Abstract
The PI3-kinase (PI3K) pathway regulates many cellular processes, especially cell metabolism, cell survival, and apoptosis. Phosphatidylinositol-3,4,5-trisphosphate (PIP3), the product of PI3K activity and a key signaling molecule, acts by recruiting pleckstrin-homology (PH) domain-containing proteins to cell membranes. Here, we describe a new structural class of nonphosphoinositide small molecule antagonists (PITenins, PITs) of PIP3–PH domain interactions (IC50 ranges from 13.4 to 31 μM in PIP3/Akt PH domain binding assay). PITs inhibit interactions of a number of PIP3-binding PH domains, including those of Akt and PDK1, without affecting several PIP2-selective PH domains. As a result, PITs suppress the PI3K-PDK1-Akt pathway and trigger metabolic stress and apoptosis. A PIT-1 analog displayed significant antitumor activity in vivo, including inhibition of tumor growth and induction of apoptosis. Overall, our studies demonstrate the feasibility of developing specific small molecule antagonists of PIP3 signaling.
Keywords: PIP3 antagonist, anticancer
Dysregulation of the phosphoinositide 3-kinase (PI3K) pathway has been implicated in many human diseases. Hyperactivation of this pathway is known to play an important role in tumorigenesis, whereas deficiencies contribute to the development of type II diabetes. Therefore, this pathway offers promising targets for the development of drugs to combat these diseases (1, 2).
Class I PI3Ks (α, β, and γ) are recruited to the plasma membrane in response to growth factor and hormone stimulation to mediate the phosphorylation of lipid phosphatidylinositol-4,5-bisphosphate (PIP2), generating phosphatidylinositol-3,4,5-trisphosphate (PIP3), which orchestrates multiple downstream intracellular signaling events (2). PIP3 signaling is terminated by the phosphatase PTEN, which dephosphorylates PIP3. Genetic alterations targeting PTEN are among the most frequent mutations in human cancers, indicating a critical role of uncontrolled signaling through PIP3 in tumorigenesis and metastasis (3). This conclusion is reinforced by transgenic studies establishing that loss of PTEN leads to tumorigenesis (1).
PIP3 controls a complex cellular signaling network regulating cell growth, proliferation, and survival. PIP3 target proteins are located in the cytosol of unstimulated cells and are recruited to the membrane through pleckstrin-homology (PH) domain-mediated binding to newly formed PIP3. Membrane translocation and activation of the PIP3 target proteins initiate a variety of local responses, including assembly of signaling complexes and priming of protein kinase cascades (1, 2). PIP3 regulates an array of PH domain-containing proteins (4), such as serine-threonine kinases Akt and PDK1, GRP1, a GDP/GTP exchange factor of ADP ribosylating factor 6, and protein tyrosine kinases of the Bruton's tyrosine kinase (Btk) and Tec families (5, 6). This diversity in PIP3 signaling makes it one of the most important second messengers downstream from growth factor and oncogene signals. A particularly important example of PIP3-dependent activation is that of serine-threonine kinase Akt. It is achieved both through the binding of Akt PH domain to PIP3 and membrane translocation of another target of PIP3, PDK1, which phosphorylates and activates Akt. The Akt family plays a fundamental role in cell survival, growth, and energy metabolism (1, 7).
Although lipid–protein interactions mediate PI3K signaling and are frequently deregulated in cancer, most therapeutic strategies targeting the PI3K pathway have focused on inhibitors for downstream targets, including PDK1 (8) and Akt (9). Phospholipid–protein interactions have not been as actively targeted, even though lipid molecules are among the most important classes of second messengers. It is surprising considering that they represent “prototypic” small molecule–protein interactions usually involving well-defined binding sites (10). Conceptually, protein–lipid interactions may be more readily targetable compared with protein–protein interactions, which frequently involve interactions of extended flat protein surfaces difficult to disrupt by small molecules. Here, we have pursued the identification of the small molecule antagonists of PIP3/protein binding. We report the discovery of selective nonphosphoinositide PIP3 inhibitors, termed PITenins (PITs). PIT-1, selected in a screen of ≈50,000 small molecules by using a PIP3/Akt PH domain binding assay, has been extensively characterized. Our studies demonstrate that PIT-1 effectively inhibits cancer cell survival and induces cell apoptosis by specifically inhibiting PIP3-dependent PI3K-PDK1-Akt signaling, resulting in significant antitumor activity in vivo. Thus, we show not only a methodology usable to identify additional classes of phospholipid–protein interactions, but also the feasibility of developing cell-permeable specific small molecule nonlipid antagonists of lipid–protein interactions.
