effect of cyclopamine was reversed by overexpressing ip6k2. This identified roles for the inositol kinase pathway in early vertebrate development and tissue morphogenesis, and in Hh signaling. We propose that IP6K2 activity is required at the level or downstream of Smoothened but upstream of the transcription activator Gli1.
Keywords: diphosphoryl inositol tetrakisphosphate, ipk2, inositol polyphosphate multikinase, inositol pentakisphosphate, inositol hexakisphosphate
Hedgehog (Hh) signaling is essential for patterning of the vertebrate skeleton during development and in adult tissue homeostasis (1, 2). Reduced Hh signaling results in developmental defects, whereas inappropriate activation contributes to the occurrence of certain cancers. The Hh signaling paradigm involves binding extracellular Hh ligands [e.g., Sonic hedgehog (Shh)] to the Patched (Ptc) receptor. This relieves the inhibition of Smoothened (Smo), allowing signaling for activation of the Gli family of transcription factors. Both Ptc and Smo are critical for proper Hh signal transduction. In the absence of Hh ligands, Ptc blocks Smo activity. This repression mechanism is not fully understood, although membrane lipid metabolism might play a role (3). Additionally, small molecule agonists could potentially bind Smo in the presence of Hh ligands and activate signaling (4).
At the cell membrane, lipid phosphoinositides are essential signaling mediators (5). Activation of phospholipase C generates inositol 1,4,5-trisphosphate (IP3) and 1,2-diacylglycerol as second messenger intracellular signals. Soluble IP3 molecule is a starting point for metabolism by inositol phosphate (IP) kinases and phosphatases. The resulting IP ensemble includes inositol tetrakisphosphate (IP4) isomers, inositol 1,3,4,5,6-pentakisphosphate (IP5), inositol hexakisphosphate (IP6), and diphosphoryl inositol phosphate (PP-IP) isomers (e.g., PP-IP4, IP7) (6, 7). Different IPs regulate vital functions, including transcription, mRNA export, RNA editing, translation, and ciliogenesis (6, 7).
The IP6 kinases (IP6K) specifically produce diphosphoryl isomers, including PP-IP4, PP-IP5, PP2-IP3, and PP2-IP4 (8, 9). Loss of IP6K activity results in different cellular defects, including altered vesicular endocytosis and telomere length in yeast, perturbed chemotaxis in Dictyostelium, and inhibited insulin exocytosis in pancreatic β-cells (9–12). Of the three documented IP6K family members, the IP6K2 isoform is linked to cell growth regulation and apoptosis in mammalian cells (13). However, IP6K2 roles in development and signaling in multicellular organisms are not fully understood.
Here we report that IP6K2 is required for development of craniofacial skeleton, somites, and slow muscle fibers in zebrafish. Moreover, loss of IP6K2 perturbs neural crest cell (NCC) development and migration, and inhibits Hh target gene expression. NCCs are migratory cells that detach from the embryonic neural epithelium along the dorsal neural tube and populate various body regions to differentiate into multiple cell types. Cranial NCCs (CNCCs) give rise to the frontonasal skeleton and pharyngeal arches, forming the maxilla, mandible, and other neck and face structures (14). Shh is required for CNCC development as well as differentiation into cartilage (15), and its loss is linked to craniofacial defects (16). We find that IP6K2 is a positive regulator of Hh signaling acting at the level or downstream of Smo but upstream of Gli transcription factors. We speculate that IP6K2 catalyzed production of PP-IP4 from IP5 is a critical step for Hh signaling during early development.
Results
ip6k2 Knockdown Alters Craniofacial and Somite Structures.
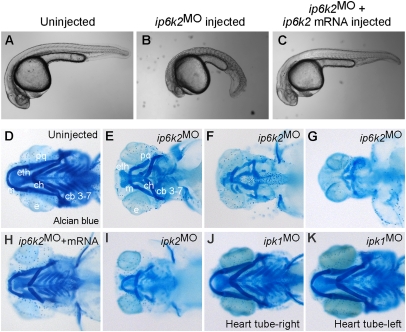
We identified the zebrafish IP6K2 coding sequence by interrogating the zebrafish GenBank database for transcripts orthologous to human IP6K2 sequence (Fig. S1), and found that the ip6k2 mRNA was maternally deposited and expressed during early embryogenesis (Fig. S2). To test for roles during early development, we depleted IP6K2 levels by injecting one-cell-stage zebrafish embryos with morpholino antisense oligonucleotides (MO) that impair translation (ip6k2AUGMO and ip6k2UTRMO) or splicing (ip6k2SPMO; Fig. S3 A and B). All these loss-of-function reagents are henceforth referred to as ip6k2MO, as they resulted in identical developmental defects. In a population of 90 ip6k2MO-injected embryos, 92% developed deformed craniofacial patterns with marked reduction of cartilage elements, as revealed by Alcian blue staining of head skeleton (Fig. 1 D–G). Less than 4% of the controls (n = 60) had a defect.
