Abstract
Development of mating preference is considered to be an early event in speciation. In this study, mating preference was achieved by dividing a population of Drosophila melanogaster and rearing one part on a molasses medium and the other on a starch medium. When the isolated populations were mixed, “molasses flies” preferred to mate with other molasses flies and “starch flies” preferred to mate with other starch flies. The mating preference appeared after only one generation and was maintained for at least 37 generations. Antibiotic treatment abolished mating preference, suggesting that the fly microbiota was responsible for the phenomenon. This was confirmed by infection experiments with microbiota obtained from the fly media (before antibiotic treatment) as well as with a mixed culture of Lactobacillus species and a pure culture of Lactobacillus plantarum isolated from starch flies. Analytical data suggest that symbiotic bacteria can influence mating preference by changing the levels of cuticular hydrocarbon sex pheromones. The results are discussed within the framework of the hologenome theory of evolution.
Keywords: holobiont, hologenome, Lactobacillus, microbiota, symbiosis
Mating preference is considered one of the mechanisms for the origin of new species (1–4). Experimentally, mating preference in flies has been shown to occur when populations were divided and reared separately for many generations under different environmental conditions (5), such as temperature and humidity (6), diet (7), and exposure to media at different pH values (8). In one of these studies, Dodd (7) reared Drosophila pseudoobscura on starch-based and on maltose-based media for more than 25 generations and discovered that “starch flies” preferred to mate with other starch flies and that “maltose flies” preferred to mate with other maltose flies (i.e., positive assortative mating). These data were surprising because there was no selection for the observed mating preference. Somehow mating preference developed as a correlated response when selection favored novel adaptation to the environment (namely, the different food sources).
Evidence presented here indicates that the “correlated response” driving mating preference is the emergence of different bacterial communities associated with the two fly populations grown on the different food sources: these bacteria, which are part of the normal fly microbiota, are responsible for the mating preference.
Results
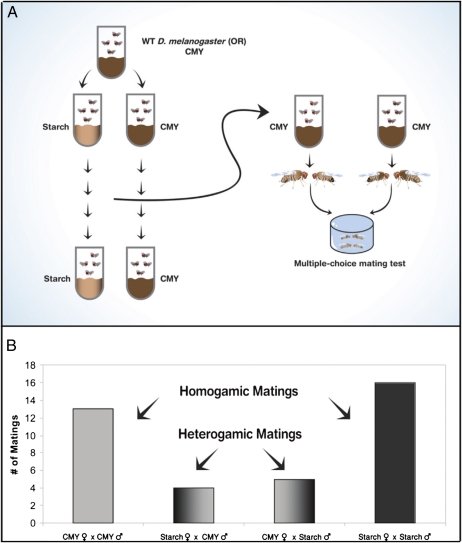
An inbred laboratory strain of WT Drosophila melanogaster maintained on standard cornmeal-molasses-yeast (CMY) agar medium, was divided in two: flies in one group continued to be grown on CMY medium whereas flies in the other group were transferred to starch medium (Fig. 1A; Materials and Methods). After a fixed number of generations, each population was grown for one generation on CMY medium (to avoid the medium itself becoming the cause for mating preference) and then tested for mating preference in mating chambers. Each chamber contained four flies: one male and one female from one population and one male and one female from the other population. Individuals of one population had the tips of their wings clipped to allow identification. Several replicates of each experiment were performed with the wing clippings alternating between populations (i.e., half of each experiment was done with wing clipping of flies from one treatment, and the other half was the reciprocal). Even though wing clipping has been previously shown not to affect mating preference in Drosophila (9, 10), we used a counterbalanced design for mating preference tests to control for any possible wing clipping effects.
Fig. 1.
(A) Schematic representation of the experimental procedure. A population of flies was divided, serially transferred in two different media, and then examined for mating preference. After rearing the flies for a number of generations on starch or CMY media, each population was grown separately for one generation on CMY medium and then tested for mating preference. The multiple-choice mating tests were performed in 24-well plastic plates; each well contained four flies: one male and one female starch-reared and one male and one female CMY-reared. Matings were recorded every 4 min for 1 h. (B) Mating preference tests of D. melanogaster after growing 11 generations on starch or CMY medium.
