Abstract
In ensemble and single-molecule experiments using the yeast proliferating cell nuclear antigen (PCNA, clamp) and replication factor C (RFC, clamp loader), we have examined the assembly of the RFC·PCNA·DNA complex and its progression to holoenzyme upon addition of polymerase δ (polδ). We obtained data that indicate (i) PCNA loading on DNA proceeds through multiple conformational intermediates and is successful after several failed attempts; (ii) RFC does not act catalytically on a primed 45-mer templated fork; (iii) the RFC·PCNA·DNA complex formed in the presence of ATP is derived from at least two kinetically distinguishable species; (iv) these species disassemble through either unloading of RFC·PCNA from DNA or dissociation of PCNA into its component subunits; and (v) in the presence of polδ only one species converts to the RFC·PCNA·DNA·polδ holoenzyme. These findings redefine and deepen our understanding of the clamp-loading process and reveal that it is surprisingly one of trial and error to arrive at a heuristic solution.
Keywords: DNA replication, replication factor C, single-molecule FRET
DNA replication is carried out by effective coordination of the activities of many proteins at the replication fork. Given the complex nature of DNA synthesis, a structural and functional conservation of the accessory proteins apparently has evolved across diverse organisms (1–3). However, eukaryotic DNA replication has diverged and become more complicated than its prokaryotic counterpart due in part to the multisubunit composition of several proteins. In yeast, lagging strand DNA synthesis requires the coordinated actions of polymerase δ (polδ), proliferating cell nuclear antigen (PCNA), and replication factor C (RFC) (4). Eukaryotic PCNA, a ring-shaped trimer of identical subunits, plays a pivotal role in both DNA replication and repair by tethering the DNA polymerase to significantly enhance the processivity of DNA synthesis (5–10). The productive assembly of a polymerase-PCNA complex bound to DNA requires loading of the PCNA clamp by RFC (11). RFC, an ATP-driven motor, is composed of a large subunit (RFC-A, 95 kDa) and four small subunits (RFC-B–RFC-E, 36–40 kDa) and forms a RFC·PCNA·ATP complex that binds specifically to primed DNA (12).
The clamp-loading process occurs via multiple steps (13–16). X-ray crystallographic and electron microscopic studies of the clamp loader complexed with the clamp revealed that PCNA was either in a closed or slightly open conformation that perhaps represents the structure of an intermediate step in PCNA loading (13, 17). PCNA, in solution, has a closed conformation and must be opened for loading on DNA and further reclosed to be retained on the DNA possibly through a specific single interface (18).
The structure of the yeast RFC·PCNA complex (13) provided the coordinates for introducing a fluorescent donor–acceptor pair within the PCNA to probe the FRET efficiencies distance information on the conformation of the PCNA subunit interface (19). In the presence of ATP, the PCNA ring was opened by RFC; upon addition of DNA, PCNA closed around the DNA through a series of steps involving out-of-plane followed by in-plane closing of the ring. In the presence of ATPγS the RFC·PCNA complex opened the clamp and bound to DNA but ring closure was not completed (19, 20). Pre-steady-state global analysis of the accompanying ATP hydrolysis showed that the ATPase reaction proceeded through multiple steps within two distinct phases; first, assembly of the RFC·PCNA·DNA complex was coupled to ATP binding and second disassembly was coupled to ATP hydrolysis (21). Completion of a functioning holoenzyme requires addition of polδ to the RFC·PCNA·DNA complex. RFC and polδ bind the same face of PCNA (22, 23); the evidence on the timing of RFC departure or whether it remains as a member of a RFC·PCNA·DNA·polδ complex is variable (24–26).
There are a number of fundamental questions that remain unanswered. We turned to a combination of single-molecule and ensemble methods to adequately identify the nature and steps anticipated along the PCNA loading–unloading pathways (27, 28). Using primarily the FRET signals from a fluorescently labeled forked-DNA and PCNA, we obtained data that indicate (i) PCNA loading on DNA proceeds through a series of conformational intermediates and is successful after multiple failed attempts; (ii) RFC loads PCNA stoichiometrically and does not act catalytically on a primed 45-mer templated fork; (iii) the RFC·PCNA·DNA complex formed in the presence of ATP consists of at least two noninterconverting populations; (iv) the species of the RFC·PCNA·DNA complex disassemble either slowly through disassociation of the PCNA into its component subunits, a process that is independent of RFC and ATP/ATPγS levels, or through a more rapid recycling of intact PCNA; and (v) in the presence of polδ, a population of the RFC·PCNA·DNA complex is converted to the RFC·PCNA·DNA·polδ holoenzyme.
Results
PCNA Complexes with DNA in the Presence of RFC and ATP.