Results
Identification of PIT-1 as a Specific PIP3 Antagonist.
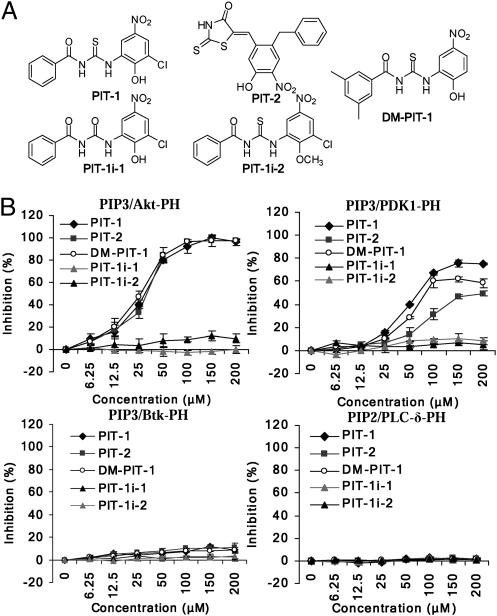
To identify compounds that disrupt the interaction between PIP3 and PH domains, we developed a high-throughput fluorescence polarization (FP)-binding assay by using recombinant 1–123 amino acid N-terminal fragment of human Akt1, encompassing the PH domain (PH123 protein), and a fluorescent NBD-labeled PIP3 molecule. The R86A mutation in the PH domain, shown to disrupt PIP3–PH interaction (11), completely abrogated fluorescent PIP3 binding by PH123 (Fig. S1A), validating the assay. Our screen of ≈50,000 diverse small molecules using this FP assay identified two distinct inhibitors of PIP3/PH domain binding, which were termed PIT-1 and PIT-2 (Fig. 1A). These molecules disrupted PIP3/Akt PH domain binding with IC50 = 31.03 and 31.52 μM for PIT-1 and PIT-2, respectively (Fig. 1B). Although giving similar results to PIT-1, PIT-2 displayed limited stability and was used primarily to confirm the major conclusions. Analysis of commercial PIT-1 analogs suggested that thiourea and hydroxyl groups are critical for the activity in vitro and revealed closely related inactive analogs of PIT-1, termed PIT-1i-1 and PIT-1i-2 (Fig. 1A), which have been used as negative controls. Besides the Akt PH domain, PIT-1 and PIT-2 also inhibited the binding of PIP3 to the PH domains of PDK1, GRP1, and ARNO with lower affinity (Fig. 1B and Fig. S1 B, C, and F) and failed to inhibit PIP3 binding to the PIP3-specific PH domain of Btk (Fig. 1B and Fig. S1G). Thus, PITs exhibited selectivity toward a distinct subset of PIP3-specific PH domains. Furthermore, PITs failed to inhibit PIP2-specific interactions between PI-4,5-P2 and PH domain of PLC-δ (4), and between PI-3,4-P2 and TAPP1 or TAPP2 (12) (Fig. 1B and Fig. S1 D and E). A lipid overlay assay, which detects binding of recombinant proteins to membrane-spotted lipids, confirmed both the inhibitory effect of PIT-1 on PIP3/Akt PH domain binding and the failure to inhibit the binding of PIP2 to PLC-δ PH domain seen in FP assay (Fig. S1I). PITs exhibit a weaker, but detectable, inhibition on the binding of PI-3,4-P2 to Akt PH domain (Fig. S1H), consistent with the notion that PITs targeting Akt PH domain binding pocket, rather than a lipid. These data suggest that PIT-1 and PIT-2 represent unique nonphosphoinositide-related selective antagonists of PIP3/PH domain binding.
Fig. 1.
The identification of PIT-1 as a specific PIP3 inhibitor. (A) Structures of the PITs and inactive analogs. (B) Specificity analysis of phosphoinositide-PH domain inhibition by PITs. TMR-conjugated PI-3,4,5-P3 (15 nM) were incubated with 5 μM Akt PH domain, 100 nM PDK1 or Btk PH domain. TMR-conjugated PI-4,5-P2 (15 nM) were incubated with 100 nM PLC-δ PH domain in the presence or absence of PITs (6.25–200 μM) for 30 min, followed by FP measurement.