In the first experiment, the mating preference test was performed after 11 generations (Fig. 1B). Of the 38 recorded matings, 29 were homogamic, i.e., “starch males” with “starch females” and “CMY males” with “CMY females,” whereas only nine were heterogamic (i.e., “starch” × “CMY”). These results demonstrate a significant positive sexual isolation index (SII) (11): SII = 0.53 ± 0.14 (SEM) and P = 0.0012 (binomial test), with the following assumptions:
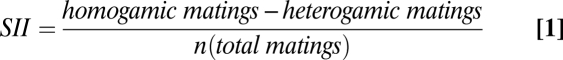 |
and
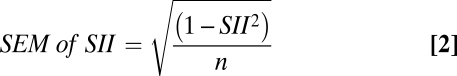 |
SII values greater than zero indicate positive assortative mating (mating preference).
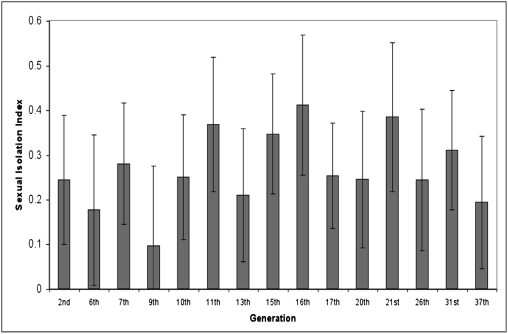
The second set of experiments was carried out exactly like the first except that the mating preference test was performed periodically, beginning after the second generation (Fig. 2). Homogamic mating occurred at all 15 times examined. Pooling all tests yielded 571 homogamic and 329 heterogamic matings: SII = 0.27 ± 0.02 (SEM), P < 0.0001. The finding that mating preference occurred after only two generations in different growth media has not been reported previously.
Fig. 2.
Mating preference tests of D. melanogaster after growth for different numbers of generations on either starch or CMY medium. The SII is plotted for each generation examined. Bars indicate the SEM.
A control experiment was performed in which two lines of CMY-reared flies, grown separately for 27 generations, were tested for mating preference. Of the 38 matings, 18 were homogamic and 20 were heterogamic (SII = −0.05, P = 0.4357 by binomial test), demonstrating no mating preference. Thus, difference in diet and not physical separation was the experimental variable responsible for the mating preferences reported here.
To test whether the addition of antibacterials affects mating preference, fly populations were reared on media supplemented with antibiotics (tetracycline, rifampicin, and streptomycin, together or separately; Materials and Methods). Following treatment with antibacterial agents, the flies’ mating preference changed from positive assortative to random: pooling the results from 10 independent experiments yielded 267 homogamic and 263 heterogamic matings (Table 1, experiment 2; SII = 0.01 ± 0.03). Clearly, there is a significant difference in mating preference between the antibiotic-treated and untreated flies, suggesting that symbiotic bacteria were responsible for the homogamic mating preference.
Table 1.
The role of bacteria in diet-induced mating preference of D. melanogaster
| Experiment | Fly treatment* | Matings | SII, mean ± SEM | P value† |
| 1 | Starch-grown × CMY-grown | 18 | 0.27 ± 0.02 | <0.0001 |
| 2 | Experiment 1 after antibiotics | 10 | 0.01 ± 0.03 | 0.4483 |
| 3 | Experiment 2 after infection with homologous bacteria‡ | 4 | 0.22 ± 0.03 | 0.0024 |
| 4 | Experiment 3 with Lactobacillus replacing homologous bacteria in starch-bred flies | 4 | 0.16 ± 0.06 | 0.0392 |
| 5 | Experiment 3 with Lactobacillus plantarum replacing homologous bacteria in starch-bred flies | 5 | 0.19 ± 0.05 | 0.0004 |
| 6 | Infection control (no added bacteria) | 4 | −0.04 ± 0.08 | 0.4052 |
*After all treatments, the flies were grown for one generation in CMY medium before performing the mating preference test.
†P value of the normal approximation to the binomial test. P < 0.05 was considered to indicate significant mating preference.
‡Antibiotic-treated starch- and CMY-grown flies were infected with bacteria isolated from their respective growth medium (before antibiotic treatment).
