Abstract
Curcuminoid synthase (CUS) from Oryza sativa is a plant-specific type III polyketide synthase (PKS) that catalyzes the remarkable one-pot formation of the C6-C7-C6 diarylheptanoid scaffold of bisdemethoxycurcumin, by the condensation of two molecules of 4-coumaroyl-CoA and one molecule of malonyl-CoA. The crystal structure of O. sativa CUS was solved at 2.5-Å resolution, which revealed a unique, downward expanding active-site architecture, previously unidentified in the known type III PKSs. The large active-site cavity is long enough to accommodate the two C6-C3 coumaroyl units and one malonyl unit. Furthermore, the crystal structure indicated the presence of a putative nucleophilic water molecule, which forms hydrogen bond networks with Ser351-Asn142-H2O-Tyr207-Glu202, neighboring the catalytic Cys174 at the active-site center. These observations suggest that CUS employs unique catalytic machinery for the one-pot formation of the C6-C7-C6 scaffold. Thus, CUS utilizes the nucleophilic water to terminate the initial polyketide chain elongation at the diketide stage. Thioester bond cleavage of the enzyme-bound intermediate generates 4-coumaroyldiketide acid, which is then kept within the downward expanding pocket for subsequent decarboxylative condensation with the second 4-coumaroyl-CoA starter, to produce bisdemethoxycurcumin. The structure-based site-directed mutants, M265L and G274F, altered the substrate and product specificities to accept 4-hydroxyphenylpropionyl-CoA as the starter to produce tetrahydrobisdemethoxycurcumin. These findings not only provide a structural basis for the catalytic machinery of CUS but also suggest further strategies toward expanding the biosynthetic repertoire of the type III PKS enzymes.
Keywords: biosynthesis, enzyme, curcumin
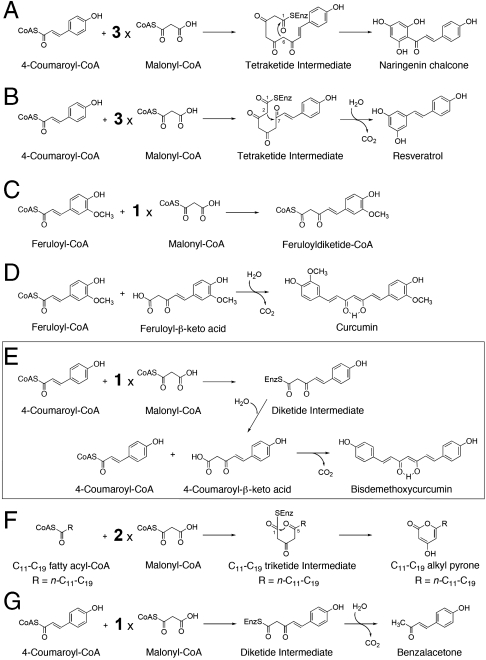
Type III polyketide synthases (PKSs) are structurally simple, homodimeric proteins that use a single active site to produce pharmaceutically and biologically important, structurally divergent natural polyketide scaffolds (1–3). For example, chalcone synthase (CHS) and stilebene synthase (STS) catalyze the sequential condensation of 4-coumaroyl-CoA as a starter substrate with three C2 units from malonyl-CoA, to produce the C6-C3-C6 scaffold of naringenin chalcone, the key intermediate in flavonoid biosynthesis, and the C6-C2-C6 scaffold of resveratrol, respectively (Fig. 1 A and B). On the other hand, it was recently reported that the C6-C7-C6 diarylheptanoid scaffold of curcumin is also produced by novel type III PKSs (Fig. 1 C and D) (4). Curcumin, the principal component of the turmeric Curcuma longa, is widely used as a food additive and in traditional Asian medicine and has been shown to possess anti-inflammatory, anticarcinogenic, and antitumor activities (5–7). Thus, in C. longa, diketide-CoA synthase (DCS) and curcumin synthase (CURS) are two key enzymes in the biosynthesis of curcumin; DCS catalyzes the condensation of feruloyl-CoA with malonyl-CoA to produce feruloyldiketide-CoA (Fig. 1C), and CURS then accepts the C6-C5 feruloyldiketide acid, the hydrolyzed product of feruloyldiketide-CoA, and catalyzes decarboxylative condensation with the second feruloyl-CoA starter to produce the C6-C7-C6 diarylheptanoid scaffold of curcumin (Fig. 1D) (4). In contrast, very interestingly, curcuminoid synthase (CUS) from the rice plant Oryza sativa is another novel type III PKS that catalyzes the “one-pot” formation of the C6-C7-C6 scaffold of bisdemethoxycurcumin (Fig. 1E). Although its physiological function remains to be elucidated, it is remarkable that a single enzyme catalyzes the condensation of two molecules of 4-coumaroyl-CoA and one molecule of malonyl-CoA through the formation of the C6-C5 4-coumaroyldiketide acid (8).