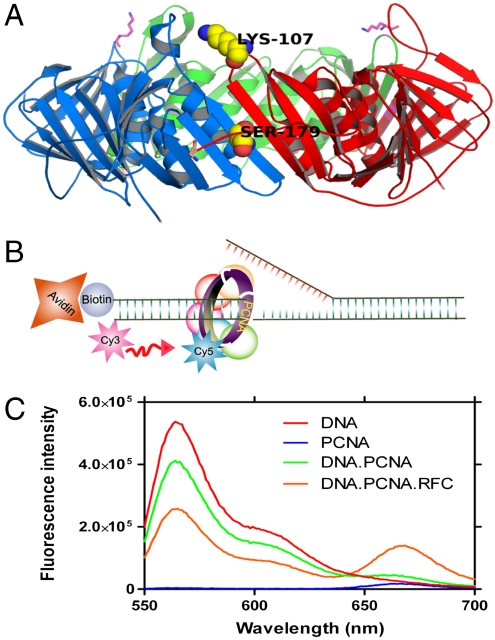
A mutant PCNA with all four native cysteines mutated to serines, and lysine 107 and serine 179 changed to cysteine and tryptophan, respectively, was labeled with Cy5-maleimide and purified by size-exclusion chromatography (Fig. 1A) (19). The labeled mutant PCNA was as active as the wild type in terms of stimulating RFC mediated ATP hydrolysis and its loading onto DNA (Fig. S1). A forked-DNA substrate was labeled with Cy3 dye that served as the FRET donor (Fig. 1B) (29–31). This forked-DNA substrate has a biotin attached at the 5′ end of the primer for surface immobilization onto a quartz microscope slide via a biotin–streptavidin interaction, which along with flap DNA also serves as a physical block to prevent the sliding-off of the loaded PCNA. A truncated form of RFC (Δ 283-RFC-A) was used because earlier experiments indicated that this form of RFC possessed catalytic activities identical to wild-type RFC in vitro and in vivo but lacked the nonspecific DNA binding attributed to the N-terminal region of RFC-A (20).
Fig. 1.
RFC mediated PCNA loading on the forked DNA. (A) Model of yeast PCNA generated using Pymol (http://www.pymol.org) from Protein Data Bank coordinates 1SXJ (13). PCNA subunits are shown in green, red, and blue. Lysine 107 was mutated to cysteine for dye labeling and serine 179 was mutated to tryptophan (K107C and S179W). (B) Schematic representation of the forked-DNA substrate, used in this study, the template had a Cy3 dye at 3′ end and a biotin was attached to 5′ end of the primer. (C) Fluorescence emission spectra obtained by exciting the Cy3-labeled DNA with a 532-nm light source. Cy5-labeled PCNA can be excited through FRET from Cy3 only when the two dyes are in close proximity (< ∼ 10 nm). Cy5 fluorescence intensity peaks at 665 nm.
The designed forked-DNA substrate was tested for PCNA loading by monitoring the FRET signal between the Cy3 and Cy5 dyes. The loading of Cy5-labeled PCNA (Cy5-PCNA) by RFC in the presence of ATP on the Cy3-labeled forked-DNA showed a distinct FRET signal (Fig. 1C). Both RFC and ATP are required for the PCNA loading, which is consistent with earlier reports (12, 19, 20). Labeled PCNA in the presence of RFC without DNA showed no FRET signal upon addition of ATP ruling out mere acceptor fluorescence variation that was caused by environmental changes upon RFC binding. No FRET signal was observed when labeled PCNA in the presence of RFC and ATP was added to the forked-DNA substrate lacking a primer, suggesting that PCNA did not load on the 5′ end of the flap strand, a process found for the PCNA-related checkpoint clamp Red 17/3/1 that loaded at the 3′ end and 5′ junctions (32).
The PCNA·RFC Complex Loading onto DNA Proceeds Through Rate-Limiting Conformational Changes Without Loss of RFC.
In order to study the kinetics of the PCNA loading, we mixed the RFC·PCNA·ATP complex with DNA in a stopped-flow instrument. An extended time course trace showed two distinct phases: a fast phase with a rate constant of 7 ± 1 s-1 and a second phase with a rate constant of 0.26 ± 0.04 s-1 (Fig. S2A). The two phases were also present when the reaction was carried out with ATPγS instead of ATP. Additionally, the overall amplitude of the signal was similar with ATP and ATPγS (Fig. S2A). Both ATP and ATPγS promote opening of the PCNA ring in the presence of RFC from 13 to 34 and 27 Å, respectively (19).
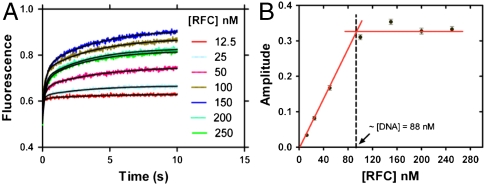
The loading experiment was also performed at varying RFC concentrations (12.5–250 nM). At all RFC concentrations, we observed the two phases mentioned above with the rates of both the fast and slow phases as well as their relative amplitudes (51 ± 5% and 49 ± 5%) being independent of the RFC concentration (Fig. 2A and Fig. S2B). Notably, the overall amplitude of the signal at different concentrations of RFC increased to the level of the DNA concentration and plateaued thereafter; at the breakpoint, the DNA and RFC concentration were equivalent (Fig. 2B). Therefore, we concluded that the RFC remained bound to the DNA and did not recycle to load the PCNA on DNA. In order to confirm this important observation, we preloaded a twofold excess of Cy3-labeled forked-DNA with unlabeled PCNA in the presence of RFC (RFC·PCNA·DNA in 1∶1∶2 ratio) and mixed this solution with Cy5-PCNA in another syringe. A very small increase in the fluorescence signal (< 10% of the observed plateau value) was observed suggesting negligible loading of labeled PCNA on the excess DNA, implying the lack of free RFC (Fig. S2C). These observations indicate that, for this forked-DNA, RFC did not dissociate upon loading of PCNA on DNA (20, 26).