Fig. 1.
IP6K2 activity is required for normal development of craniofacial and somite structures. (A–C) Lateral view of uninjected (A), ip6k2SPMO-injected (B), and ip6k2SPMO + ip6k2 mRNA-coinjected (C) live embryos at 24 hpf. Eighty percent of the ip6k2SPMO embryos had a small head and reduced trunk; both the defects were rescued by ip6k2 mRNA coinjection. Effective ip6k2SPMO suppression of splicing was confirmed by RT-PCR (Fig. S3 A and B). (D–K) Ventral view of Alcian blue stained head skeleton of 5-d-old uninjected control (D), ip6k2SPMO (E–G), ip6k2SPMO + ip6k2 mRNA (H), ipk2SPMO (I), and ipk1MO1 (J and K)-injected embryos. The anterior neurocranium was mostly maintained in 40% of ip6k2MO embryos, but distorted or reduced in 60% of embryos (F and G). The pharyngeal skeleton was affected in 92% embryos injected (n = 90). ip6k2 mRNA coinjection restored normal craniofacial skeleton (H). Craniofacial skeleton was altered in ipk2SPMO embryos (I). No craniofacial deformity was evident in ipk1MO1 embryos (J and K). e, eye; m, Meckel's cartilage; pq, palatoquadrate; ch, ceratohyal; cb 3–7, ceratobranchial arches 3–7; eth, ethmoid plate.
The anterior neurocranium, consisting of the trabeculae and ethmoid plate, was mostly maintained in ip6k2MO embryos. However, the pharyngeal skeleton was distinctively affected. Cartilages were shortened, including mandibular (Meckel's and palatoquadrate) and hyoid arches (Fig. 1 E–G), and loss of pharyngeal apparatus (Fig. 1G). Similar results were obtained by ip6k2MO injection into transgenic Tg(sox10(7.2):mrfp) (17) embryos, in which primarily NCCs express membrane-tethered red fluorescent protein (RFP) under control of sox10 promoter (Fig. S4 A and B). Interestingly, ip6k2MO embryos were also defective in somite and slow muscle development (Fig. 1 A and B and Fig. S4 C and D). In contrast to chevron-shaped somites in wild-type embryos, ip6k2MO embryos had U-shaped somites (Fig. 1 A and B). ip6k2SPMO defects were mostly reversed by coinjecting with nontargeted mRNA encoding IP6K2 (Fig. 1 C and H and Fig. S3 C–E), confirming specificity of the ip6k2MO. Overall, these findings revealed a requirement for IP6K2 action in craniofacial, pharyngeal arch, and somite development.
PP-IP4 Synthesis Pathway Is Critical for Craniofacial Development.
To determine which IP6K2 catalyzed step is essential for craniofacial development, we performed loss-of-function studies targeting different enzymes in the IP pathway. Ipk2 phosphorylates IP4 to produce IP5, which is a substrate for either IP6K2 to produce PP-IP4, or Ipk1 to produce IP6 (6, 7). IP6 is the precursor for synthesis of IP7 and IP8 isomers by IP6K2 and Vip1 (18, 19). We speculated that depletion of an upstream enzyme (Ipk2) would result in additional phenotypes, including those resulting from depletion of downstream enzymes (IP6K2, Ipk1). We found that ipk2SPMO injection resulted in craniofacial defects that were strikingly similar to perturbations in ip6k2MO embryos (Fig. 1I; in 65% of ipk2SPMO-injected embryos, n = 52). In addition, ipk2SPMO embryos were defective in left-right asymmetric placement of the heart tube, with 35% showing a perturbation (n = 52). This was expected based on our prior studies showing left-right asymmetry defects in ipk1MO embyros (20). In contrast, none of the ip6k2MO defects were observed in ipk1MO1 embryos (Fig. 1 J and K). Because depletion of Ipk2 and IP6K2 resulted in shared perturbations that were absent in ipk1MO embryos, we concluded that the ip6k2MO defects result from inhibition of the IP pathway upstream of IP6.
ip6k2 Knockdown Inhibits the Development and Migration of Neural Crest Cells.