Fig. 1.
Proposed mechanisms for the formation of polyketides. (A) Naringenin chalcone from 4-coumaroyl-CoA and three molecules of malonyl-CoA by CHS, (B) resveratrol from 4-coumaroyl-CoA and three molecules of malonyl-CoA by STS, (C) feruloyldiketide-CoA from feruloyl-CoA and one molecule of malonyl-CoA by DCS, (D) curcumin from feruloyl-CoA and feruloyldiketide acid by CURS1, (E) bisdemethoxycurcumin from two molecules of 4-coumaroyl-CoA and one molecule of malonyl-CoA by CUS, (F) triketide alkyl pyrones from C11-C19 fatty acyl-CoA and two molecules of malonyl-CoA by PKS18, and (G) benzalacetone from 4-coumaroyl-CoA and one molecule of malonyl-CoA by BAS.
Recent crystallographic and site-directed mutagenesis studies on plant and bacterial type III PKSs have revealed that the functional diversity of the type III PKSs is principally derived from modifications of the active-site cavity (1, 2). The amino acid sequence of O. sativa CUS shares 40–51% identity with other plant type III PKSs; 49% identity with M. sativa CHS, 51% identity with C. longa DCS, and 45% identity with C. longa CURS (Fig. S1). Sequence comparisons revealed that the Cys-His-Asn catalytic triad and most of the characteristic active-site residues of the CHSs are well conserved in CUS. However, Thr132, Thr197, Gly256, and Phe265, lining the active-site cavity of M. sativa CHS, are characteristically substituted with Asn, Tyr, Met, and Gly, respectively, which may account for the catalytic activity of CUS. The first three of these residues are altered in a number of functionally divergent type III PKSs and play crucial roles in controlling the substrate and product specificities of the type III PKS enzyme reactions (1, 2). Indeed, recent structural analyses of Pinus sylvestris STS have demonstrated that the backbone change of the loop leads to the subtle displacement of Thr132, resulting in the rearrangement of the hydrogen bond networks including Thr132 and a nucleophilic water molecule at the active-site center (9). On the other hand, the “gatekeeper” Phe265 is located at the junction between the active-site cavity and the CoA binding tunnel in M. sativa CHS and is well conserved in a number of type III PKSs (Fig. S1). This residue is thought to control the substrate entry into the active site by forming part of the wall in the active-site cavity (10).
To clarify the three-dimensional structural details of the CUS-catalyzed enzyme reaction and to elucidate the structure-function relationship of the type III PKS enzymes, we now present the crystal structure of CUS at 2.5-Å resolution. The crystal structure revealed a unique, downward expanding active-site architecture, previously unidentified in the known type III PKSs (1, 2), and indicated the presence of a putative nucleophilic water molecule that forms hydrogen bond networks at the active-site center. These findings have led to the proposal that CUS utilizes the nucleophillic water to terminate the polyketide chain elongation at the diketide stage, and that the decarboxylative condensation of the second coumaroyl starter with the 4-coumaroyldiketide acid intermediate proceeds within the active-site cavity to produce the C6-C7-C6 diarylheptanoid scaffold. In addition, structure-based mutagenesis studies clearly identified the crucial residues for controlling the substrate and product specificities of the enzyme.
Results and Discussion
Overall Structure of CUS.
The CUS crystal structure was solved by the molecular replacement method and refined to 2.5-Å resolution. The asymmetric unit contained four nearly identical monomers (A, B, C, and D), with root-mean-square deviations (rmsds) of 0.36–0.46 Å for the Cα-atoms, where A/B and C/D form two sets of biologically active, symmetric dimers. Significant backbone changes were not observed between the monomers. The overall structure of CUS recapitulated the thiolase fold observed in other type III PKSs (Fig. S2A). Upon dimerization, each A/B and C/D dimer buries 2,330 Å2 and 2,230 Å2 of the surface, respectively, and Val147, corresponding to Met137 of M. sativa CHS, protrudes into the other monomer by the formation of a cis-peptide bond between Val147 and Pro148 on a loop. However, unlike many of the typical type III PKSs, Val147 is not involved in the formation of part of the wall of the active site, as observed in the bacterial type III PKSs, such as Mycobacterium tuberculosis alkylpyrone synthase PKS18 (Fig. 1F) (11) and Neurospora crassa 2′-oxoalkylresorcylic acid synthase (ORAS) (12). The catalytic triad of Cys174, His316, and Asn349 is buried deep within each monomer and sits at the intersection of a traditional 16-Å-long CoA binding tunnel and a large internal cavity, in a location and orientation very similar to those of the other plant type III PKSs. A structure-based similarity search using the Dali program (13) suggested that the overall structure of CUS is highly homologous to those of the previously reported plant type III PKSs (rmsds 1.0–1.2 Å and 2.0–2.2 Å for plant and bacterial type III PKSs, respectively), in which the most closely related structural homologue is that of Rheum palmatum benzalacetone synthase (BAS) (PDB entry 3A5Q, Z score = 57.2, rmsd of 1.0 Å over 360 residues aligned with a sequence identity of 53%) (14). R. palmatum BAS produces the diketide benzalacetone by the one-step condensation of 4-coumaroyl-CoA and malonyl-CoA through the formation of 4-coumaroyldiketide acid (Fig. 1G).