Further evidence that bacteria were responsible for the mating preference came from four independent infection experiments. Microbiota, isolated from starch or CMY media upon which the flies had grown, were inoculated into separate vials containing sterile CMY medium. Virgin, antibiotic-treated flies were then introduced into these vials (containing the starch-derived or CMY-derived bacteria on CMY medium) so that each antibiotic-treated fly population was exposed to its specific corresponding microbiota. After one generation of growth with the corresponding microbiota, positive assortative mating was regained (SII = 0.22 ± 0.03, P = 0.0024; Table 1, experiment 3). Thus, the reinfected flies showed similar mating preference to flies before antibiotic treatment.
The most abundant bacteria associated with flies reared on the different media were characterized by analysis of the 16S rRNA gene sequences of the third-generation flies (Table 2). D. melanogaster reared on the standard CMY medium contained the endosymbiont Wolbachia plus a diverse bacterial community. However, the flies transferred to starch medium contained Wolbachia plus 26% Lactobacillus plantarum. CMY-reared flies contained only 3% L. plantarum. Viable counts of fly homogenates showed that starch flies contained 2.3 × 105 Lactobacillus sp. per fly, whereas CMY flies contained only 2.6 × 104 Lactobacillus sp. per fly. Although this is only a limited analysis of the microbiota, it demonstrates that the associated bacteria of the CMY and starch populations are already different by the third generation and points to L. plantarum as a likely candidate for inducing the mating preference phenomenon. Wolbachia species could not have been responsible for the homogamic mating preference for two reasons: (i) both fly populations contained the same Wolbachia wMel strain, as verified by sequencing the wsp gene, and (ii) Wolbachia was not present in flies that were antibiotic-treated and subsequently infected. The presence of Wolbachia before antibiotic treatment and its absence after treatment and infection was demonstrated by PCR using primers specific for both its 16S rRNA and wsp genes (Fig. S1). Accordingly, we next isolated Lactobacillus and other species from starch-reared flies, used them to infect antibiotic-treated, starch-reared flies, and then tested these flies for mating preference against antibiotic-treated, CMY-reared files infected with CMY-derived bacteria. More than 80% of the culturable bacteria from starch-bred flies were Lactobacillus spp. (Wolbachia is not culturable). Similar percentages of Lactobacilli were found in the starch medium after fly growth.
Table 2.
Bacterial communities in D. melanogaster grown on CMY or starch
| Representation in clone library, %* |
|||
| Closest match (accession no.) | Identity, % | CMY† | Starch‡ |
| Acetobacter pomorum strain EW816 (EU096229.1) | 89.81 | 1.49 | — |
| Acetobacter pomorum strain EW816 (EU096229.1) | 93.99 | 1.49 | — |
| Acetobacter pomorum strain EW816 (EU096229.1) | 94.00 | 1.49 | — |
| Acetobacter pomorum strain EW816 (EU096229.1) | 100.00 | 14.93 | — |
| Bacillus firmus strain XJSL2-8 (GQ903397.1) | 100.00 | 11.94 | — |
| Enterococcus faecalis strain 3–12 (GU177628.1) | 100.00 | 1.49 | — |
| Lactobacillus plantarum strain IMAU:10272 (GU138600.1) | 100.00 | 2.99 | 26.09 |
| Low G+C Gram-positive bacterium T135 (AB116139.1) | 99.44 | 5.97 | — |
| Weissella paramesenteroides strain CTSPL5 (EU855224.1) | 97.24 | 1.49 | — |
| Weissella paramesenteroides strain CTSPL5 (EU855224.1) | 99.64 | 4.48 | — |
| Wolbachia endosymbiont of Drosophila melanogaster (AB360385.1) | 100.00 | 47.77 | 73.91 |
16S rRNA gene analysis was performed on files (third generation) grown on CMY medium for one generation before mating preference tests.
*Sequences with ≥99% identity were clustered by DOTUR (9).
†Based on 64 clones.
‡Based on 23 clones.
Infection with a mixture of isolated Lactobacillus spp. caused a significant increase in mating preference (Table 1, experiment 4), compared with antibiotic-treated controls without added bacteria (Table 1, experiment 6). When a pure culture of L. plantarum, obtained from starch-bred flies, was used to infect antibiotic-treated flies, significant homogamic mating preference was obtained (Table 1, experiment 5). In the latter experiment, the data clearly demonstrate that a single bacterial species can induce mating preference. Parallel experiments using other bacterial species isolated from starch-bred flies, e.g., Providencia rettgeri and a mixture of 41 bacterial strains isolated from starch-bred flies had no effect on mating preference.