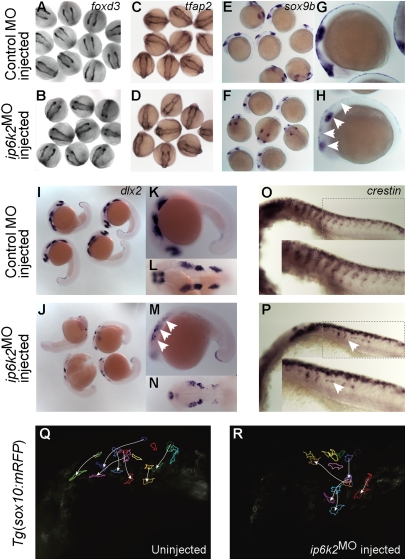
Because much of the craniofacial skeleton is derived from CNCC, we investigated if NCC development is defective in ip6k2MO embryos. Early NC development occurs in three distinct stages—induction or specification, maintenance, and migration—each marked by expression of stage-specific genes. In ip6k2MO embryos, expression of foxd3 (21) and tfap2 (22) was normal at 11 h postfertilization (hpf), suggesting NC specification was unaltered (Fig. 2 A–D). However, at 14 hpf, sox9b expression domain (23) was smaller in ip6k2MO embryos compared with controls (Fig. 2 E–H), revealing a potential defect in NCC maintenance. Strikingly, at 22 hpf, expression of dlx2 and crestin were altered (Fig. 2 I–P). dlx2 encodes a transcription factor that is essential for pharyngeal arch development and expressed in migratory NC streams and pharyngeal primordia (24), whereas crestin is a retroelement that is expressed in premigratory cranial and trunk NCC and later in actively migrating NCC (25). The dlx2 expression domain at 22 hpf was reduced in ip6k2MO embryos, consistent with a deficit in migratory cranial NC streams (Fig. 2 I–N). As sox9b determines the dlx2-expressing cell population size (23), reduced sox9b expression might directly impact dlx2 expression in ip6k2MO embryos. Streams of crestin-expressing NCC that normally migrate ventrally from the neural tube and populate the trunk were also disrupted in ip6k2MO embryos (Fig. 2 O and P). We observed both reduced migratory NCC numbers and abnormally migrated NCCs. Thus, several aspects of NCC development and migration were affected in ip6k2MO embryos.
Fig. 2.
IP6K2 is critical for NCC development and migration. (A–P) Whole-mount in situ hybridizations for NC marker gene expression specifying distinct stages of NC development: (A–D) specification (foxd3 and tfap2 at 11 hpf), (E and H) maintenance (sox9b at 14 hpf), and (I–P) migration (dlx2 and crestin, 22 hpf) in control MO (A, C, E, G, I, K, L, and O) and ip6k2ATGMO (B, D, F, H, J, M, N, and P)-injected embryos. Arrowheads indicate distinct differences in the expression of sox9b (E–H), dlx2 (I–N), and crestin (O and P) between control MO and ip6k2ATGMO embryos. (Q and R) Analysis of NCC migration in real time using Tg(sox10(7.2):mrfp) fish (17) (see SI Materials and Methods for details). Fluorescence images (frames) for the hindbrain and eye regions were captured from a 6-h time-lapse sequence, with dorsal toward the top and anterior to the left. The 0-min time point represents 16-hpf stage. Images of uninjected (Q) and ip6k2ATGMO-injected (R) embryos show an overlay of first and last images onto which the tracings of migratory paths of CNCC were superimposed. Arrowheads indicate end point of each cell's trajectory.
We also investigated NCC migration in real time using Tg(sox10(7.2):mrfp) fish (17). In the controls, at 16 hpf, NCC migrated from mid- and hindbrain regions interiorly into the head, as well as ventrally toward the optic vesicle (Fig. 2Q and Movie S1). By 22 hpf, NCC migrated to the anteriormost region of the embryo. However, in ip6k2MO embryos, NCC migrated aberrantly (Fig. 2R and Movie S2). We measured velocity (μm/s) of NCC migration either by net distance traveled in 6 h or average distance traveled in 15-min intervals over 6 h. Velocity of NCC migration, when accounting for net distance traveled, was significantly less in ip6k2MO embryos compared with controls (ip6k2MO = 0.00151 ± 0.00042, n = 5; control = 0.00530 ± 0.00176, n = 5; P value: 0.0016). In contrast, there was no significant difference in NCC velocity when averaged in 15-min intervals (ip6k2MO = 0.00751 ± 0.00263, n = 11; control = 0.00919 ± 0.00365, n = 14; P value: 0.2118). This suggested that NCC in ip6k2MO embryos were motile but had altered directional migration.
IP6K2 Depletion Perturbs Hh-Induced Gene Expression During Embryogenesis.