Active-Site Architecture of CUS.
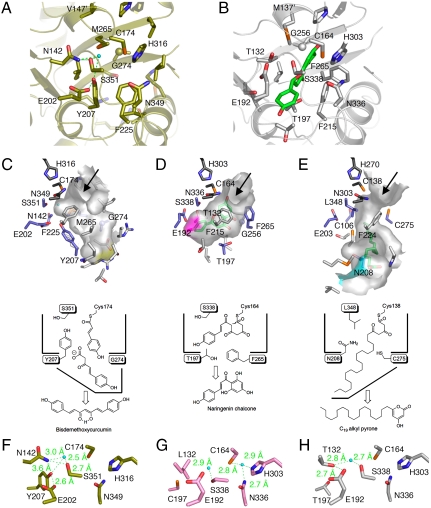
A comparison of the active-site structure of O. sativa CUS with that of M. sativa CHS (10) revealed that in the crystal structure of CUS, the backbone torsion angle of Asn142(-92,-23), as compared with that of Thr132(-112,6) of M. sativa CHS, is slightly shifted by a ϕ angle of -20° and a ψ angle of +29°, and the amino moiety of Asn142 forms a hydrogen bond with Ser351, corresponding to Ser338 of M. sativa CHS (Fig. 2 A and B). Furthermore, the backbone torsion angle of Tyr207(-92,-40), as compared with that of Thr197(-111,-22) of M. sativa CHS, is shifted by a ϕ angle of -19° and a ψ angle of +18°, with a slight displacement of its Cα-atom, and the side chain of Tyr207 protrudes toward Ser351. The conformational differences of Asn142 and Tyr207 cause the loss of the so-called CHS coumaroyl binding pocket that thought to bind the aromatic moiety of the tetraketide intermediate (10) from the active-site cavity of CUS (Fig. 2 C and D). The coumaroyl binding pocket is also absent in the active-site cavity of R. palmatum BAS, but in this case, the contraction is caused by the simultaneous substitution of the CHS’s conserved Thr132 and Phe215 with Leu, along with the conformational change of Ser338, and as a result, the enzyme catalyzes only one condensation with malonyl-CoA to produce the diketide benzalacetone (14).
Fig. 2.
Comparison of the active-site structures of O. sativa CUS and other type III PKSs. (A and B) Comparison of the active-site structures of (A) O. sativa CUS and (B) M. sativa CHS. The naringenin molecule is shown as a green stick model. The water molecule and the hydrogen bonds are indicated with a light-blue sphere and green dotted lines, respectively. (C–E) Surface and schematic representations of (C) O. sativa CUS, (D) M. sativa CHS, and (E) M. tuberculosis PKS18. The bottom of the long pocket in CUS is indicated as a yellow surface. The bottom of the coumaroyl binding pocket and part of the side of the long-chain binding tunnel are highlighted as purple and light blue surfaces, respectively. The naringen and myristic acid bound to the active-site cavities of M. sativa CHS and M. tuberculosis PKS18, respectively, are shown as green stick models. (F–H) Close-up views of the electronic hydrogen bond networks of (F) O. sativa CUS, (G) R. palmatum BAS, and (H) P. sylvestris STS. The water molecules and the hydrogen bonds are indicated with a light-blue sphere and green dotted lines, respectively.
As mentioned above, Val147, corresponding to Met137 of M. sativa CHS, protrudes into the other monomer by the formation of a cis-peptide bond between Val147 and Pro148 on a loop. The highly conserved Met137 in the plant type III PKSs (Leu in P. sylvestris STS) usually participates in forming part of the wall of the active site. However, in O. sativa CUS, Met265, corresponding to Gly256 of M. sativa CHS, blocks the cross-subunit interaction of Val147′ by placing the side chain of Met265 in front of Val147′ and precludes Val147′ from participating in the formation of the CUS active-site cavity (Fig. 2 A and B). In addition, the side chain of Met265 protrudes toward the catalytic Cys174 with a slight positional change, thereby contributing to the altered shape and size of the active-site cavity (Fig. 2C).