To discover the origin of L. plantarum in starch-bred flies, CMY-bred flies were treated with antibiotics before dividing them into CMY and starch media. No homogamic mating preference was observed for the three generations that were tested. In the no-antibiotic control, significant homogamic mating preference was observed already in the first generation. As the flies were reared on sterile media, these data indicate that the bacterium responsible for the mating preference, L. plantarum, was already present in CMY-bred flies, albeit in low numbers (consistent with the data in Table 2), and, when transferred to starch medium, were amplified and induced mating preference.
To gain some insight at a biochemical level into how the fly microbiota may influence mating behavior, we analyzed the cuticular hydrocarbon (CH) composition of antibiotic-treated and untreated CMY and starch flies (Table 3, Figs. S2 and S3, and Tables S1 and S2). In nontreated female flies, there was a significant difference between the two populations in at least four of the major CHs. In nontreated male flies, there was a significant difference between CMY and starch-bred flies in at least three CHs. In most cases, antibiotic treatment reduced the level of CHs and decreased the differences between the two populations. These data suggest that symbiotic bacteria can influence the levels of fly sex pheromones.
Table 3.
Major differences in CH profiles of CMY and starch bred flies
| Mean CH per fly ± SEM, ng |
||||||
| No antibiotic treatment |
Treated with antibiotics |
|||||
| Peak name | Retention time (min) | Identified compounda | CMY (n = 3) | Starch (n = 2) | CMY (n = 3) | Starch (n = 3) |
| Females | ||||||
| F16 | 17.92 | 7-Tricosene | 44.7 ± 10.1 | 22.6 ± 0.5 | 25.8 ± 2.3 | 16.2 ± 0.8 |
| F24 | 21.08 | 7-Pentacosene | 37.1 ± 7.7 | 11.1 ± 2.3 | 20.3 ± 2.5 | 10.5 ± 0.8 |
| FF12 | 23.77 | 7,11-Heptacosadiene | 117.4 ± 21.8 | 172.0 ± 5.6 | 50.6 ± 8.1 | 59.4 ± 8.8 |
| F40 | 26.54 | 2-Methyloctacosane | 87.2 ± 2.5 | 136.2 ± 4.3 | 34.2 ± 4.3 | 49.0 ± 6.1 |
| Males | ||||||
| M12 | 16.4 | Cis-vaccenyl acetate | 9.9 ± 6.2 | 51.6 ± 7.8 | 16.2 ± 5.7 | 32.4 ± 14.0 |
| M16 | 17.92 | 7-Tricosene | 259.1 ± 59 | 495.4 ± 39.9 | 181.2 ± 15.8 | 415.0 ± 10.5 |
| M24 | 21.08 | 7-Pentacosene | 146 ± 29.8 | 61.5 ± 8.4 | 93.1 ± 3.5 | 69.7 ± 4.8 |
aBased on the GC CH profiles of D. melanogaster (12) and on GC-MS analyses.
Discussion
The major findings of this study are (i) diet-induced mating preference occurred in D. melanogaster after only one generation on different growth media and was maintained under these conditions for at least 37 generations, (ii) fly-associated commensal bacteria were responsible for the mating preference, (iii) L. plantarum was responsible, at least in part, for the mating preference, and (iv) the source of L. plantarum was the commensal microbiota of the CMY-bred flies, which were amplified in starch medium. There are abundant data demonstrating that changes in the diet of an animal result in changes in its microbiota, especially microorganisms associated with the digestive tract (13, 14). In the present case of a shift from molasses-based CMY to starch-based media, one would expect that bacteria secreting amylases would come to the fore. This is consistent with the report that L. plantarum is amylase-positive (15).
Mating preference has previously been achieved in long-term laboratory experimental populations by exposure to different selection pressures. In one of these studies, D. melanogaster populations were maintained for 30 y on media supplemented with either heavy metals or ethanol, and an SII of 0.34 was obtained (16). Part of the mating preference was a result of multiple genetic factors distributed over the chromosomes. The other part was a result of cytoplasmic factors the authors (16) attributed to Wolbachia, because tetracycline treatment reduced the mating preference by approximately 50%. However, the cytoplasmic factor could be any of the tetracycline-sensitive bacteria associated with the flies. In the short-term experiments described here, amplification of L. plantarum was responsible for mating preference, as demonstrated by direct infection experiments. It is also possible that the diverse microbiota in CMY-bred flies compared with starch-bred flies contributed to mating preference.