Fig. 2.
RFC loads stoichiometric amounts of PCNA with rates independent of RFC concentration. (A) Labeled PCNA (200 nM) was incubated with RFC (12.5–250 nM) in the presence of excess ATP (5 mM). This preformed RFC·PCNA·ATP complex was mixed with a fixed concentration of DNA (88 nM) in a stopped-flow instrument and the FRET signal was followed at 665 nm. The loading traces were fit to a double-exponential equation with the rate constants of 7 ± 1 s-1 and 0.26 ± 0.04 s-1 for fast and slow phases. The respective amplitudes of the fitted fast and slow phases as a function of [RFC] are presented in Fig. S2B. (B) The overall amplitude of the signal (sum of the fast and slow amplitudes) was plotted against the concentration of RFC and showed saturation at a concentration equivalent to the concentration of DNA used in the reaction. The relative amplitudes of the fast and slow phases were 51 ± 5% and 49 ± 5% at different concentrations of RFC.
There are reports about multiple loadings of PCNA on DNA during nucleotide incorporation (25), consequently, we wished to confirm the results of stoichiometric PCNA loading by RFC by a nonkinetic method. Using the fluorescence from the Cy3- and Cy5-labeled DNA and PCNA, respectively, we designed a colorimetric assay to assess the stoichiometry of the loaded PCNA. Our results found that the PCNA is loaded on the forked-DNA substrate by RFC in a 1∶1 ratio (Fig. S3).
We investigated the effect of the PCNA concentration on its loading rate to determine whether the above fast phase corresponds to a bimolecular association by varying the concentration of the RFC·PCNA complex relative to a fixed concentration of DNA (Fig. S2D). The ratio of PCNA to RFC was adjusted to achieve 95% formation of the PCNA·RFC complex; higher concentrations of RFC were avoided to preclude binding of free RFC to the DNA. Neither the fast nor the slow phase rate constants (7 ± 1 s-1, 0.28 ± 0.03 s-1) or the relative amplitudes (55 ± 1%, 45 ± 1%) for the loading showed any significant change. We conclude that the observed two-phase loading did not represent the bimolecular association of the RFC·PCNA·ATP complex with DNA.
PCNA Subunit Exchange.
To study PCNA stability, we constructed a mutant PCNA with a FRET pair across the subunit interface (Fig. 1A). PCNA was labeled with a 5-[2(acetyl) aminoethyl] aminonaphthalene- 1-sulfonate (AEDANS) fluorophore at C107 that does not affect the ATPase activity of RFC (19). The FRET donor (W179) and acceptor (C107-AEDANS) pair allowed us to follow the PCNA subunit exchange associated with dissociation of the trimer into its subunits. Excitation of PCNA-K107C-AEDANS-S179W at 290-nm wavelength resulted in a fluorescence emission spectrum with two maxima (Fig. S4A). The specific tryptophan fluorescence (330 nm) of the PCNA-K107C-AEDANS-S179W was quenched by approximately 40% in comparison with the unlabeled protein (PCNA-K107C-S179W); the peak at wavelength 488 nm is produced by FRET between the tryptophan and AEDANS fluorophore.
Kinetics of the PCNA subunit exchange in solution was monitored by mixing the above FRET complex with PCNA lacking the AEDANS-tryptophan pair as trap. The decrease in the AEDANS fluorescence (Fig. S4B) that occurred upon mixing was well approximated by a single-exponential function to yield a rate constant of 0.0023 s-1 (Table S1). The PCNA subunit exchange rate constant was found to be independent of the trap concentration over a 30-fold excess. The PCNA subunit exchange rate constant was measured while complexed to RFC and decreased by a factor of 2 (Table S1). The addition of DNA to the mixture containing ATP and RFC did not affect the observed PCNA subunit exchange rate constant (Table S1). These findings rule out any active role of RFC in PCNA subunit exchange.
Disassembly of the RFC·PCNA·DNA Complex.