Furthermore, a structural comparison revealed significant backbone changes in residues 272–280 in CUS and residues 263–271 in M. sativa CHS (rmsds of 1.7 Å for the Cα-atoms, Fig. S2B). These conformational changes are caused by various neighboring amino acid substitutions and especially by the replacement of the gatekeeper Phe265 of M. sativa CHS with the smaller Gly274 residue in CUS. As a result, the Cα-atom of Gly274 in CUS, as compared with that of Phe265 in M. sativa CHS, is displaced toward the protein surface at a 3.1-Å distance. The large-to-small replacement in CUS not only widens the active-site entrance further but also opens a gate to a downward expanding, 11-Å long pocket (Fig. 2C and Fig. S2C). It is remarkable that the location and the orientation of the downward expanding pocket are significantly different from those of the CHS’s coumaroyl binding pocket (Fig. 2D) or those of the long-chain binding tunnel of the bacterial type III PKSs, such as M. tuberculosis PKS18 (Fig. 2E), that accommodate the long-chain fatty acyl moieties (C12-C20) of substrates and intermediates (11, 12, 15, 16). The estimated total cavity volume is 642 Å3, which is 1.9 times larger than that of the C6-C4 diketide-producing R. palmatum BAS (345 Å3) and almost as large as that of the C6-C3-C6 chalcone-producing M. sativa CHS (754 Å3). Manual docking studies predicted that the expanded active-site cavity of CUS is long enough to accommodate the two C6-C3 coumaroyl units and one malonyl unit, to produce the C6-C7-C6 scaffold of bisdemethoxycurucumin (Fig. S3). Presumably, CUS utilizes the unique downward expanding pocket to accommodate the coumaroyldiketide acid intermediate for subsequent decarboxylative condensation with the second 4-coumaroyl-CoA starter molecule to produce bisdemethoxycurcumin (Fig. 1E). In contrast, in the case of R. palmatum BAS, the coumaroyldiketide acid produced by the thioester bond cleavage of the enzyme-bound diketide intermediate subsequently undergoes decarboxylation to yield the C6-C4 benzalacetone (Fig. 1G). It should be noted that neither the diketide acid nor benzalacetone is detected in the CUS enzyme reaction (8).
Thioesterase-Like Electronic Hydrogen Bond Network in CUS.
One of the crucial points for the catalytic mechanism of CUS is the termination of the polyketide chain elongation at the diketide stage and the generation of the coumaroyldiketide acid intermediate for the second condensation reaction. It is possible that the diketide acid is produced by nonenzymatic, spontaneous hydrolysis of the 4-coumaroyldiketide-CoA, once it is formed and released from the enzyme. However, another possibility is the enzymatic formation of the diketide acid by the thioester bond cleavage of the enzyme-bound intermediate, as in the case of R. palmatum BAS (14). Our previous crystallographic studies on R. palmatum BAS revealed that BAS utilizes a nucleophilic water molecule, which forms hydrogen bond networks with the Cys-His-Asn catalytic triad, for the thioester bond cleavage of the diketide intermediate bound to the catalytic Cys164, to produce the coumaroyldiketide acid intermediate (Figs. 1G and 2G) (14). On the other hand, crystallographic studies of P. sylvestris STS revealed the so-called “aldol-switch” hydrogen bond network, including Thr132 and a nucleophilic water molecule neighboring the catalytic Cys164 at the active-site center of STS (numbering in M. sativa CHS) (Figs. 1B and 2H) (9). Austin et al. thus proposed that the nucleophilic water activated by the hydrogen bond network cleaves the C-S bond of the enzyme-bound, linear tetraketide intermediate, to produce the stilbene scaffold (9).
Interestingly, the CUS crystal structure revealed the presence of a putative nucleophilic water molecule forming hydrogen bonds with Asn142 and Ser351 (Fig. 2F and Fig. S2D), similar to that of the aldol switch in STS (Fig. 2H). Furthermore, the crystal structure demonstrated that the side chain of Glu202, behind Tyr207, is isometric to those of the corresponding Glu192 residues in M. sativa CHS, R. palmatum BAS, and P. sylvestris STS (Fig. 2). Remarkably, in the CUS structure, the Cγ carbon of Glu202 rotates by nearly 114°, thereby forming a hydrogen bond between the carboxyl group of Glu202 and the hydroxyl group of Tyr207. As a result, the water molecule forms hydrogen bond networks with Ser351-Asn142-H2O-Tyr207-Glu202, neighboring the catalytic Cys174 at the active-site center. We propose that this unique steric arrangement in the active-site of CUS can be regarded as a configurational analogue of the aldol-switch in P. sylvestris STS (9). In contrast, the hydrogen bond networks of CUS were not conserved in similar positions in the crystal structure of R. palmatum BAS (14), which would account for the functional differences between CUS and BAS. Moreover, the previously reported crystal structure of R. palmatum BAS revealed that the coumaroyl monoketide intermediate covalently bound to the catalytic Cys164 occupies either the CoA-binding tunnel or the active-site cavity, depending on the conformation of the gatekeeper Phe265 (14), which is substituted with Gly274 in O. sativa CUS. The large-to-small substitution with Gly274 in O. sativa CUS would thus not only widen the active-site entrance but also facilitate the movement of the enzyme-bound second coumarate for subsequent decarboxylative condensation with the C6-C5 diketide acid intermediate to produce the C6-C7-C6 bisdemethoxycurucumin (Fig. 3E and Fig. S3).