The data presented here provide further support for the hologenome theory of evolution (17), which posits that the holobiont (host plus its associated microorganisms) acts as a unit of selection in evolutionary change. The hologenome is defined as the sum of the genetic material of the host and its microbiota. One of the principles of the theory is that variation, an important factor in evolution, can be brought about by modification in either the host or the microbiota genomes. In this study, changes in the fly diet led to a rapid amplification of certain bacteria, especially L. plantarum, which was shown to be responsible for mating preference. According to the hologenome theory, microbial amplification is equivalent to gene multiplication and leads to variation in the holobiont. In a review by Andersson and Simmons (18), the authors state: “As experimental evidence accumulated, mate choice became widely recognized, but the genetic mechanisms underlying its evolution remain the subject of debate.” Clearly, a rapid change in host microbiota is one mechanism for mate choice.
The mechanism by which bacteria induce mating preference remains to be elucidated. In principle, a bacterially induced mating signal could be a volatile compound emitted by the fly holobiont or a detectable compound on its surface. The odor of many animals results from microbial modification of compounds secreted by the host (19, 20) or compounds released by the microorganisms themselves (21); moreover, odor is known to influence sexual behavior (22–24). In the case of Drosophila, odorant receptors are located in the antennae and maxillary palps. At least five of the CHs, which have been shown to play a major role in fly mating (23), showed significant differences between starch- and CMY-bred flies (Table 3). These differences were reduced after antibiotic treatment, suggesting that symbiotic bacteria can influence the levels of fly sex pheromones and, by doing so, modify fly behavior.
How can a bacterially induced mating preference, as described here, contribute to speciation and evolution in nature? One possibility is that, in the natural world, multiple environmental factors act synergistically to differentiate the microbiota and strengthen the homogamic mating preference. For example, it is reasonable to assume that fly populations living on different nutrients will be, at least to some extent, geographically separated. The combination of partial geographic separation and diet-induced mating preference would reduce interbreeding of the populations. Slower changes in the host genome could further enhance the mating preference. The stronger the mating preference, the greater the chance that two populations will become sexually isolated, and many evolution biologists (25–28) have argued that the emergence of sexual isolation is the central event in the evolution of species.
Materials and Methods
Drosophila Stocks and Culture Conditions.
A WT strain of D. melanogaster Oregon R was used in all experiments. Each population was maintained in three 50-mL vials, with 10 mL solid medium, at 25 °C on (i) CMY medium (0.65% agar, 7.6% cornmeal, 7.6% molasses, 5% inactivated yeast, 0.1% methyl-4-hydroxybenzoate, 0.76% ethanol, and 0.4% propionic acid) or (ii) starch medium (3% starch, 5% inactivated yeast, 1.5% agar, and 0.5% propionic acid).
Adaptation to Starch Medium.
A population of flies maintained on CMY medium was transferred to a medium that contained one-half concentration CMY medium and one-half concentration of starch medium. After this one-generation adaptation step, the flies were then grown on starch medium for the rest of the experiment. Flies were transferred for one generation on CMY (regardless of their origin) before mating tests were performed.
Antibiotic Treatment.
Flies were transferred into 50-mL vials containing 10 mL CMY medium, supplemented with 50 μg/mL tetracycline, 200 μg/mL rifampicin, 100 μg/mL streptomycin, or a mixture of the three antibiotic agents. Random mating data were obtained when testing antibiotic-treated flies (with each individual antibiotic, as well as the mixture) for mating preference. Controls—namely fly lines that were reared on their respective media without antibiotics—showed homogamic mating preference when tested.
Microbial Infection Experiments.
Bacteria were extracted from the medium upon which the flies were reared for a single generation (either CMY or starch) by adding 10 mL of sterile PBS solution to the used medium (from one growth vial) and mixing vigorously. After allowing the solids to settle, 5 mL of the fluid was collected and centrifuged at low speed (100 × g) for 10 min to further remove solid debris. The resulting supernatant was then centrifuged at high speed (16,000 × g) for 2 min to pellet bacteria. The bacterial pellet was resuspended and washed twice in PBS solution. The final pellet was resuspended in 5 mL of PBS solution.