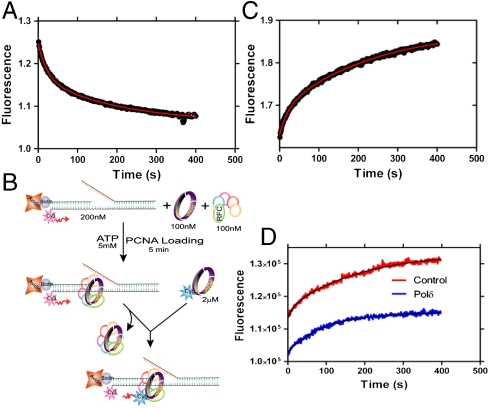
In order to study the mechanism for dissociation of PCNA from the assembled complex, the complex of RFC·PCNA·DNA containing Cy3-labeled PCNA and Cy5-labeled DNA was mixed with an excess of unlabeled PCNA in the presence of ATP (Fig. 3A). The disappearance of the FRET signal showed two distinct exponential decay phases with rate constants of 0.048 ± 0.004 s-1 for the fast phase and 0.005 ± 0.001 s-1 for the slow phase with amplitudes of 33 ± 8% and 67 ± 8%, respectively. The presence of two distinct decay rates suggests either multistep unloading or the presence of at least two populations of the ternary complex. We ruled out multistep unloading as the source of biphasic decay because (i) we did not observe any multistep FRET decrease during the unloading of a stable complex in our single-molecule measurements (described in a later section) and (ii) the disappearance of the FRET signal by PCNA trapping requires labeled PCNA dissociation. Interestingly, for the unloading reaction of the complex formed in the presence of ATPγS, only the slow phase was observed (Fig. S5). Subsequently, we measured the dependence on ATP concentration for the unloading reaction.
Fig. 3.
Unloading of PCNA. (A) PCNA (100 nM) loaded onto DNA (100 nM) was mixed with unlabeled trap PCNA at 2 μM concentration and the FRET signal was followed at 665 nm. The data were fit to a double-exponential equation (0.048 ± 0.004 s-1 and 0.005 ± 0.001 s-1). (B) Schematic presentation of experimental procedure for Fig. 3C. (C) Experiment performed as depicted in Fig. 3B. RFC loads Cy5-labeled PCNA at a rate equal to its off-rate from DNA. (D) A preassembled complex of unlabeled PCNA (100 nM) with RFC (100 nM) and DNA (200 nM) was mixed with Cy5-labeled PCNA (2 μM) in the absence and presence of polδ (100 nM). In the absence of polδ (control) the traces were fitted to a double-exponential equation with a fast rate constant 0.06 ± 0.005 s-1 and a slow rate constant 0.01 ± 0.003 s-1. In the presence of polδ, the traces were fitted to a single-exponential equation with a rate constant 0.01 ± 0.003 s-1. The amplitude of the signal with polδ is 60.5 ± 0.5% of that observed for the control.
At all concentrations of ATP, we observed the two phases with rate constants of 0.05 ± 0.01 s-1 for the fast phase and 0.005 ± 0.001 s-1 for the slow phase with relative amplitudes of 38 ± 9% and 62 ± 9%, respectively, in satisfactory agreement with the data from Fig. 3A (Fig. S6). These results suggest ATP independent unloading of PCNA in at least two different conformational states on DNA. Moreover, the coincidence of the slower rate constant for disassembly with that for subunit exchange suggests that the latter process may control dissociation in this population. The difference in the relative amplitudes of the two kinetic phases in the loading and unloading of PCNA, however, should not be interpreted as reflective of their relative concentrations given the absence of representative, defined FRET complexes.
We next established whether RFC dissociated along with PCNA from the RFC·PCNA·DNA complex. To answer this question, we carried out the trapping experiment illustrated in Fig. 3B. In brief, if RFC·PCNA dissociates simultaneously from the DNA and exchanges with Cy5-PCNA, the rate of appearance of the FRET signal in the DNA should be the same as the rate previously observed for the disappearance of the FRET signal in Fig. 3A. A FRET acceptor fluorescence time trace shown in Fig. 3C was essentially the mirror image of Fig. 3A, indicating the presence of two phases with rate constants of 0.05 ± 0.01 s-1 and 0.004 ± 0.001 s-1 and amplitudes of 32 ± 7% and 68 ± 7% that are within error identical to those reported above. Consequently, RFC is released along with PCNA in the dissociation of both populations of the RFC·PCNA·DNA complex.
A similar experiment can be carried out with polδ to ascertain whether one or both populations form a holoenzyme complex. In the presence of polδ, the kinetic FRET trace (Fig. 3D) is monophasic with a rate constant of 0.013 ± 0.003 s-1 and an amplitude of 60.5 ± 0.5% compared to the control experiment in Fig. 3C. We conclude that the displaced population represents the slower unloading species in Fig. 3A (rate constant of 0.005 s-1, relative amplitude 68 ± 7%) and is not capable of holoenzyme function; the other population characterized by decay rate constant of 0.05 s-1 proceeds to form a holoenzyme complex whose composition is RFC·PCNA·DNA·polδ.
Real-Time Monitoring of RFC Mediated PCNA Loading in the Single-Molecule Level.