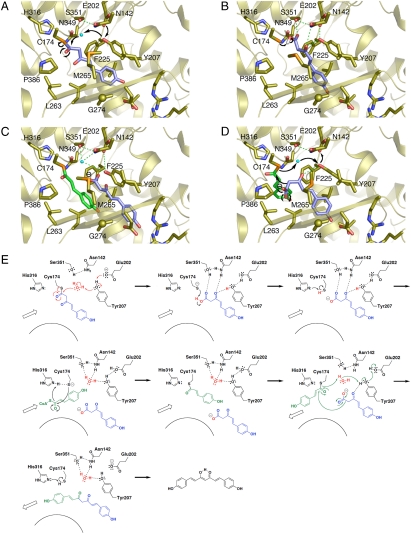
Fig. 3.
Proposed mechanism for the CUS enzyme reaction. (A–D) Three-dimensional model of (A) hydrolysis of the diketide intermediate covalently bound to the catalytic Cys174, (B) conversion of the carboxylic intermediate into the carboxylate, (C) loading of the second coumaroyl-CoA into the catalytic Cys174, and (D) coupling reaction between the second monoketide and the extender intermediates. (E) Schematic representation of the proposed mechanism.
On the other hand, C. longa CURS catalyzes formation of the C6-C7-C6 curcumin by decarboxylative condensation of the C6-C5 feruloyldiketide acid with feruloyl-CoA (Fig. 1D) (4). Interestingly, sequence comparison revealed that Ser351, Asn142, and Tyr207, the three residues involved in the hydrogen bond networks in O. sativa CUS, are altered with Gln, Ile, and Ser, in C. longa CURS, respectively (Fig. S1). Further, Gly265 in CUS is substituted with the gatekeeper Phe in C. longa CURS (Fig. S1). These differences indicate that the active-site architecture including the hydrogen bond networks observed in CUS is not conserved in the active-site of CURS and suggest that CUS and CURS utilize different catalytic mechanism to generate the C6-C7-C6 diarylheptanoid scaffold.
Proposed Mechanism of the CUS Enzyme Reaction.
On the basis of these findings, we propose that O. sativa CUS employs unique catalytic machinery for the remarkable one-pot formation of the C6-C7-C6 diarylheptanoid scaffold of bisdemethoxycurucumin (Fig. 3). Thus, CUS utilizes the nucleophilic water activated by the hydrogen bond networks to terminate the initial polyketide chain elongation at the diketide stage by the cleavage of the thioester bond of the enzyme-bound coumaroyl diketide intermediate to generate 4-coumaroyldiketide acid (Fig. 3 A and E). Asn142 and Tyr207 then orient the α,β-unsaturated carbonyl of the diketide acid, and Ser351 hydrogen bonds with the acid carbonyl, which is then converted into a carboxylate anion through proton abstraction by the thiolate anion of the catalytic Cys174 (Fig. 3 B and E). The second 4-coumaroyl-CoA starter is subsequently loaded into the active site at the catalytic center Cys174, which is reactivated by His316, resulting in the accommodation of the diketide intermediate into the downward expanding pocket of the active-site cavity (Fig. 3 C and E). Reorientation of the enzyme-bound coumaroyl monoketide then follows, to facilitate the final “tail-to-tail” coupling with the coumaroyl diketide acid (Fig. 3 D and E). Here, the nonpolar environment for the carboxylate intermediates, presumably provided by Tyr207, Leu263, and Pro386, assists the decarboxylative condensation reaction to produce and release the C6-C7-C6 bisdemethoxycurcumin, with restoration of the hydrogen bond networks at the active site (Fig. 3 D and E).
Structure-Based Mutagenesis of CUS.