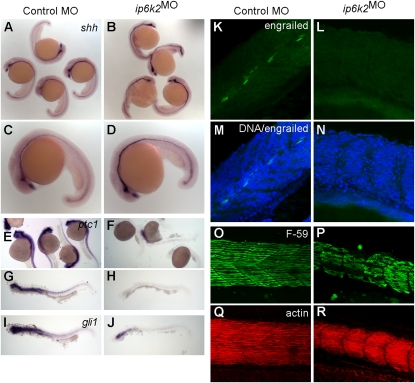
The ip6k2MO embryo defects in somite, slow muscle, craniofacial, and NCC development are all shared hallmarks of disrupted Hh signaling (15, 26–30). To test whether Hh signaling was inhibited in ip6k2MO embryos, we analyzed Hh target gene expression. In zebrafish, shh is expressed in the central nervous system ventral region and along the spinal cord, in a single row of floor plate cells (31) and in the notochord, which is essential for somite development. Hh pathway activation induces transcription of a number of genes, including ptc (32) and gli1 (33). Knockdown of ip6k2 clearly reduced ptc1 and gli1 expression in somites (Fig. 3 E–J), even as shh expression was slightly elevated in floor plate cells (Fig. 3 A–D). In somites, development of muscle pioneer cells and slow muscle fibers, but not fast muscle fibers, require Hh signaling (26, 27). In controls, expression of the homeodomain containing engrailed protein (34) in muscle pioneer cells at the horizontal myoseptum was clearly visible. However, no engrailed expression was detected in ip6k2MO embryos (Fig. 3 K–N).
Fig. 3.
IP6K2 is required for Hh target gene expression during development. (A–J) Whole-mount in situ hybridizations showing expression of Hh target genes in zebrafish embryos (24-hpf stage) injected with either control MO (column 1) or ip6k2SPMO (column 2). Expression of shh (A–D) was slightly elevated, whereas ptc1 (E–H) and gli1 (I and J) expression was down-regulated in ip6k2SPMO (B, D, F, H, and J) compared with controls (A, C, E, G, and I). (K–N) Whole-mount immunohistochemistry showing engrailed protein levels (detected by 4D9 antibody) in control MO (K) or ip6k2SPMO (L) embryos at 24 hpf. Engrailed was not detected in ip6k2SPMO embryos (L). M and N overlay engrailed immunohistochemistry and TO-PRO3-stained DNA. (O–R) Lateral views of embryos at 24 hpf stained for myosin heavy chain using F59 antibody to visualize slow muscle cells (O and P) and F-actin using rhodamine-phalloidin to reveal both fast and slow muscle cells (Q and R). Dorsal is toward the top and anterior to the left.
To visualize slow muscle fibers and somite boundaries, localization of myosin heavy chain and F-actin was analyzed. At 24 hpf, controls had organized development of slow muscle fibers and chevron-shaped somite boundaries (Fig. 3O). In contrast, in ip6k2MO embryos, slow muscle fibers were malformed and decreased in number (Fig. 3P). U-shaped somite boundaries were also visible in ip6k2MO embryos. Interestingly, there was no apparent change in F-actin staining (Fig. 3 Q and R), suggesting that other muscle cell-types populated somite spaces in the absence of slow muscle fibers. It is known that slow muscle regulates the pattern of trunk NCC migration (35). Importantly, coinjection of mRNA encoding zebrafish IP6K2 with ip6k2SPMO reversed the defects in Hh signaling, including restoration of somite shape and ptc1 expression (Fig. S3 C–E and K–M). In addition, coinjection of ip6k2 mRNA rescued NCC migration defects in ip6k2SPMO embryos (Fig. S3 G–I). Injection of ip6k2 mRNA alone did not cause any apparent defects (Fig. S3 F, J, and N). This confirmed the specificity of the ip6k2 knockdown perturbations. Taken together, these findings provided evidence for an IP6K2 role in the zebrafish Hh signaling pathway.
Altering IP6K2 Activity in Mammalian Cells Impacts Hh Signaling.