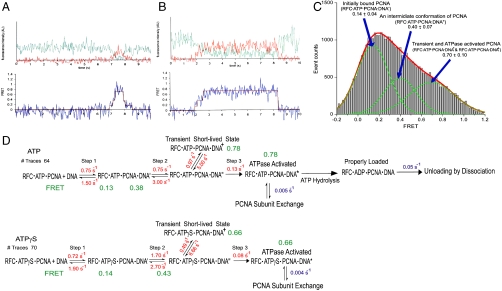
In order to study the multistep conformational changes of RFC·PCNA complex seen in the ensemble measurement (Fig. 2A), we monitored the loading process in a single-molecule FRET setup. The preformed RFC·Cy5-PCNA·ATP complex was delivered into a sample chamber containing the surface-immobilized forked-Cy3-DNA substrate while continuously recording the FRET signal. The Cy3-label on the forked-DNA was excited with a 532-nm laser and the fluorescence intensities from Cy3 and Cy5 were simultaneously recorded with an electron multiplying CCD camera to obtain continuous FRET efficiency time traces with 35-ms time resolution until most of the Cy3 dye molecules were photobleached. The collected data were analyzed with a hidden Markov model (HMM) to determine the rates between the various FRET states. The errors in rate are typically within 20% given the number of single-molecule traces used in the analysis (33). To determine the number of FRET states in the model, we explored the number of states from three to six and concluded that four is the minimum number of states to describe the system properly. A further increase in the number of states simply added states with zero FRET efficiencies, transitions between which are not observable in a FRET trace. The model used in the final analyses consisted of three nonzero FRET states in series where transitions take place only between adjacent states, which agreed well with our visual inspection of the FRET traces (Fig. 4 A and B). Due to the limited time resolution of the FRET traces, some FRET traces showed fast transitions that bypass the intermediate step(s) between a beginning and ending state, but there was no evidence of a more complicated kinetic scheme that required more than three nonzero FRET states. Transition to an additional zero FRET state from each of the nonzero FRET states was allowed to accommodate single-step photobleaching. We did not observe PCNA loading on DNA in the absence of RFC or ATP, which confirmed that PCNA loading on the forked-DNA substrate required both RFC and ATP.
Fig. 4.
Intermediate steps and rates during the process of the PCNA loading. (A and B) Single-molecule fluorescence intensity and FRET time traces showing various types of transitions between the intermediate steps in the PCNA loading process. The traces [fluorescence intensity (Upper) and FRET (Lower)] shown here are from PCNA loading in the presence of ATP. The green line in the intensity trace represents Cy3 fluorescence intensity and the red line represents Cy5 intensity. (C) FRET efficiency histogram of PCNA loading on the substrate DNA in the presence of RFC and ATP or ATPγS. Parts of FRET traces showing nonzero FRET efficiency, selected by visual inspection, were used to construct the histogram. At least three nonzero FRET states are apparent from the histogram as seen in the sample traces. Three Gaussian curves were used to fit the histogram. The lowest FRET state around 0.14 corresponds to the initially bound state of PCNA on the DNA (RFC·ATP·PCNA·DNA′ in Fig. 4D). The middle FRET state around 0.40 corresponds to the next intermediate conformation of PCNA on DNA (RFC·ATP·PCNA·DNA′′ in Fig. 4D). The highest FRET state around 0.70 includes the properly loaded PCNA conformation, improperly loaded PCNA conformation, and the other two transient conformations (RFC·ATP·PCNA·DNA+ and RFC·ATP·PCNA·DNA) as shown in Fig. 4D. (D) Kinetics of PCNA loading acquired from single-molecule (depicted in red) and ensemble (depicted in blue) studies in the presence of ATP or ATPγS. The model consists of FRET states in series where transitions take place only between adjacent states. The states (RFC·ATP·PCNA + DNA and RFC·ATPγS·PCNA + DNA) include an initial bound state RFC·PCNA on DNA (zero FRET), two intermediate states, RFC·PCNA·DNA′ and RFC·PCNA·DNA′′ (medium FRET), and a loaded state (high FRET). The latter consists of the transient species RFC·PCNA·DNA+, ATPase activated state, properly loaded state, and improperly loaded state as implicated by the PCNA unloading ensemble kinetics results. Unloading of PCNA takes place by dissociation or subunit exchange.
One unexpected observation in the single-molecule loading experiment was the unloading of some PCNA complexes from DNA in a fairly short time (< 1 s), whereas some others lasted longer than the acceptor dye photobleaching lifetime (> 5 s) (Fig. 4 A and B). A FRET efficiency histogram of PCNA loading is exhibited in Fig. 4C. We built the lifetime histograms of the loaded final state and fit the histograms with exponential decays to discover two distinct unloading timescales indicating two different loaded states of PCNA (Fig. S7 A and B). We suggest that the short-lived loaded state represents poorly arranged attempts of loading, whereas the long-lived state represents stably loaded PCNA on the DNA (Fig. 4D). From the double-exponential fits of the histograms, we found that about 10–15% of the PCNA complex is stably loaded on DNA—i.e., RFC must make eight or nine attempts before it loads PCNA stably on DNA. We analyzed the data in a different way to confirm this result. We fit the histograms with a single-exponential decay corresponding to the shorter lifetime events and obtained the counts for the rest of the events that do not belong to the single-exponential decay curve (threshold = 0.5 event count) and attributed those to longer lifetime events. This alternative analysis also yielded ∼10% of longer lifetime components. Due to the limiting dye photobleaching lifetime and blinking, the unloading timescale of stably loaded PCNA could not be determined from the single-molecule experiments. Three-step binding was also observed when the PCNA loading was performed in the presence of ATPγS instead of ATP with no noticeable difference in the rates. Similar experiments as described above were carried out in the presence of polδ. However, we observed insignificant differences in the rates of the initial three steps of PCNA loading. A kinetic sequence that combines the results of the single-molecule and ensemble studies is presented in Fig. 4D.