Flies treated with antibiotics (either CMY-reared or starch-reared) were allowed to propagate on CMY medium that was supplemented with bacteria extracted from CMY medium, starch medium, or with PBS solution (as a control). Multiple-choice mating tests were performed using CMY flies reared on CMY medium supplemented with bacteria extracted from CMY, and starch flies reared on starch medium supplemented with bacteria from starch medium. Similar mating preference tests were performed with CMY flies reared on CMY medium with PBS solution and starch flies reared on CMY medium supplemented with PBS solution as controls.
Multiple Choice Mating Tests.
For mating tests, virgin male and female flies from each treatment were anesthetized with CO2, pooled from all relevant vials, and kept separately under a 12 h light/12 h dark cycle for 4 d on CMY medium. To test for assortative mating, a 24-cell plate was used. Each cell had a 2.5-mm-diameter hole that was capped (for fly introduction) and was lined with a 2 × 2 mm no. 3 filter paper (Whatman) soaked with deionized water. The entire apparatus was sealed with Parafilm M (Pechiney). Wings of flies were clipped for identification of source. Previous studies found no clipping effect (9, 10). To control for any clipping effects, a counterbalanced design was used: each cell had two clipped-wing flies (one male and one female) from the same population and two nonclipped flies (one male and one female) from another population. To transfer flies into the mating chamber without anesthesia, an aspirator was used to transfer them one by one. Males were introduced first, and then females, one after the other (the clipped female first).
Analysis of the Bacterial Community from CMY- and Starch-Reared Flies Using 16S rRNA Gene Clone Libraries.
DNA was extracted from 10 third-generation flies (five males and five females) with the UltraClean Soil DNA kit (MoBio), according to the manufacturer's protocol. Primers 16S8F and 16S1492R (29) were used for amplification of the 16S rRNA genes from extracted flies DNA. 16S rRNA genes were amplified in a 25-μL reaction mixture consisting of 2.5 μL of 10× buffer, 0.5 μL of a 2.5-mM total deoxynucleoside triphosphate mixture, each primer at 5 μM, 10 ng of template DNA, and 1.25 U of Ex Taq DNA polymerase (TaKaRa). Amplification conditions for the PCR included an initial denaturation step of 94 °C for 3 min, followed by 30 cycles of 94 °C for 35 s, 56 °C for 35 s, and 72 °C for 45 s, and a final extension step of 72 °C for 3 min. Reaction products were checked for size and purity on 1% agarose gel.
Amplified DNA from fly samples was ligated into the pGEM-T Easy vector (Promega) according to the protocol of the manufacturer. The ligated vector and insert were transformed into competent Escherichia coli TG1 cells. Each clone was amplified by colony PCR with M13 forward and reverse primers in a 25-μL reaction mixture consisting of 2.5 μL of 10× buffer, 0.5 μL of a 2.5-mM total deoxynucleoside triphosphate mixture, each primer at 5 μM, and 1.25 U of Ex Taq (TaKaRa) or Extensor DNA polymerase (Thermo Fisher Scientific). Amplification conditions for the colony PCR included an initial denaturation step of 95 °C for 4.5 min, followed by 30 cycles of 95 °C for 0.5 min, 59.5 °C for 0.5 min, and 72 °C for 1 min, and a final extension step of 72 °C for 10 min. Reaction products were checked for size and purity on 1% agarose gel. PCR products were cleaned with ExoSAP-IT (USB). DNA sequencing was performed by the chain termination method using an ABI Prism sequencer (model 377, version 2.1.1; Applied Biosystems).
Sequences were aligned with ClustalX (30), and a DNA distance matrix was created with BioEdit. Sequences that had greater than 99% identity were clustered together with DOTUR (31). BLASTN (32) was then used to characterize each sequence cluster.
Lactobacillus Isolation and Infection Experiments.