To test the hypothesis that Tyr207 and Gly274 play a crucial role for the formation of bisdemethoxycurcumin, and to further clarify the intimate structural details of the enzyme reaction, we constructed Y207F and G274F mutants of O. sativa CUS and investigated the effects of the mutagenesis on the enzyme activities. These mutations were designed to disrupt the hydrogen bond networks and to restore the gatekeeper Phe, as in the other plant type III PKSs, respectively. We also constructed the M265L mutant, in which Met265, corresponding to Gly256 of M. sativa CHS, which is thought to be important for controlling the substrate and product specificities of the enzyme reactions, is replaced with the bulkier Leu, as in the functionally divergent plant type III PKSs (1, 2). Considering the absence of the typical gatekeeper at position 274 in CUS, Met265 was thought to be crucial for the enzyme reaction. All the mutant proteins were expressed in Escherichia coli at levels comparable with wild-type enzyme and purified to homogeneity. Comparison of the circular dichroism (CD) spectra revealed that they retained the native conformation of the wild-type enzyme (Fig. S4).
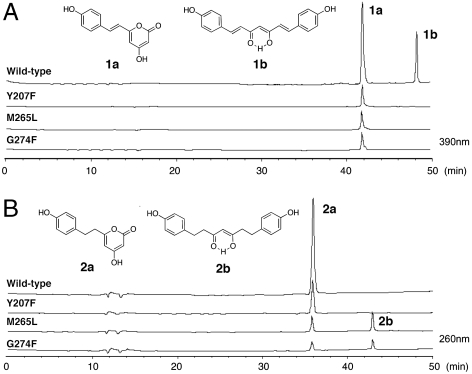
When 4-coumaroyl-CoA and malonyl-CoA are used as the substrates, the wild-type CUS produces bisdemethoxycurcumin (1b in Fig. 4A, 8.9% yield), along with bisnoryangonin (BNY) (1a in Fig. 4A, 15% yield), the triketide pyrone formed by condensation of 4-coumaroyl-CoA with two molecules of malonyl-CoA. Consistent with our hypothesis, the removal of the hydroxyl group from Tyr207 by the Y207F mutation, and the small-to-large substitutions of G274F and M265L, resulted in the complete loss of the bisdemethoxycurcumin-forming activity and only afforded BNY (1.8% and 2.1% yields, respectively, Fig. 4A). These results supported that the three residues, Tyr207, Met265, and Gly274, indeed play important roles in the formation of the C6-C7-C6 scaffold of bisdemethoxycurcumin. Further, it was remarkable that the G274F and M265L point mutants altered the substrate specificity for the aromatic substrate, and accepted 4-hydroxyphenylpropionyl-CoA (= dihydro-4-coumaroyl-CoA) as a starter substrate to produce tetrahydrobisdemethoxycurcumin (2b in Fig. 4B, 0.2% and 1.2% yields, respectively), along with the triketide pyrone dihydro-BNY (2a in Fig. 4B, 1.5% and 8% yields, respectively). This is a previously undescribed demonstration of the enzymatic formation of the C6-C7-C6 tetrahydrodiarylheptanoid scaffold. In contrast, the wild-type CUS and the Y207F mutant did not accept 4-hydroxyphenylpropionyl-CoA to produce the curcuminoid but only yielded the BNY-type triketide pyrone (35% and 5.2% yields, Fig. 4B). On the other hand, when benzoyl-CoA and phenylacetyl-CoA were incubated with the wild-type and mutant enzymes as starter substrates, no curcuminoid products were identified. As mentioned above, one of the critical steps for the CUS enzyme reaction is the termination of the polyketide chain elongation at the diketide stage and the formation and accommodation of the diketide intermediate for the second round condensation reaction (Fig. 1E). Presumably, because of the conformational flexibility of the 4-hydroxyphenylpropionyl-CoA molecule with the reduced double-bond, the polyketide elongation reaction is not effectively terminated at the diketide stage but further proceeds to yield the triketide pyrone, instead of the formation of the C6-C7-C6 tetrahydrodiarylheptanoid as a major product.
Fig. 4.
HPLC elution profiles of enzyme reaction products of the wild-type CUS and its Y207F, M265L, and G274F mutant enzymes. The enzyme reaction products from (A) 4-coumaroyl-CoA and malonyl-CoA and from (B) 4-hydroxyphenylpropionyl-CoA and malonyl-CoA as the substrates.
Finally, homology modeling of the CUS Y207F and M265L mutant predicted that the Y207F substitution only disrupts the hydrogen bond networks at the active site (Fig. S5B), whereas the M265L replacement narrows the gate of the downward expanding pocket with constriction of the active-site wall near the entrance but maintains the pocket and the hydrogen bond networks in the active-site cavity (Fig. S5C). These observations strongly suggest that the hydroxyl group of Tyr207 is indeed involved in the Ser351-Asn142-H2O-Tyr207-Glu202 hydrogen bond networks to activate the water molecule for the bisdemethoxycurcumin production, and that Met265 controls the substrate and product specificities of the enzyme reactions by the steric modulation of the active-site cavity. On the other hand, a homology model of the G274F mutant revealed that the small-to-large substitution further narrows the gate of the pocket, as compared with that of the M265L substitution (Fig. S5D). These observations suggest that the unique, downward expanding pocket indeed plays a crucial role, and that the more flexible 4-hydroxyphenylpropionyl-CoA starter substrate, instead of 4-coumaroyl-CoA, was accepted in the constricted active-site of the mutants to produce the C6-C7-C6 tetrahydrodiarylheptanoid.