Discussion
PCNA Loading.
The single-molecule and ensemble kinetics reveal kinetic signatures for the various RFC·PCNA·DNA species in the PCNA loading process designated in Fig. 4D, although we do not know their molecular identities. Our results from following the time course of the FRET signal during PCNA loading indicated the presence of two distinct phases. The bimolecular association of RFC·PCNA with DNA was not observed in our measurements as the signal did not show a PCNA concentration dependence (Fig. S2D). The fact that we did not see significant changes in the amplitude or rate constants when we used ATP or ATPγS (Fig. S2A) suggests that ATP hydrolysis is not required for this aspect of clamp loading. However, it is required for clamp closure (19). Importantly, the biphasic nature of clamp loading was not due to multiple loading of PCNA molecules on the DNA as confirmed by the 1∶1 stoichiometry of PCNA∶DNA (Fig. 2A and Fig. S3).
The interpretation that RFC acts stoichiometrically and not catalytically in loading of PCNA is supported by the fact that (i) RFC does not dissociate within the experimental time frame from the PCNA loaded onto the DNA (Fig. 2A); (ii) once dissociated from the complex, RFC remains competent for subsequent binding to a new PCNA molecule and loading it onto DNA (Fig. 3); and (iii) direct gel analysis of the PCNA·DNA ratio (Fig. S3). We propose that RFC remains bound to the PCNA upon loading of the clamp and that this ATPase activated RFC·PCNA·DNA∗ complex partitions into two populations in the proportions 1∶2 as revealed by the biphasic nature of PCNA dissociation from the complex. However, there is evidence that in the loading of PCNA onto longer nicked plasmids RFC acts catalytically (34) suggesting that the behavior of RFC is dictated by the available spacing on the DNA substrate.
Further parsing of the clamp-loading process was furnished by our single-molecule data. There are at least three conformational changes of PCNA during the PCNA loading. Inspection of the traces in Fig. 4 A and B as well as the histogram analysis in Fig. 4C reveals the presence of three states in the PCNA loading process that yield a stably loaded state (Fig. 4D) that occurs only on average after approximately eight or nine attempts from the last intermediate conformation (0.97 s-1 vs. 0.13 s-1). This kinetic sequence does not change when ATPγS is substituted for ATP, suggesting that in accord with ensemble kinetics the hydrolysis of ATP is not required to achieve a stable RFC·PCNA·DNA complex. Previous ensemble FRET studies (19) found that the opening of the PCNA in the RFC·PCNA·DNA complex achieved by ATP and ATPγS differed only slightly in the extent to which the distance between the subunit interface was open (34 vs. 27 Å, respectively).
The appearance of these steps revealed the stochastic nature of the clamp-loading process. Simulation of the kinetic sequence in Fig. 4D showed the single-molecule data to be biphasic with the rates and amplitudes within the same order of magnitude as the ensemble kinetics. It has been suggested that the clamp-loading process is completed in multiple stages, but it was not heretofore shown experimentally (19). Structural and molecular dynamics simulation studies on the clamp-loading process have described the possible conformational changes in the clamp and the clamp loader where PCNA is reported to be opened out-of plane and closed in-plane (35–37). Indeed some of the states in Fig. 4D could represent such species.
PCNA Unloading.
We found biphasic kinetics for PCNA unloading from DNA when the clamp was loaded in the presence of ATP, whereas only a single phase was observed with ATPγS. The kinetics of unloading were not altered when excess ATP was mixed with a fluorescently silent PCNA trap during the reaction that eliminated the possibility that ATP levels were limiting. Because ATP but not ATPγS led to a completely closed form of PCNA on DNA (19), we anticipated that increased ATP levels through hydrolysis would drive the RFC·PCNA·DNA complex into one state. The RFC·ATP·PCNA·DNA∗ partitions to dissociate from the DNA with rates of 0.048 ± 0.004 s-1 and 0.005 ± 0.001 s-1, respectively. These two rate constants are in remarkably good agreement with those from an earlier study that found rate constants by surface plasmon resonance of 0.06 s-1 and 0.007 s-1 for the two phases for PCNA dissociation from a primed DNA template in the presence of ATP (20). Likewise with ATPγS, a single phase and single rate constant of 0.007 s-1 was reported.