To characterize the mode of action of PIT-1, we performed NMR-based analysis of PIT-1 binding to the PH domain of Akt. We found PIT-1 induces significant changes in the 2D HSQC 1H-15N correlation spectrum of Akt PH123, indicative of the direct PIT-1/Akt PH domain binding (Fig. S2A). We titrated Akt PH123 with PIT-1 to identify the residues affected by binding. Using published Akt2 HSQC peak assignments (13), we observed that the residues W22, Y26, and N54, located in the PIP3 binding pocket of the Akt PH domain, are affected by PIT-1. We found that PIT-1 had no effect on the R86A PH domain mutant, which lacks PIP3 binding activity, confirming the PIP3-mimetic nature of PIT-1. Using this information, we propose a binding model of PIT-1 on the surface of the Akt PH domain in which the PIT-1 binding site overlaps with the PIP3 binding site. Consistent with NMR data, W22 and Y26 flank the RPR motif that interacts with the 3-phosphate of PIP3 and the nitro group of PIT-1, whereas N54 packs against the terminal phenyl group of PIT-1 and does not interact with PIP3 (Fig. S2 B and C). We confirmed the direct binding of PIT-1 to Akt PH domain by using SPR, which showed that the molecule is a reversible binder with K d of ≈43.2 μM (Fig. S1J).
PIT-1 Inhibits the PIP3-Mediated PI3K-PDK1-Akt Signaling Pathway.
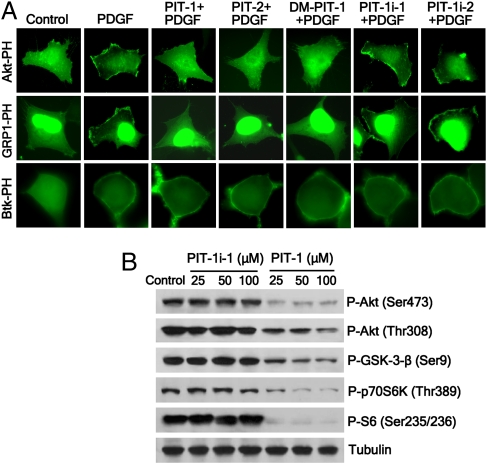
To begin characterizing cellular activities of PITs, we performed a PH domain translocation assay that measures PIP3-mediated association of the GFP-fused PH domains with the plasma membrane (5). As shown in Fig. 2A, both PIT-1 and PIT-2 significantly inhibited the translocation of PIP3-specific Akt and GRP1 PH domains in response to PDGF stimulation. Neither PIT-1i-1 nor PIT-1i-2 inhibited the translocation of Akt and GRP1 PH domains (Fig. 2A and Fig. S3B). No inhibition of translocation was observed with either the Btk PH domain, which does not bind PITs, or PIP2-specific PLC-δ, TAPP1, and TAPP2 PH domains (Fig. 2A and Fig. S3 A and B). These results, like the FP analysis, indicate that PITs are specific PIP3 antagonists with selectivity toward a distinct subset of PIP3-specific PH domains.
Fig. 2.
PIT-1 inhibits PI3K/PDK1/Akt signaling in cancer cells. (A) PITs inhibit growth factor-induced plasma membrane translocation of the PIP3-specific Akt and GRP1, but not that of the Btk PH domain. SUM159 cells were transfected with the GFP-fusion vectors, serum starved overnight, and incubated with 100 μM PITs or PIT-1is for 1 h, followed by stimulation with 100 ng/mL PDGF for 5 min. The representative fluorescent images are shown. (B) PIT-1 inhibits Akt signaling pathway in U87MG cells. U87MG cells were treated with PIT-1 or PIT-1i-1 (25, 50, 100 μM) for 12 h, and the indicated PIP3-dependent phosphorylation events were evaluated by Western blot.