When L. plantarum had been identified as the major component of the starch fly microbiota, we isolated bacteria from starch flies, grown for one generation on CMY, on Brain-Heart Infusion (BHI) and de Man–Rogosa–Sharpe media (Difco). Flies were homogenized and the homogenate was serially diluted and plated on BHI agar and de Man–Rogosa–Sharpe agar plates. Plates were then incubated at 30 °C for 48 to 72 h. Morphologically different colonies were then picked and streaked on BHI agar plates and incubated at 30 °C for 48 to 72 h. This process was repeated twice to ensure cultures are pure. The different isolates were then identified according to their 16S rRNA genes, obtained by colony-PCR using 16S8F and 16S1492R primers (29). 16S rRNA genes of the isolated strains were amplified in a 25-μL reaction mixture consisting of 2.5 μL of 10× buffer, 0.5 μL of a 2.5-mM total deoxynucleoside triphosphate mixture, each primer at 5 μM, 10 ng of template DNA, and 1.25 U of Ex Taq DNA polymerase (TaKaRa). Amplification conditions for the PCR included an initial denaturation step of 94 °C for 3 min, followed by 30 cycles of 94 °C for 35 s, 56 °C for 35 s, and 72 °C for 45 s and a final extension step of 72 °C for 3 min. Reaction products were checked for size and purity on 1% agarose gel. PCR products were cleaned with ExoSAP-IT (USB). DNA sequencing was performed by the chain termination method in an ABI Prism sequencer (model 377, version 2.1.1; Applied Biosystems). Of the 25 isolates that were screened, four were identified as Lactobacillus spp., two of which were identified by BLASTN (32) as L. plantarum strain IMAU80181 (accession no. GU125601.1).
At the first stage, all Lactobacillus spp. isolates (identified as L. plantarum and Lactobacillus brevis) were used for infection experiments, in which the isolates were grown for 48 h in liquid BHI medium at 30 °C on a rotary shaker (model 4300; New Brunswick Scientific) at 150 rpm. These cultures were then mixed and centrifuged at 16,000 × g. The pellet was resuspended in PBS solution and diluted to approximately 108 cells per mL, which were then applied on the fly medium for infection (as described earlier). A subsequent infection experiment was done in which only one L. plantarum isolate was used to infect antibiotic-treated starch flies for subsequent mating preference tests.
Wolbachia Identification.
To test whether Wolbachia spp. were present in the extracted DNA samples, PCR reactions using specific primers for Wolbachia sp. 16S rRNA and for the Wolbachia surface protein (wsp) genes were performed, using primers and PCR amplification conditions as previously described by Zhou et al. (33). Briefly, 16S rRNA and wsp genes were amplified in a 25-μL reaction mixture consisting of 2.5 μL of 10× buffer, 0.5 μL of a 2.5-mM total deoxynucleoside triphosphate mixture, each primer at 5 mM, 10 ng of template DNA, and 1.25 U of Ex-Taq DNA polymerase (TaKaRa). Amplification conditions for the PCR included an initial denaturation step at 94 °C for 35 min, followed by 30 cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 45 s, and a final extension step at 72 °C for 35 min. Reaction products were checked for size and purity on 1% agarose gels.
Chemical Analysis of CHs.
Starch or CMY flies, reared for one generation on CMY, were collected as pupae from culture vials, separated by sex, and placed for 4 d in empty vials lined with no. 3 Whatmann paper soaked in 4% sucrose. Flies were then transferred to empty glass vials and were kept at −20 °C. For CH extraction, frozen flies were divided into three groups of three flies, each in a glass vial, to which 200 μL of pentane containing 100 ng n-octadecane (C-18), as an internal standard, were added. Vials were then shaken at room temperature for 5 min at 150 rpm. One to two microliters of the extract were injected into the gas chromatography apparatus (CP 3900; Varian). Quantitative analyses of CHs were conducted by using a DB-1 fused silica column that was temperature-programmed from 150 °C (1 min of initial hold) at 5 °C/min to 300 °C. Compound quantification was done by peak integration in comparison with the internal standard. Peaks identity was verified by GC-MS as well as by comparison with the retention time data presented by Everaerts et al. (12).
Supplementary Material
Acknowledgments
We thank the Ayelet Fishman laboratory for help with the GC/MS analyses and Ralph Martinez for useful comments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009906107/-/DCSupplemental.