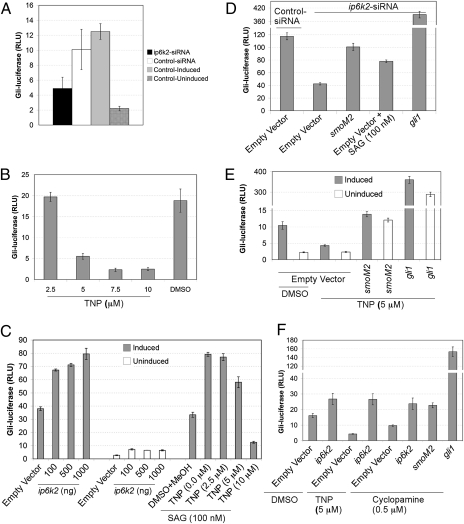
To investigate whether the ip6k2 role in Hh signaling is conserved in mammalian cells, we assayed the effect of ip6k2 knockdown on Hh pathway activation in NIH 3T3 cells transiently transfected with Gli-dependent firefly luciferase (8× gli-luciferase) and constitutive β-galactosidase reporters (36). Strikingly, Gli-luciferase activity was ∼50% reduced in ip6k2-siRNA cells in comparison with control-siRNA cells (Fig. 4A). We also used Shh-LIGHT2 cells [clonal NIH 3T3 cell line with Gli-dependent firefly luciferase stably incorporated and transiently transfected with a β-galactosidase reporter (36)] (Fig. S5A), and Shh-LIGHT Z3 cells [NIH 3T3 cell line with stably incorporated Gli-dependent firefly luciferase and constitutive β-galactosidase reporters (37)] (Fig. S5B). siRNA-mediated depletion of ip6k2 transcript levels in these cells resulted in a 45–55% decrease in Shh-induced Gli-luciferase activity in comparison with control-siRNA cells. These findings were consistent with our findings in zebrafish showing that depletion of IP6K2 levels diminished the Hh signaling response.
Fig. 4.
IP6K2 positively regulates the Hh response pathway. (A) Depletion of ip6k2 reduces Shh-dependent Gli-luciferase activity. Bar graphs showing Gli-luciferase activity expressed as relative luciferase units (RLU) in NIH 3T3 cells cotransfected with 8× gli-luciferase and constitutive β-galactosidase reporter plasmids, and additionally subjected to ip6k2 or control siRNA treatment for 72 h. Cells were induced with Hh ligand (Shh-N conditioned medium) for 24 h, and Gli-luciferase activity was measured (normalized by β-galactosidase activity). Non–siRNA-treated cells (control induced) and non–siRNA-treated uninduced cells (control uninduced) were controls. (B) TNP inhibits Hh pathway activity. Shh-LIGHT Z3 cells were treated with TNP (2.5–10 μM) and Hh ligand induction for 24 h, and Gli-luciferase activity was quantified. DMSO treatment served as control. (C) IP6K2 acts as an effector of the Hh response pathway. Shh-LIGHT Z3 cells were transfected with an empty plasmid (1 μg) or a plasmid expressing ip6k2 ORF (0.1–1 μg) and assayed Gli-luciferase activity with and without Hh ligand induction after 24 h. Additionally, Shh-LIGHT Z3 cells were subjected to 100 nM SAG and increasing TNP concentrations (0.0–10 μM), with Gli-luciferase activity determined after 24 h induction with Hh ligand. (D) Epistasis analysis indicating IP6K2 acts at the level or downstream of Smo. Shh-LIGHT Z3 cells were cotransfected with control or ip6k2 siRNA and empty plasmid or plasmid expressing smoM2 or gli1, and 72 h later treated with Hh ligand alone or with SAG for 24 h. Gli-luciferase activity was determined. (E) smoM2 or gli1 overexpression rescues the inhibitory effect of TNP. NIH 3T3 cells were cotransfected with reporter plasmids, and either an empty plasmid or plasmid expressing smoM2 or gli1, grown for 24 h. Gli-luciferase activity was determined after 24-h treatment with TNP (5 μM) or DMSO, with or without Hh ligand induction. (F) ip6k2 overexpression reverses inhibitory effects of TNP and cyclopamine. NIH 3T3 cells were cotransfected with reporter plasmids and either an empty plasmid or plasmid expressing ip6k2, smoM2, or gli1, and grown for 24 h. Cells were subjected to Hh ligand induction in presence of DMSO, TNP (5 μM), or cyclopamine (0.5 μM), and Gli-luciferase activity was determined. Data are the mean ± SD for three independent assays.
Recently, a specific inhibitor for IP6K enzymes was reported, designated N2-(m-(trifluoromethy)lbenzyl) N6-(p-nitrobenzyl)purine (TNP) (38). When Shh-LIGHT Z3 cells were treated with TNP, a dose-dependent decrease was observed in Gli-luciferase activity (Fig. 4B). Notably, Gli-luciferase activity for cells treated with 7.5 μM TNP was 2.33 ± 0.37, whereas it was 18.83 ± 2.77 for DMSO-treated cells, reflecting 88% inhibition. Control β-galactosidase activity in both cell types was 0.419 ± 0.022 and 0.459 ± 0.018 relative units, respectively, showing TNP did not cause any significant growth or transcriptional inhibition. Together, TNP inhibition of IP6K2 activity blocked activation of Hh signaling and mimicked ip6k2 knockdown defects. This directly linked the Hh pathway with IP6K2 activity.