Next, we examined the effect of PITs on PIP3-mediated PI3K-PDK1-Akt signaling. We first analyzed U87MG glioblastoma cells, which show elevated basal levels of PIP3 because of the loss of PTEN (14). Human gliomas frequently display increased PIP3 synthesis due to PI3K and PTEN mutations, and inhibition of PI3K signaling has emerged as a promising new therapeutic strategy against this cancer (15). Both PIT-1 and PIT-2 suppressed PI3K-PDK1-Akt-dependent phosphorylation events in U87MG cells. Inactive analogs PIT-1i-1 and PIT-1i-2 failed to inhibit PDK1/Akt signaling (Fig. 2B and Fig. S3C). Similarly, PIT-1 significantly suppressed PI3K-PDK1-Akt signaling in growth factor-stimulated breast carcinoma SUM159 cells (Fig. S3D). These data confirm that PIT-1 and PIT-2 inhibit PI3K-PDK1-Akt signaling. Moreover, PITs equally inhibited phosphorylation of Akt1, Akt2, and Akt3 (Fig. S3E), consistent with >70% identity between their PH domains. We also demonstrated that PITs treatment failed to induce changes of PIP3 levels in cells, suggesting the inhibition of Akt phosphorylation in cell-based assays was not from inhibition of PI3-K itself (Fig. S3G).
Comparison of PIT-1 with several previously reported Akt inhibitors showed differences in mechanism. Neither Akt inhibitor X nor allosteric inhibitor VIII blocked membrane translocation of Akt PH domain (Fig. S4 A and B). Furthermore, PIT-1 and the Akt kinase inhibitors acted additively in attenuating Akt activation, phosphorylation of downstream targets, and inhibition of cell survival. Conversely, no additive effect was observed when Akt inhibitor VIII and X were combined (Fig. S4 C and D). These data emphasize that PITs represent a distinct class of Akt inhibitors and suggest mechanistic differences could be exploited to achieve efficient inhibition of Akt signaling by combining PH domain-targeting PITs with inhibitors of the Akt kinase activity.
PIT-1 Preferentially Reduces Viability of PTEN-Deficient Glioblastoma Cells.
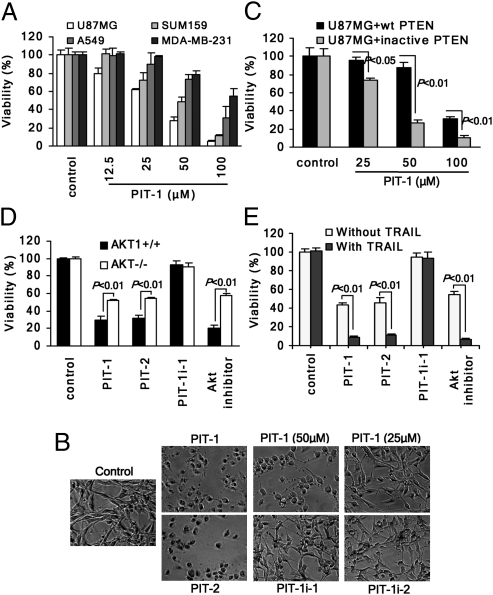
Given the critical role of PIP3-dependent Akt signaling in promoting cell survival, we tested the effect of PITs on cancer cell viability. Both PIT-1 and PIT-2 reduced viability of multiple cancer cell lines in a dose- and time-dependent manner (Fig. 3A and Fig. S5 A and B). PIT-1 consistently suppressed long-term colony formation by U87MG cells (Fig. S5C). Exposure of cells to PITs resulted in eventual cell rounding, loss of adhesion, and cell death (Fig. 3B). In contrast, inactive analogs PIT-1i-1 and PIT-1i-2 caused no loss of viability (Fig. 3B and Fig. S5 A and B). Previous reports (16) suggested that constitutive exposure of transformed cells to oncogenic signaling may render cancer cells hypersensitive to disruption of oncogenic signals (“oncogene addiction”). Therefore, we next examined whether PIT-1 may cause preferential cell death in the cells containing elevated PIP3 levels. We found the toxicity of PIT-1 was more pronounced in U87MG cells expressing inactive PTEN than in cells expressing wild-type PTEN (Fig. 3C). This result is consistent with changes in Akt phosphorylation observed in PTEN-expressing and -deficient cells. Akt phosphorylation was significantly higher in PTEN-deficient cells, and PIT-1 inhibited it to a much greater extent in these cells compared with WT PTEN cells (Fig. S5H). These data are consistent with the notion that PTEN-deficient cancer cells become overly dependent on PIP3 signaling and, therefore, are hypersensitive to the disruption of this pathway compared with that of wild-type PTEN-expressing cells. In addition to U87MG cells, we have also examined MCF7 cells, which express E545K PI3KCA mutant, and confirmed that PITs are able to inhibit Akt activation in the presence of a constitutively active PI3K (Fig. S3F).
