Enzymes that use energy gained by ATP-hydrolysis to alter nucleosomes, the building blocks of chromatin, are involved in all processes occurring on DNA (1, 2). These ATP-dependent chromatin remodeling factors regulate access to DNA either by moving nucleosomes away from a transcription factor binding site or into such a site, occluding further access (1, 2). All known ATP-dependent nucleosome remodeling factors contain a protein with a highly conserved SWI/SNF-type ATPase core domain. This ATPase is usually imbedded in a complex with other subunits that regulate its function. Despite the importance of these enzymes, we know little of how they operate in the living cell. In a paper published in PNAS, Rippe and coworkers (3) present a study on the mobility of ATP-dependent chromatin remodeling factors in living cells and propose a mechanism by which these factors rapidly identify target sites in the nucleus.
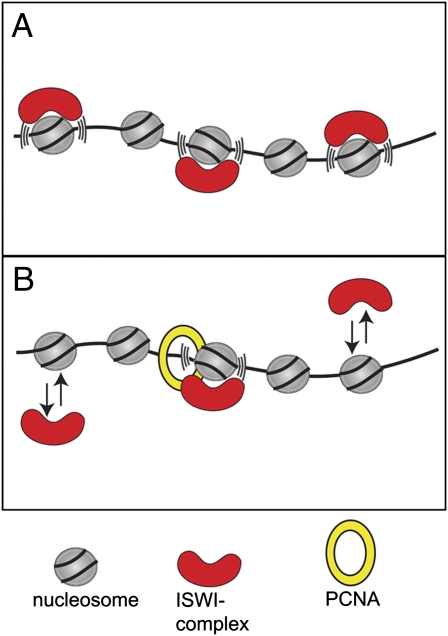
Chromatin remodeling factors are surprisingly abundant, given that they are enzymes, wherein one molecule can usually successively remodel multiple nucleosomes (4, 5). Observations with chromatin that was assembled in the test tube from Drosophila embryo extracts have led to the hypothesis that nucleosomes might be constantly remodeled in the cell (e.g., shuffled back and forth) to allow access to the underlying DNA for incoming factors (6–8) (Fig. 1A). Rippe and coworkers (3) present data that lead to an alternative model called a “continuous sampling mechanism,” wherein an abundance of remodeling complexes sample nucleosomes constantly but transiently without causing remodeling. Only after recognition of a specific cue (e.g., targeting molecule, posttranslational histone modification) is a stable interaction with chromatin formed, allowing processive chromatin remodeling. Once a nucleosome is highlighted to be remodeled, it will be rapidly bound because of the abundance of remodeling factors (Fig. 1B). Therefore, the combination of a large concentration of remodelers with transient binding reactions allows for rapid sampling of the entire genome and a fast but tightly regulated response (within seconds to minutes) on activation by special triggers.
Fig. 1.
Two possible modes by which ATP-dependent nucleosome remodeling factors interact with chromatin. (A) These factors may engage with nucleosome most of the time, continuously shoving them back and forth, and because they are also abundant, they would create a highly “transparent” and plastic chromatin, allowing rapid access to incoming factors. (B) These factors may interact very transiently with nucleosomes and engage productively in nucleosome remodeling only once their interactions are stabilized [e.g., by binding to proliferating cell nuclear antigen (PCNA) at chromatin replication or DNA repair sites].
Rippe and coworkers (3) focus on the Imitation Switch (ISWI) class of SWI/SNF factors, namely, the highly similar proteins SNF2H and SNF2L and ACF1, which binds SNF2H in the ATP-utilizing chromatin assembly and remodeling factor (ACF) complex. SNF2H and SNF2L form multiple complexes even in the same cell type with diverse biological roles, including gene activation and repression, DNA replication, and repair (9–11). ISWI complexes move nucleosomes along the DNA without major disruption of the nucleosomes (reviewed in 1). The ACF complex is involved in creating regular nucleosomal arrays (referred to as nucleosome spacing) (reviewed in 1). The authors use imaging of fluorescently tagged chromatin remodelers to study their dynamics in living human and mouse cells in culture. They use fluorescence fluctuation microscopy techniques, such as fluorescence recovery after photobleaching, that give insights into the mobility and interactions of proteins in living cells.