In conclusion, the crystal structure of O. sativa CUS revealed a unique, downward expanding active-site architecture, previously unidentified in the known type III PKSs, and the presence of a putative nucleophilic water molecule, which forms hydrogen bond networks at the active-site center. These observations suggest that CUS utilizes the nucleophilic water to terminate the initial polyketide chain elongation at the diketide stage. Thioester bond cleavage of the enzyme-bound intermediate generates the coumaroyldiketide acid, which is then kept within the downward expanding pocket for subsequent decarboxylative condensation with the second 4-coumaroyl-CoA, to produce the C6-C7-C6 diarylheptanoid scaffold. Furthermore, the structure-based site-directed mutants, M265L and G274F, altered the substrate and product specificities to accept a 4-hydroxyphenylpropionyl-CoA starter to produce tetrahydrobisdemethoxycurcumin. These findings provide insights into not only a structural basis for the catalytic machinery of CUS but also for further strategies toward expanding the biosynthetic repertoire of the type III PKS enzymes.
Methods
Chemicals.
4-Coumaroyl-CoA and 4-hydroxyphenylpropionyl-CoA were synthesized as described previously (14). Tetrahydrobisdemethoxycurcumin was synthesized according to the published method (17). The spectroscopic data (NMR, UV, and MS) were completely identical to the published data. Dihydro-BNY was enzymatically synthesized according to the published method (18), using Scutellaria baicalensis CHS (19). Oligonucleotides were obtained from Invitrogen. Bisdemethoxycurcumin was obtained from Tokyo Chemical Industry Co., Ltd. Standard chemicals were obtained from Sigma-Aldrich and Hampton Research.
Structure Determination.
The recombinant CUS, with a His6-tag at the N terminus, was overexpressed in E. coli BL21(DE3)pLysS, purified, and crystallized, as previously reported (20). Diffraction datasets were collected at 100 K by using a Rigaku RA-Micro7 with a Cu target and a Rigaku R-AXIS VII imaging-plate area detector. Data were indexed, integrated, and scaled using the HKL-2000 program.
The initial phases of the CUS structure were determined by molecular replacement using the CUS structure generated by the SWISS-MODEL package (http://expasy.ch/spdpv/), based on the crystal structure of R. palmatum BAS (PDB entry 3A5Q) as a search model. The molecular replacement was performed with Molrep (21). Crystallographic refinement and model building were performed with CNS (22) and MIFit (23), respectively. Each refinement cycle was followed by model building using the σA-weighted 2Fo-Fc and Fo-Fc electron density maps. The water molecules were automatically placed into the difference electron density maps with MIFit and were retained or rejected based on geometric criteria as well as their refined B-factors. After several rounds of model building and refinement, the final model was obtained. The final model consists of residues 19–399 of monomer A, 20–399 of monomer B, 21–399 of monomer C, and 21–398 of monomer D. Residues 301–305 and 368–374 of monomer A, 298–305 and 368–374 of monomer B, 300–306 and 368–374 of monomer C, and 212–214, 299–306, and 368–374 of monomer D were not well defined in the electron density map, and these regions are thus not modeled in the crystal structure. The qualities of the final models were assessed with PROCHECK (24). A total of 89.1% of the residues in CUS are in the most favored regions of Ramachandran plot, 10.0% in the additional allowed regions, and 0.9% in generously allowed region. Details of the data collection, processing and structure refinement are summarized in Table S1. The cavity volume was calculated by CASTP (http://cast.engr.uic.edu/cast/). All crystallographic figures were prepared with PyMOL (DeLano Scientific, http://www.pymol.org).
Enzyme Reaction.
The reaction mixture contained 81 μM of 4-coumaroyl-CoA or 4-hydroxyphenylpropionyl-CoA, 81 μM of malonyl-CoA, and 20 μg of the purified enzyme, in a final volume of 100 μL of 100 mM KPB buffer (pH 7.0), containing 1 mM EDTA. The reactions were incubated at 37 °C for 3 h and stopped by the addition of 5 μL of 20% HCl. The products were then extracted twice with 300 μL of ethyl acetate. The products were separated by reverse-phase HPLC (JASCO 880) on a TSK-gel ODS-80Ts column (4.6ϕ × 150 mm, TOSOH), at a flow rate of 0.8 mL/ min. Gradient elution was performed with H2O and AcCN, both containing 1% AcOH: 0–40 min, linear gradient from 10 to 100% AcCN, and 40–50 min, 100% AcCN. Elutions were monitored by a multichannel UV detector (MULTI 340, JASCO) at 280 or 360 nm; UV spectra (198–400 nm) were recorded every 0.4 s. Online LC-ESIMS spectral analyses were performed as previously described (14), except for the use of the elution program described above. Identification of the enzyme reaction products was performed by direct comparison with the authentic compounds, either obtained in our previous studies (BNY), chemically or enzymatically synthesized as described above (tetrahydrobisdemethoxycurcumin and dihydro-BNY), or purchased (bisdemethoxycurcumin).