Fig. 3.
PIT-1 reduces cell viability, displays preferential effect in PTEN-deficient cells, and synergizes with TRAIL. (A) PIT-1 reduces the viability of multiple cancer cells. Different cancer cell lines were incubated with PIT-1 (12.5–100 μM) for 48 h. Cell viability was determined by using ATP assay. (B) PITs, but not the inactive derivatives, reduce viability of U87MG cells. U87MG cells were incubated with compounds for 48 h, and the representative bright field images of cells are shown. (C) The toxicity of PIT-1 is more pronounced in PTEN-deficient U87MG cells expressing R130M inactive mutant of PTEN compared with the cells expressing wild-type PTEN. After treatment with PIT-1 (25, 50, 100 μM) for 48 h, cell death was analyzed by using Sytox cell death assay. (D) Loss of cell viability caused by PITs, as well as Akt inhibitor, is more pronounced in Akt1-expressing cells compared with the triple knockout cells deficient in all Akt isoforms. Cells were incubated with 100 μM PIT-1, PIT-2, PIT-1i-1, or 10 μM Akt inhibitor VIII for 24 h, followed by analysis of cell viability by using ATP assay. (E) PITs, as well as Akt inhibitor, sensitize U87MG cells to killing by TRAIL. U87MG cells were incubated with 100 μM PIT-1, PIT-2, PIT-1i-1, or 10 μM Akt inhibitor VIII in the presence or absence of 10 ng/mL TRAIL for 24 h, followed by cell viability analysis by using ATP assay.
Up-regulation of PI3K/Akt signaling is an important mechanism of chemoresistance of cancer cells (17). In particular, inhibition of PI3K and Akt was shown to sensitize cancer cells to killing by a TNFα family member, TRAIL. TRAIL represents a particularly promising anticancer agent because of its intrinsic selectivity toward cancer cells (18). At the same time, overactivation of intracellular antiapoptotic mechanisms has been shown to attenuate cancer cell sensitivity to TRAIL (19). Although U87MG cells are indeed resistant to TRAIL, we found that PIT-1 substantially sensitized U87MG cells to killing by TRAIL (Fig. 3E and Fig. S5D). Similar results were obtained by using PIT-2 as well as Akt kinase inhibitor and PI3K inhibitor, LY294002, but not PIT-1i-1 (Fig. 3E and Fig. S5 D–G). Furthermore, this effect was again significantly more pronounced in U87MG cells expressing inactive PTEN than in cells expressing wild-type PTEN (Fig. S5J).
To confirm the mode of action of PIT-1 in the cells is directly related to the inhibition of PIP3/Akt PH domain interaction, we tested induction of cell death in the cells overexpressing constitutively active Akt lacking PH domain, but containing a membrane-targeting myristoylation signal. We found that overexpression of PH domain-deficient Akt indeed significantly protected U87MG cells from PIT-1 killing (P < 0.05, IC50 changed from 37.4 to 103.6 μM; Fig. S5K). A similar result was also obtained with PIT-2 (IC50 changed from 38.7 to 112.5 μM; Fig. S5L).
However, inhibition of cell death by Myr-Akt was not complete. This result could be explained by inhibition of another PIT-1 target, PDK1 (Fig. 1B and Table S1), which regulates cell survival both through Akt as well as independently (20). Akt Thr308 phosphorylation is mediated by PDK1, so PIP3-dependent membrane colocalization of both Akt and PDK1 contribute to Akt activation and cell survival. We indeed observed that PIT-1 and PIT-2 inhibited phosphorylation of Myr-Akt (Fig. S5N), further suggesting that PDK1 is also targeted by PITs in the cells. In concert with the possible role of PDK1 in PIT-induced toxicity, a significantly more pronounced inhibition of cell death by Myr-Akt was observed in cells treated with PIT-6 (IC50 changed from 10.6 to 168.8 μM; Fig. S5M and Table S1). This analog of PIT-1 is selective for Akt PH domain versus PDK1 PH domain (Table S1). Finally, the highest cellular activity displayed by PIT-7, which targets Akt more efficiently and retains PDK1 binding (Table S1), also supports the notion that both PDK1 and Akt mediate PITs’ toxicity.