IP6K2 Is a Positive Effector of the Hh Pathway, Acting Between Smo and Gli.
Next we tested for IP6K2 gain-of-function effects by exogenously overexpressing ip6k2. Shh-LIGHT Z3 cells were transfected with a plasmid expressing ip6k2 and assayed for Gli-luciferase activity with and without induction by Hh ligand (from Shh-N conditioned medium; SI Materials and Methods). In comparison with control (empty vector)-transfected cells, exogenous ip6k2 expression elevated Gli-luciferase activity with induction by Hh ligand, whereas only a marginal change in activity was detected under uninduced conditions (Fig. 4C). Thus, IP6K2 functions as a positive effector, facilitating Hh pathway response upon ligand stimulation.
To pinpoint the step requiring IP6K2 activity, we used a dominantly active form of Smo (SmoM2) (36), a Smo agonist SAG (39), and gli1 overexpression (40) to stimulate Hh signaling independent of Hh ligand. Shh-LIGHT Z3 cells were cotransfected with siRNA and either an empty plasmid or a plasmid expressing smoM2 or gli1. Gli-luciferase activity was determined. Strikingly, ip6k2-siRNA inhibition of the Hh response was reversed with constitutively active forms of Smo (SmoM2 or SAG) and with gli1 overexpression (Fig. 4D). Both smoM2 and gli1 rescued the TNP inhibitory effect (Fig. 4E). We reasoned if IP6K2 acts on or downstream of Smo, effects of a Smo agonist would be compromised by IP6K2 inhibition. Indeed, activation as initiated by SAG was reduced in cells treated with increasing TNP concentrations (Fig. 4C).
Others have shown that ip6k2 overexpression restores the PP-IP production inhibited by TNP treatment (38). Here, ip6k2 overexpression reversed the inhibitory TNP effect on Hh response (Fig. 4F). We also tested whether ip6k2 overexpression reversed the inhibitory effect of cyclopamine, a plant steroidal alkaloid that inactivates Smo (36, 41, 42). Similar to gli1 or smoM2, overexpression of ip6k2 reversed cyclopamine inhibition of the Hh response (Fig. 4F). Overall, this indicated that IP6K2 acts at the level or downstream of Smo in the Hh pathway.
IP6K2 and Hh Signaling Are Linked to Changes in IP Flux.
To investigate if Shh induction of the Hh pathway triggers changes in IP levels, total soluble IP profiles were evaluated after different cell treatments. NIH 3T3 cells were metabolically labeled with myo(1,2)[3H] inositol and treated with DMSO alone, TNP alone, or Hh ligand with DMSO, TNP, or cyclopamine (Fig. S6). In control cells, IP3, IP5, IP6, and modest IP7 levels were detected (Fig. S6B). As reported (38), IP5 and IP6 levels were elevated with TNP treatment (Fig. S6C). Interestingly, changes in PP-IP4 and IP8 levels were observed with Hh ligand induction (Fig. S6D). However, treatment with TNP or cyclopamine blocked the effect of Hh ligand, with PP-IP4 and IP8 not readily detected (Fig. S6 E and F). In parallel, we analyzed IP profiles for cells treated with ip6k2 siRNA. Differences were detected in the relative IP5 and IP6 peaks compared with control cells (Fig. S6 G and H). Thus, modulating Hh signaling or loss of IP6K2 activity altered IP levels.
Discussion
This study identifies a previously unknown function of the IP kinase IP6K2 in Hh signaling during embryonic development and tissue patterning. Evidence supporting this conclusion comes from the strengths of two unique model systems, zebrafish embryos and mammalian cultured cells. Loss of ip6k2 function results in zebrafish craniofacial and somite malformations, as well as defective NCC and slow muscle development. Strikingly, the expression of Hh target genes is down-regulated with IP6K2 inhibition or depletion. Moreover, TNP, a pharmacological inhibitor of IP6K activity, also blocks Hh signaling. Together, these findings reveal a role for a novel diphosphoryl inositol phosphate effector in the Hh pathway acting on or downstream of Smo.