Rippe and coworkers (3) provide several pieces of evidence in favor of the continuous sampling mechanism. They determine the concentrations of the ISWI factors in the nucleus to be roughly one SNF2H molecule for every 140 nucleosomes. Extrapolating to the total concentration of all known SWI/SNF-type remodeling complexes, they come up with a value of one remodeler per ∼14 nucleosomes, confirming the previous notion that remodeling factors are fairly abundant molecules in the cell. They then demonstrate that SNF2H and SNF2L are relatively immobile at DNA replication and repair foci (residence times of several seconds). This observation is consistent with studies that report roles for ISWI complexes in chromatin replication and DNA repair (11–14). In G1 or G2, the majority (>95%) of the remodeler molecules were shown to be highly mobile (millisecond residence time) with diffusion rates not much different from those found in the cytoplasm. Such a time scale seems incompatible with the time required for nucleosome remodeling by ACF, as measured by single-molecule analysis (15). A SNF2L form lacking an active ATP-binding site exhibited only slightly increased mobility, suggesting that only a small fraction of the remodelers is actively involved in chromatin remodeling during G1 or G2. This conclusion was corroborated by experiments in which ATP was depleted in the cell by addition of azide. Together, these data are consistent with the scenario that the ISWI complexes transiently interact with chromatin unless a specific feature, such as found in replication foci, causes these factors to engage with the chromatin more permanently.
The proposed continuous sampling mechanism puts special importance on the “trigger” that recruits chromatin remodeling enzymes. At replication and repair foci, this trigger is, at least in part, the sliding clamp proliferating cell nuclear antigen, a key molecule of DNA replication and repair with which SNF2H interacts (13). Histone modifications also play a crucial role in targeting and regulating nucleosome remodeling factors (16). For example, histone H3 lysine 4 trimethylation is required to stabilize the interaction of the ISWI-containing nuclesome remodeling factor complex with chromatin (17). Interaction with sequence-specific transcription factors is also key for targeting chromatin remodeling factors to specific sites, including ISWI complexes (9).
It will be important to test if the reported observations can be extended to other SWI/SNF-type remodeling factors. Although there are common aspects in their mechanism, their interaction with and action on nucleosomes differ significantly in detail (1). Furthermore, it will be exciting to test the modes of function of chromatin remodeling factors in various developmental states, especially stem cells. These cells are characterized by a highly dynamic plastic chromatin (18). This dynamic chromatin could, at least in part, be possible through the promiscuous actions of ATP-dependent chromatin remodeling factors.
The work of Rippe and coworkers (3) highlights the important contribution that in vivo imaging approaches can provide to our understanding of mechanisms that shape chromatin. Constant technological improvements and breakthroughs, such as superresolution microscopy, are going to make this type of approach an incredibly powerful way to reveal processes in the cell.
Acknowledgments
Work in my laboratory is funded by the Biotechnology and Biological Sciences Research Council (BBSRC), Medical Research Council, and European Union Epigenome Network of Excellence.
Footnotes
The author declares no conflict of interest.
See companion article on page 19873.
References
- 1.Becker PB, Hörz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 2.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 3.Erdel F, Schubert T, Marth C, Längst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci USA. 2010;107:19873–19878. doi: 10.1073/pnas.1003438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fyodorov DV, Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- 5.Varga-Weisz PD, Becker PB. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev. 2006;16:151–156. doi: 10.1016/j.gde.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Becker PB. The chromatin accessibility complex: Chromatin dynamics through nucleosome sliding. Cold Spring Harb Symp Quant Biol. 2004;69:281–287. doi: 10.1101/sqb.2004.69.281. [DOI] [PubMed] [Google Scholar]
- 7.Varga-Weisz PD, Blank TA, Becker PB. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varga-Weisz PD, et al. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 9.Dirscherl SS, Krebs JE. Functional diversity of ISWI complexes. Biochem Cell Biol. 2004;82:482–489. doi: 10.1139/o04-044. [DOI] [PubMed] [Google Scholar]
- 10.Corona DF, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta. 2004;1677:113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimura K, et al. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci USA. 2009;106:9280–9285. doi: 10.1073/pnas.0901184106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Collins N, et al. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- 13.Poot RA, et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 14.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan C, Varga-Weisz P. The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res. 2007;618:41–51. doi: 10.1016/j.mrfmmm.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 18.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]