Finally, the loss of viability caused by both PIT-1 and PIT-2 was more pronounced (P < 0.01) in Akt1-expressing cells compared with triple knockout fibroblasts deficient in all Akt isoforms (21) (Fig. 3D and Fig. S5I). Importantly, Akt kinase inhibitor displayed a similar effect (Fig. 3D and Fig. S5I). Overall, these data confirm the contribution of Akt to the PIT-1-induced cell death.
PIT-1 Induces Apoptosis and Metabolic Stress.
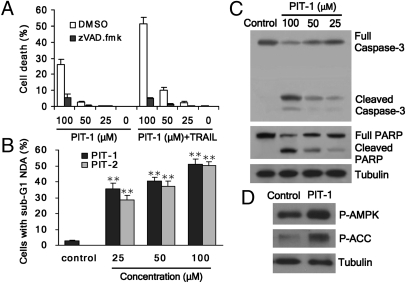
Because PI3K/PIP3/Akt signaling plays a critical role in the inhibition of apoptosis (7), we examined whether PITs induce apoptosis. Cell death triggered by PIT-1 and PIT-2 alone or in combination with TRAIL was inhibited by a pan-caspase inhibitor zVAD.fmk (Fig. 4A and Fig. S6A). We also found that killing of cells by PIT-1 and PIT-2 was reduced in cells lacking two important apoptosis mediators, Bax and Bak (Fig. S6B). Cell death induced by PIT-1 showed multiple apoptotic features. Namely, PIT-1 and PIT-2 treatment caused a significant increase in subG1 DNA content in cells (P < 0.01) (Fig. 4B). In addition, the nuclei of treated cells had a condensed and fragmented morphology, characteristic for apoptosis (Fig. S6C). Finally, Western blot analysis showed that PIT-1 induced the cleavage/activation of a critical apoptosis executioner molecule, caspase-3, and its substrate, PARP (Fig. 4C). These data suggest the activation of apoptosis by PIT molecules, consistent with inhibition of antiapoptotic activity by the Akt-mediated signaling pathway.
Fig. 4.
PIT-1 induces apoptosis and metabolic stress in cancer cells. (A) Cell death triggered by PIT-1 alone or in combination with TRAIL is inhibited by pan-caspase inhibitor zVAD.fmk. U87MG cells were incubated with 100 μM zVAD.fmk together with PIT-1 (25, 50, 100 μM) in the presence or absence of 10 ng/mL TRAIL for 24 h and analyzed with Sytox cell death assay. (B) PITs treatment causes a significant increase in subG1 DNA content in U87MG cells. U87MG cells were incubated with PIT-1 or PIT-2 (25, 50, 100 μM) for 48 h, fixed, and stained with PI for 30 min. The percentage of cells with hypodiploid DNA was determined by FACS. (C) PIT-1 induces caspase-3 activation. U87MG cells were treated with PIT-1 (25, 50, 100 μM) for 48 h. Cleavage of caspase-3 and PARP was analyzed by Western blot. (D) Treatment of U87MG cells with 100 μM PIT-1 for 48 h induces an increase in phosphorylation of AMPK and its main substrate, ACC. **P < 0.01 compared with control group.
Changes in cellular metabolism have recently emerged as an important component of the PI3K/PIP3/Akt signaling, contributing to regulation of cell viability (7). Thus, we next investigated whether PIT-1 causes dysregulation of energy homeostasis and induction of metabolic stress in cancer cells. PIT-1 indeed induced significant increases in phosphorylation of AMP-activated protein kinase (AMPK), a key factor in regulation of energy homeostasis activated by metabolic stress (22), as well as increased phosphorylation of its main substrate acetyl-CoA carboxylase (ACC) (Fig. 4D). Furthermore, we observed increased formation of the autophagic marker LC3-II (Fig. S6D), which showed characteristic perinuclear punctuate staining (Fig. S6E). Electron microscopy showed the appearance of characteristic double membrane-enclosed autophagic vesicles even at the early stages of PIT-1 treatment (Fig. S6F). This result indicates the induction of autophagy, a large-scale catabolic process playing a key role in mediating cellular adaptation to metabolic stress (23), consistent with the induction of metabolic stress by PIT-1. It is also consistent with the established role of Akt as an inhibitor of autophagy (24).