We also linked different developmental defects to distinct enzymatic steps in the IP pathway, all of which depend on Ipk2, the most upstream enzyme examined. Depleting Ipk2 levels (for IP5 production) in zebrafish embryos results in perturbations that reflect the combined defects resulting from IP6K2 and Ipk1 depletions. However, the developmental defects associated with Ipk1 depletion are not identical to those caused by IP6K2 depletion and show no links to perturbations of Hh signaling. This indicates that the IP6K2 activity upstream of Ipk1 is critical for craniofacial development, and pinpoints the requirements in the Hh pathway to the IP6K2 enzymatic step between IP5 and PP-IP4.
Interestingly, a recent study in mice reported that an ip6k2 knockout predisposes animals to aerodigestive tract carcinoma (43). However, in contrast to our findings in zebrafish, these ip6k2-knockout mice show apparently normal embryonic development and growth. The sharp distinction between the mouse and zebrafish IP6K2 depletion defects is likely attributed to inherent differences in the number of IP6K isoforms and their relative ability for functional compensation. In zebrafish, only two IP6K isoforms exist, whereas there are three in mouse and humans (44). Targeted deletion of ip6k1 in mice results in growth retardation and decreased insulin release from pancreatic β-cells (45). A mouse ip6k3 knockout has not been reported. Thus, in different vertebrates, the relative roles for isoforms that generate PP-IPs might be distinct. Of note, our mammalian cell culture experiments directly show a role for IP6K2 in Hh signaling and correlate with the zebrafish results.
Discovering a functional linkage between IP6K2 and the Hh signaling pathways has multiple mechanistic consequences. Our studies suggest that IP6K2 acts as an activator to regulate Shh-induced Hh pathway response at the level or downstream of Smo. IPs are known to regulate target protein activity through direct IP binding (reviewed in ref. 6). Indeed, in vitro studies have reported that IP6 binding is a negative regulator of β-arrestin (46, 47). Others have recently shown that β-arrestin is required for in vivo Smo function in Hh signaling (48). Given that ipk1-depleted embryos do not show defects associated with altered Hh signaling, we speculate the PP-IP4 role is distinct from this potential IP6 regulation. However, both the PP-IP4 role and the IP6 role could converge, and both act in primary cilium (44, 48, 49).
We propose that IP6K2-catalyzed PP-IP4 production positively regulates the activity of an Hh pathway component. Exogenous small molecules, e.g., cyclopamine, can inactivate mammalian Smo through direct binding (36, 41, 42). However, various small-molecule agonists can compete with cyclopamine to bind Smo and activate Hh pathway (39). Together, these findings support the hypothesis that an endogenous small-molecule ligand for Smo might exist. PP-IP4 is a promising candidate for such an endogenous effector, and a potentiator of the Hh response. As IP6K2 itself is insufficient to reverse inhibition by Ptc, it is possible that Ptc directly inhibits IP6K2 activity in the absence of Hh pathway activation, analogous to a recently proposed mechanism where Ptc functions to transport an agonist away from the Smo activator (4). The link between IP6K2 activity and the Hh pathway is particularly significant, because ligand-dependent pathway activation is important for survival and growth of a number of cancers (2). Inhibiting IP6K2 activity might provide new therapeutic avenues for targeting aberrant Hh pathway activation.
Materials and Methods
For zebrafish studies, three independent MOs were used: ip6k2AUGMO (5′-CATCCCTTCGATGGCTGGGCTCATC-3′) targeted translational start codon region (−1 to +24 position), ip6k2UTRMO (5′-GGGCATTAGACTCTCACTCCTGGTC-3′) targeted 5′UTR region (−32 to −8 position), and ip6k2SPMO (5′-ATAATATCTCTCTTACCTCTTGTCC-3′) targeted the exon2-intron2 splice junction. The control ip6k25-mispair-AUGMO (mismatches in lowercase; 5′-CATgCCTTCcATGcCTGGcCTgATC-3′) contains 5 mismatches that disrupt targeting to the translational start codon region. ipk2SPMO (5′-AAGACATAAACATACCCTCATGCCC-3′) targeted the exon4-intron4 splice junction. ipk1MO1 was previously reported (20). MOs were from Gene Tools, except ip6k2ATGMO from Open Biosystems. For ip6k2 mRNA, zebrafish ip6k2 (catalog no. MDR1734-7601034; Open Biosystems) was linearized with XhoI and used as template. We injected 1–2 nL of 2.5-μg/μL ip6k2MO, 0.2-μg/μL ip6k2 mRNA, both ip6k2MO and ip6k2 mRNA, 1 nL of 7.5-μg/μL ipk1MO or 1 nL of 10-μg/μL ipk2SPMO into the yolk just below the single cell of fertilized embryos. Embryos were raised in embryo medium at 28.5 °C. Methods for histology, whole-mount in situ hybridization, immunohistochemistry, and time-lapse microscopy are in SI Materials and Methods. NIH 3T3 and COS-7 cells used for mammalian cell experiments were cultured in DMEM supplemented with 10% FCS and antibiotics. The ip6k2 siRNA (catalog no. sc-39072) and control siRNAs (sc-37007) were purchased from Santa Cruz Biotechnology. Methods for cell culture, transfection, drug treatment, IP labeling, and assays for Hh pathway activation are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. B. Appel (University of Colorado, Denver, CO), C. Chiang (Vanderbilt University, Nashville, TN), M. Cooper (Vanderbilt University, Nashville, TN), L.-E. Jao (Vanderbilt University, Nashville, TN), E. Knapik (Vanderbilt University, Nashville, TN), and L. Solnica–Krezel (Washington University, St. Louis, MO) for reagents and critical discussions; Dr. S. Sarmah for assisting with zebrafish experiments and microscopy; V. Grover for help with Hh assays, and Dr. T. R. Dawson for technical assistance. This work was supported by a grant from the American Heart Association (to B.S.), a Vanderbilt Zebrafish Pilot Project Grant (to S.R.W.), and the Vanderbilt Cell Imaging Shared Resource (to S.R.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007256107/-/DCSupplemental.