Site-Directed Mutagenesis and Purification of Mutant Enzymes.
The plasmids expressing the CUS mutants (Y207F, M265L, and G274F) were constructed with a QuikChange Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer’s protocol, using the following pairs of primers (mutated codons are underlined): Y207F (5′-CGTCGCCGCCGAGCTCACCCTCATGTTCTTCACCGGGCCCGA-3′ and 5′-GCAGCCCTCGTCGGGCCCGGTGAAGAACATGAGGGTGAGCTCG-3′), M265L (5′-GGAGTCGGACCACGCCCTGAACTTGCGGTTCACGGAGCC-3′ and 5′-GTCCAAGCGGCGCTCCGTGAACCGCAAGTTCAGGGCGTGGT-3′), G274F (5′-CGGTTCACGGAGCGCCGCTTGGACTTCGTCCTCGGGCGGCAG-3′ and 5′-GACCTGCCGCCCGAGGACGAAGTCCAAGCGGCGCTCCGT-3′). The mutant enzymes were expressed, extracted, and purified by chromatography on a Ni Sepharose 6 Fast Flow column (GE Healthcare) as previously reported. After the imidazole was removed with an Amicon Ultra-15 filter (Millipore), the purified enzymes were used in the enzyme assay.
Supplementary Material
Acknowledgments.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to I.A. and H.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011499107/-/DCSupplemental.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ALE).
References
- 1.Abe I, Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27:809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- 2.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 3.Schröder J. Comprehensive Natural Products Chemistry. Vol 1. Oxford: Elsevier; 1999. pp. 749–771. [Google Scholar]
- 4.Katsuyama Y, Kita T, Funa N, Horinouchi S. Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J Biol Chem. 2009;284:11160–11170. doi: 10.1074/jbc.M900070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: A short review. Life Sci. 2006;78:2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Sharma RA, Gescher AJ, Steward WP. Curcumin: The strory so far. Eur J Cancer. 2005;41:1955–1968. doi: 10.1016/j.ejca.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Duvoix A, et al. Chemopreventive and therapeutic effects of curcumin. Cancer Lett. 2005;223:181–190. doi: 10.1016/j.canlet.2004.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Katsuyama Y, Matsuzawa M, Funa N, Horinouchi S. In vitro synthesis of curcuminoids by type III polyketide synthase from Oryza sativa. J Biol Chem. 2007;282:37702–37709. doi: 10.1074/jbc.M707569200. [DOI] [PubMed] [Google Scholar]
- 9.Austin MB, Bowman ME, Ferrer JL, Schröder J, Noel JP. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 11.Sankaranarayanan R, et al. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol. 2004;11:894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 12.Rubin-Pitel SB, et al. Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem Biol. 2008;15:1079–1090. doi: 10.1016/j.chembiol.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 14.Morita H, et al. A structure-based mechanism for benzalacetone synthase from Rheum palmatum. Proc Natl Acad Sci USA. 2010;107:669–673. doi: 10.1073/pnas.0909982107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Austin MB, et al. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 16.Austin MB, et al. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat Chem Biol. 2006;2:494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Portes E, Gardrat C, Castellan A. A comparative study on the antioxidant properties of tetrahydrocurcuminoids and curcuminoids. Tetrahedron. 2007;63:9092–9099. [Google Scholar]
- 18.Eckermann C, et al. Stilbenecarboxylate biosynthesis: A new function in the family of chalcone synthase-related proteins. Phytochemistry. 2003;62:271–286. doi: 10.1016/s0031-9422(02)00554-x. [DOI] [PubMed] [Google Scholar]
- 19.Abe I, Morita H, Nomura A, Noguchi H. Substrate specificity of chalcone synthase: Enzymatic formation of unnatural polyketides from synthetic cinnamoyl-CoA analogues. J Am Chem Soc. 2000;122:11242–11243. [Google Scholar]
- 20.Morita H, Wanibuchi K, Kato R, Sugio S, Abe I. Expression, purification and crystallization of plant type III polyketide synthase that produces diarylheptanoids. Acta Crystallogr F. 2010;66:948–950. doi: 10.1107/S1744309110023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 22.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.