PIT-1 and Its Analogs, the Inhibition of in Vivo Tumor Growth.
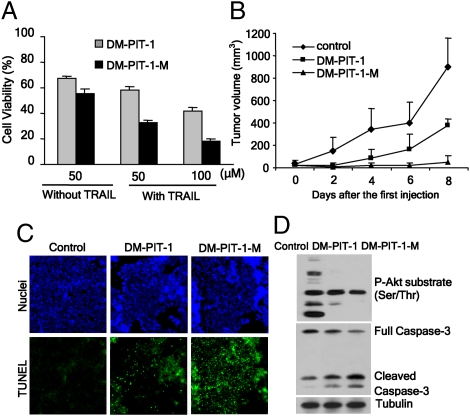
Given the significant inhibition of PI3K/PIP3/Akt signaling and cancer cell survival by PIT-1 in vitro, we examined its anticancer activity in vivo. Because of limited aqueous solubility of PIT-1, we used a dimethyl analog of PIT-1, DM-PIT-1 (Fig. 1A) for in vivo analysis, because we found that it can be effectively incorporated (unlike PIT-1) into the long circulating polyethylene glycol-phosphoethanolamine (PEG-PE) mixed micelles. We have confirmed that DM-PIT-1 did not display any significant differences from PIT-1 across the range of assays used to characterize PIT-1 (Figs. 1B and 2A, Fig. S1 B–D, and Fig. S3 A and B). DM-PIT-1 displayed somewhat increased activity compared with PIT-1 in inhibiting PI3K-PDK1-Akt signaling (Fig. S7A) and inducing cell death (Fig. S7B), which may be associated with better cell permeability arising from its increased lipophilicity. The incorporation of DM-PIT-1 into PEG-PE mixed micelles (DM-PIT-1-M) significantly increased solubility (up to 1 mM) and had no influence on induction of cell death (Fig. 5A).
Fig. 5.
DM-PIT-1 inhibits in vivo tumor growth. (A) DM-PIT-1 encapsulation into PEG-PE micelles facilitates cytotoxicity of DM-PIT-1. U87MG cells were incubated with DM-PIT-1 or DM-PIT-1-M with or without 10 ng/mL TRAIL for 24 h, followed by cell viability analysis using ATP assay. (B) Administration of DM-PIT-1 or DM-PIT-1-M for 8 d attenuates syngeneic 4T1 beast tumor growth in BALB/c mice. After inoculation of tumor cells, DM-PIT-1 or DM-PIT-1-M was administered i.v. daily at doses of 0.4 and 1 mg/kg, respectively, for 8 d. The tumor volumes were measured every two days and are shown. (C) DM-PIT-1 and DM-PIT-1-M administration induces apoptosis of cancer cells in vivo measured with TUNEL assay (green), and nuclei were stained with Hoechst (blue). (D) DM-PIT-1 and DM-PIT-1-M administration suppresses PI3K/PIP3/Akt signaling, and increases cleavage of caspase-3 in tumor cells evaluated by using Western blot.
We next examined the effect of i.v. administration of both unencapsulated DM-PIT-1 and DM-PIT-1-M on syngeneic 4T1 breast cancer growth in BALB/c mice. We found both of them significantly attenuated tumor growth in vivo compared with the control after 8-d administration (P < 0.01). Using the micellar form of DM-PIT-1 allowed a substantially higher dose (1 mg/kg per day) of the drug compared with the free drug (0.4 mg/kg per day) because of the increased solubility, leading to a more pronounced inhibition of tumor growth (P < 0.01). In particular, on day 8 of the treatment, free DM-PIT-1 and DM-PIT-1-M reduced tumor volume by 58.1 and 95.2%, respectively, compared with controls (Fig. 5B). Administration of both DM-PIT-1 formulations was well tolerated by healthy mice without any signs of overt toxicity or loss of weight (Fig. S7C). Consistent with the in vitro data, analysis of the tumors from DM-PIT-1-treated animals showed widespread induction of apoptosis measured by using TUNEL assay. In addition, the nuclei of treated cells displayed characteristic nuclear fragmentation (Fig. 5C and Fig. S7D). Western blot results showed that compound treatment caused a significant increase in the cleavage/activation of caspase-3, as well as cleavage of PARP, again, indicative of apoptosis. Furthermore, consistent with the in vitro mechanism of action, administration of the compound efficiently suppressed the PI3K/PIP3/Akt signaling in the tumor cells (Fig. 5D and Fig. S7E).