References
- 1.Varjosalo M, Taipale J. Hedgehog: Functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9:437–445. doi: 10.1038/nrm2414. [DOI] [PubMed] [Google Scholar]
- 4.Hausmann G, von Mering C, Basler K. The hedgehog signaling pathway: Where did it come from? PLoS Biol. 2009;7:e1000146. doi: 10.1371/journal.pbio.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balla T, Szentpetery Z, Kim YJ. Phosphoinositide signaling: New tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcázar-Román AR, Wente SR. Inositol polyphosphates: A new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- 7.York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- 9.York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- 10.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo HR, et al. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 12.Illies C, et al. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- 13.Nagata E, et al. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- 14.Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- 15.Wada N, et al. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- 16.Cordero D, et al. Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J Clin Invest. 2004;114:485–494. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucenas S, et al. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat Neurosci. 2008;11:143–151. doi: 10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin H, et al. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J Biol Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulugu S, et al. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 20.Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- 22.Knight RD, et al. Lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- 23.Yan YL, et al. A pair of Sox: Distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 24.Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: Part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev Dyn. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Barresi MJ, Stickney HL, Devoto SH. The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development. 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- 27.Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberhart JK, Swartz ME, Crump JG, Kimmel CB. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Burgess S, Hopkins N. Analysis of the zebrafish Smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development. 2001;128:2385–2396. doi: 10.1242/dev.128.12.2385. [DOI] [PubMed] [Google Scholar]
- 30.van Eeden FJ, et al. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- 31.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 32.Concordet JP, et al. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- 33.Karlstrom RO, et al. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development. 2003;130:1549–1564. doi: 10.1242/dev.00364. [DOI] [PubMed] [Google Scholar]
- 34.Hatta K, Bremiller R, Westerfield M, Kimmel CB. Diversity of expression of engrailed-like antigens in zebrafish. Development. 1991;112:821–832. doi: 10.1242/dev.112.3.821. [DOI] [PubMed] [Google Scholar]
- 35.Honjo Y, Eisen JS. Slow muscle regulates the pattern of trunk neural crest migration in zebrafish. Development. 2005;132:4461–4470. doi: 10.1242/dev.02026. [DOI] [PubMed] [Google Scholar]
- 36.Taipale J, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 37.Cooper MK, et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- 38.Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexandre C, Jacinto A, Ingham PW. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- 41.Cooper MK, Porter JA, Young KE, Beachy PA. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- 42.Incardona JP, Gaffield W, Kapur RP, Roelink H. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development. 1998;125:3553–3562. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- 43.Morrison BH, et al. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarmah B, Wente SR. Zebrafish inositol polyphosphate kinases: New effectors of cilia and developmental signaling. Adv Enzyme Regul. 2010;50:309–323. doi: 10.1016/j.advenzreg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci USA. 2008;105:2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaidarov I, Krupnick JG, Falck JR, Benovic JL, Keen JH. Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 1999;18:871–881. doi: 10.1093/emboj/18.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J Biol Chem. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- 48.Kovacs JJ, et al. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarmah B, Winfrey VP, Olson GE, Appel B, Wente SR. A role for the inositol kinase Ipk1 in ciliary beating and length maintenance. Proc Natl Acad Sci USA. 2007;104:19843–19848. doi: 10.1073/pnas.0706934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.