References
- 1.Mayr E. Animal Species and Evolution. Cambridge, MA: Harvard Univ Press; 1963. pp xiv, 797 pp. [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. pp xiii, 545 pp. [Google Scholar]
- 3.McKinnon JS, et al. Evidence for ecology's role in speciation. Nature. 2004;429:294–298. doi: 10.1038/nature02556. [DOI] [PubMed] [Google Scholar]
- 4.Spieth HT, Ringo JM. Mating behavior and sexual isolation. In: Ashburner M, Carson HL, Thompson JN Jr, editors. The Genetics and Biology of Drosophila. 3c. London: Academic Press; 1983. pp. 224–270. [Google Scholar]
- 5.Rice WR, Hostert EE. Laboratory experiments on speciation: What have we learned in 40 years? Evolution. 1993;47:1637–1653. doi: 10.1111/j.1558-5646.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 6.Kilias G, Alahiotis SN, Pelecanos M. A multifactorial genetic investigation of speciation theory using Drosophila-melanogaster. Evolution. 1980;34:730–737. doi: 10.1111/j.1558-5646.1980.tb04012.x. [DOI] [PubMed] [Google Scholar]
- 7.Dodd DMB. Reproductive isolation as a consequence of adaptive divergence in Drosophila-pseudoobscura. Evolution. 1989;43:1308–1311. doi: 10.1111/j.1558-5646.1989.tb02577.x. [DOI] [PubMed] [Google Scholar]
- 8.de Oliveira AK, Cordeiro AR. Adaptation of Drosophila-Willistoni experimental populations to extreme ph medium. I. Changes in viability and developmental rate. Heredity. 1980;44:111–122. [Google Scholar]
- 9.van den Berg MJ, Thomas G, Hendriks H, van Delden W. A reexamination of the negative assortative mating phenomenon and its underlying mechanism in Drosophila melanogaster. Behav Genet. 1984;14:45–61. doi: 10.1007/BF01066068. [DOI] [PubMed] [Google Scholar]
- 10.Dodd DMB, Powell JR. Founder-flush speciation: An update of experimental results with Drosophila. Evolution. 1985;39:1388–1392. doi: 10.1111/j.1558-5646.1985.tb05704.x. [DOI] [PubMed] [Google Scholar]
- 11.Malogolowkincohen C, Simmons AS, Levene H. A study of sexual isolation between certain strains of Drosophila paulistorum. Evolution. 1965;19:95–103. [Google Scholar]
- 12.Everaerts C, Farine JP, Cobb M, Ferveur JF. Drosophila cuticular hydrocarbons revisited: Mating status alters cuticular profiles. PLoS ONE. 2010;5:e9607. doi: 10.1371/journal.pone.0009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud E, Champailler A, Raimbault M. Degradation of raw starch by a wild amylolytic strain of Lactobacillus plantarum. Appl Environ Microbiol. 1994;60:4319–4323. doi: 10.1128/aem.60.12.4319-4323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koukou K, et al. Influence of antibiotic treatment and Wolbachia curing on sexual isolation among Drosophila melanogaster cage populations. Evolution. 2006;60:87–96. [PubMed] [Google Scholar]
- 17.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 18.Andersson M, Simmons LW. Sexual selection and mate choice. Trends Ecol Evol. 2006;21:296–302. doi: 10.1016/j.tree.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Hunt DWA, Borden JH. Terpene alcohol pheromone production by dendroctonus-ponderosae and Ips-paraconfusus (Coleoptera, Scolytidae) in the absence of readily culturable microorganisms. J Chem Ecol. 1989;15:1433–1463. doi: 10.1007/BF01012375. [DOI] [PubMed] [Google Scholar]
- 20.Leyden JJ, McGinley KJ, Hölzle E, Labows JN, Kligman AM. The microbiology of the human axilla and its relationship to axillary odor. J Invest Dermatol. 1981;77:413–416. doi: 10.1111/1523-1747.ep12494624. [DOI] [PubMed] [Google Scholar]
- 21.Persson S, Edlund MB, Claesson R, Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol. 1990;5:195–201. doi: 10.1111/j.1399-302x.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 22.Dickson BJ. Wired for sex: The neurobiology of Drosophila mating decisions. Science. 2008;322:904–909. doi: 10.1126/science.1159276. [DOI] [PubMed] [Google Scholar]
- 23.Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- 24.Benton R. Sensitivity and specificity in Drosophila pheromone perception. Trends Neurosci. 2007;30:512–519. doi: 10.1016/j.tins.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Coyne JA. Genetics and speciation. Nature. 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 26.Mayr E. Systematics and the Origin of Species from the Viewpoint of a Zoologist. New York: Columbia Univ Press; 1942. [Google Scholar]
- 27.Dobzhansky TG. Genetics and the Origin of Species. New York: Columbia Univ. Press; 1937. [Google Scholar]
- 28.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 29.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhou WG, Rousset F, O'Neil S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.