A key insight into the mode of PCNA unloading is the coincidence between the rate constant for PCNA subunit exchange (Fig. S4 and Table S1) and those for PCNA dissociation for clamp loaded in the presence of ATPγS and the slower phase for ATP. Based on this observation, we conclude that the slow unloading is due to subunit exchange. However, that does not mean the RFC·ATP·PCNA·DNA∗ complexes (Fig. 4D) attained by RFC in the presence of ATPγS and ATP necessarily have the same configuration as noted above. In fact, this ATPase activated state must consist of at least two populations. In the case of ATP, we propose that ATP hydrolysis by RFC converts one of these populations to a properly loaded RFC·PCNA complex that can generate a functional holoenzyme as discussed below. In the absence of polδ, this population of the two dissociates more rapidly. The other is not converted and is unloaded at a slower rate (∼0.005–0.010 s-1) through subunit dissociation as found for ATPγS. Because neither of the two unloading rates depend on ATP concentration, RFC does not appear to catalyze the unloading process (Fig. S6) although the trapping experiments (Fig. 3) revealed that RFC dissociated simultaneously. In addition, the subunit exchange rate was unaffected by the presence of RFC (Table S1). The finding that RFC does not require ATP hydrolysis to unload PCNA is also consistent with an earlier report (38).
In the presence of polδ, the ATPγS assembled RFC·PCNA·DNA complex has been shown to support primer extension although the processivity of the replication was severely limited—tens of nucleotides vs. > 5,000 nucleotides incorporated (24, 39). With ATP, the key issue is which of these two populations supports holoenzyme function. Importantly, whereas the addition of polδ had a negligible effect on the rate constants (0.013 s-1 versus 0.005 s-1) or fraction (61% versus 67%) of the population characterized by the slower dissociation rate, its presence suppressed the dissociation of PCNA and RFC from DNA in the population characterized by the faster rate of dissociation (Fig. 3D). We conclude that the holoenzyme has the composition of RFC·PCNA·DNA·polδ. Given the structure of the RFC·PCNA complex, repositioning of RFC would appear necessary to allow polδ to bind to the same face of the clamp (40). It remains to be determined whether the processivity of replication is terminated by the loss of RFC or PCNA or polδ or their combination from the holoenzyme complex.
Without actual structures the various states in our revised model for PCNA loading (Fig. S8) should be viewed as merely representative. The major point is that clamp loading, despite evolution, remains largely a heuristic process proceeding through a number of intermediate states. The clamp loader through its binding of ATP and ATPase activity not only facilitates clamp opening and closure but also probably acts to recycle incorrectly loaded clamps. Rather than following a highly confined, highly efficient set of steps, clamp loading is achieved stochastically by multiple steps of trial and error and ultimately successful holoenzyme formation.
Materials and Methods
Proteins.
The mutant yeast PCNA (K107C) was expressed and purified from Escherichia coli as described before (19). Yeast clamp loader constructs for individual RFC subunits were generously provided by Mike O’Donnell (Rockefeller University, New York). RFC-B, C, and D were cloned into a pET11a vector, ΔN 283-RFC-A and RFC-E were cloned into pLANT vector, and the expression and purification were carried out as described earlier (41). Polymerase delta was generously provided by Satya Prakash (University of Texas).
Labeled DNA Substrate.
High-pressure liquid chromatography (HPLC) purified oligonucleotides (5′-CTC GGA CTG CAC GTG CCG CGT GGG CAG CTC GAG CAG GCT GAC CAG-Cy3- 3′, 5′-CGT GGT GGT AGG TGA GGG CGG CAC GTG CAG TCC GAG-3′, and 5′-/5′ Biotin-CTG GTC AGC CTG CTC-3′) were purchased from Integrated DNA Technologies. To make the forked-DNA substrate, equimolar amounts of the above primers were mixed and heated at 92 °C for 2 min and gradually cooled down to room temperature for annealing.
Labeling of Mutant PCNA.
The mutant yeast PCNA (K107C) was labeled with Cy5 and checked for activity as described before (19). Detailed procedure is given in SI Materials and Methods.
Steady-State and Stopped-Flow Measurements.
Steady-state fluorescence spectroscopy was measured on a Jobin Yvon FluoroMax-4 fluorimeter and stopped-flow studies were performed on a Kintek stopped-flow machine equipped with a fluorescence detector. Conditions for each experiment are detailed in the figure captions and detailed procedures are available in SI Materials and Methods.
Real-Time Single-Molecule Fluorescence Measurements.
We used PEG-coated quartz microscope slides to monitor fluorescence signals from single Cy3 and Cy5 fluorophores. Cy3-attached forked-DNA substrate was immobilized on a quartz slide through biotin–streptavidin interaction. The RFC·PCNA complex in the presence of ATP or ATPγS was delivered into the channel containing the DNA substrate. Cy3-labeled forked-DNA substrate was excited with a 532-nm laser and fluorescence intensities from Cy3 and Cy5 were recorded simultaneously. FRET time traces were analyzed with an HMM using a home-built HMM optimization code (SI Materials and Methods).
Supplementary Material
Acknowledgments.
We thank Dr. Zhihao Zhuang for PCNA mutants constructs. We thank Prof. Mike O’Donnell (Rockefeller University, New York) and Prof. Satya Prakash (University of Texas) for generously providing us the RFC constructs and polymerase δ protein. We also thank the Benkovic lab members for their valuable comments. This work was financially supported by National Institutes of Health (NIH) Grant GM013306 (to S.J.B.) and NIH Pathway to Independence Award (R00 GM079960), Dreyfus New Faculty Award, and Searle Scholarship (T.H.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014139107/-/DCSupplemental.
References
- 1.Bartlett R, Nurse P. Yeast as a model system for understanding the control of DNA replication in Eukaryotes. Bioessays. 1990;12:457–463. doi: 10.1002/bies.950121002. [DOI] [PubMed] [Google Scholar]
- 2.Bell S, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 3.Nossal NG. Protein-protein interactions at a DNA replication fork: Bacteriophage T4 as a model. FASEB J. 1992;6:871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- 4.Nick McElhinny S, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer G, Burgers PM. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990;18:261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 7.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 8.Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 9.Prelich G, et al. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 10.Warbrick E. The puzzle of PCNA’s many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 11.Tsurimoto T, Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: Functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes X, Burgers PM. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem. 2001;276:34768–34775. doi: 10.1074/jbc.M011631200. [DOI] [PubMed] [Google Scholar]
- 13.Bowman G, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 14.Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem. 2006;281:35531–35543. doi: 10.1074/jbc.M606090200. [DOI] [PubMed] [Google Scholar]
- 15.Miyata T, et al. The clamp-loading complex for processive DNA replication. Nat Struct Mol Biol. 2004;11:632–636. doi: 10.1038/nsmb788. [DOI] [PubMed] [Google Scholar]
- 16.Venclovas C, Colvin ME, Thelen MP. Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 2002;11:2403–2416. doi: 10.1110/ps.0214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyata T, et al. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci USA. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dionne I, Brown NJ, Woodgate R, Bell SD. On the mechanism of loading the PCNA sliding clamp by RFC. Mol Microbiol. 2008;68:216–222. doi: 10.1111/j.1365-2958.2008.06150.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci USA. 2006;103:2546–2551. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes X, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Levin MK, Sakato M, Zhou Y, Hingorani MM. Mechanism of ATP-driven PCNA clamp loading by S. cerevisiae RFC. J Mol Biol. 2009;388:431–442. doi: 10.1016/j.jmb.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossi R, Jonsson ZO, Allen BL, Hardin SH, Hubscher U. Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen. J Biol Chem. 1997;272:1769–1776. doi: 10.1074/jbc.272.3.1769. [DOI] [PubMed] [Google Scholar]
- 23.Oku T, et al. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells. 1998;3:357–369. doi: 10.1046/j.1365-2443.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 24.Burgers PM. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 25.Masuda Y, et al. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 2007;35:6904–6916. doi: 10.1093/nar/gkm822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podust V, Tiwari N, Stephan S, Fanning E. Replication factor C disengages from proliferating cell nuclear antigen (PCNA) upon sliding clamp formation, and PCNA itself tethers DNA polymerase delta to DNA. J Biol Chem. 1998;273:31992–31999. doi: 10.1074/jbc.273.48.31992. [DOI] [PubMed] [Google Scholar]
- 27.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 28.Lee T, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaboord B, Benkovic SJ. Accessory proteins function as matchmakers in the assembly of the T4 DNA polymerase holoenzyme. Curr Biol. 1995;5:149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 30.Smiley R, Zhuang Z, Benkovic SJ, Hammes GG. Single-molecule investigation of the T4 bacteriophage DNA polymerase holoenzyme: Multiple pathways of holoenzyme formation. Biochemistry. 2006;45:7990–7997. doi: 10.1021/bi0603322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhuang Z, Berdis AJ, Benkovic SJ. An alternative clamp loading pathway via the T4 clamp loader gp44/62-DNA complex. Biochemistry. 2006;45:7976–7989. doi: 10.1021/bi0601205. [DOI] [PubMed] [Google Scholar]
- 32.Majka J, Binz SK, Wold MS, Burgers PM. Replication protien A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 33.Lee TH. Extracting kinetics information from single-molecule fluorescence resonance energy transfer data using hidden markov models. J Phys Chem B. 2009;113:11535–11542. doi: 10.1021/jp903831z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bylund G, Burgers PM. Replication proetin A-directed unloading of PCNA by the Ctf18 cohision establishment complex. Mol Cell Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgescu RE, et al. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov I, Chapados BR, McCammon JA, Tainer JA. Proliferating cell nuclear antigen loaded onto double-stranded DNA: Dynamics, minor groove interactions and functional implications. Nucleic Acids Res. 2006;34:6023–6033. doi: 10.1093/nar/gkl744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazmirski S, Zhao Y, Bowman GD, O’Donnell M, Kuriyan J. Out-of-plane motions in open sliding clamps: Molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2005;102:13801–13806. doi: 10.1073/pnas.0506430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao N, Johnson A, Bowman GD, Kuriyan J, O’Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J Biol Chem. 2006;281:17528–17539. doi: 10.1074/jbc.M601273200. [DOI] [PubMed] [Google Scholar]
- 39.Langston L, O’Donnell M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finkelstein J, Antony E, Hingorani MM, O’Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.