Modulation of Selectivity of PIT-1 Toward Akt PH Domain.
PIT-1 and DM-PIT-1 display selectivity toward a subset of PIP3-binding PH domains. To explore whether we could achieve improved selectivity toward a particular PH domain, we have compared the crystal structures of the PH domains of Akt, PDK1, and GRP1 (Protein Data Bank ID codes: 1UNQ, 1W1D, and 1FHX). Specifically we aimed to take advantage of the nonconservative substitution in PIP3 binding pocket of Leu52 of Akt1 for structurally equivalent Arg305 of GRP1 and Lys495 of PDK1 (Fig. S2C). These residues were modeled to be in the proximity of the nitro group of PIT-1, and we hypothesized that hydrophobic groups in R2 and R3 positions of PIT-1 would be preferred for Akt and disfavored for PDK1 and GRP1 binding. Indeed, PIT-4, -5, and -6 exhibit improved Akt potency and selectivity (Fig. S8 A, E, and F and Table S1) compared with PIT-1. Consistently, SPR analysis showed that PIT-6 has an increased binding to Akt PH domain (Kd is ≈20.3 μM) compared with PIT-1 (K d is ≈43.2 μM) (Fig. S1 J and K). Importantly, consistent with the proposed mechanism of cellular activity of PIT-1, increased affinity of PIT-1 analogs toward Akt led to a substantially higher inhibition of Akt signaling in U87MG cells (Fig. S8B) as well as increased cytotoxicity (Fig. S8C and Table S1). Further, targeting of PDK1 may provide additional benefit, as PIT-7, displaying increased Akt binding and some PDK1 binding, exhibited the strongest activity in suppressing Akt signaling, and reducing cancer cell viability (Fig. S8 A–C, E, G, and H and Table S1). Toxicity of new PIT-1 analogs was significantly (P < 0.05) attenuated by overexpression of PH domain-deficient activated Akt, consistent with the contribution of PITs/Akt PH domain interaction to cell death (Fig. S8D). The new PIT-1 analogs also remained inactive toward PIP3/Btk PH and PIP2/PLC-δ/TAPP1/TAPP2 PH domain. Overall, these data suggest that a rational approach can be used for selectively targeting PH domains. Specifically, activity of PIT-1 toward Akt and induction of cell death can be significantly increased by targeted chemotype modifications coupled with elimination of features that can present metabolic liabilities.
Discussion
Here, we describe PITs, a new class of specific nonphosphoinositide small molecule PIP3 antagonists (IC50 ranges from 13.4 to 31 μM in PIP3/Akt PH domain binding assay and from 6.6 to 39.9 μM in cell viability assay). These molecules showed activity against PIP3-dependent PI3K/PDK1/Akt signaling in vitro and significant antitumor activity in vivo. PITs can trigger an array of cellular responses including inhibition of cell survival, induction of apoptosis, restoration of cellular sensitivity to TRAIL, and activation of metabolic stress. Importantly, these effects are preferentially induced in PTEN-deficient U87MG cells, suggesting inhibition of PIP3 and Akt signaling as a promising strategy against human tumors characterized by elevated PIP3 levels such as glioblastomas (15). In vitro activities of PIT-1 translated into pronounced inhibition of tumor growth in vivo by PIT-1 analog, DM-PIT-1. At the same time, DM-PIT-1 is well tolerated upon systemic administration in mice. Overall, our studies establish the feasibility of developing anticancer agents, targeting the PI3K pathway through specific, selective disruption of PIP3/PH domain interactions.
Multiple lines of evidence argue that the effects of PITs are specific for PH domains and the PI3K/PDK1/Akt pathway. First, two structurally distinct molecules, PIT-1 and PIT-2, isolated based on the inhibition of the PIP3 interaction with the Akt PH domain in vitro, displayed very similar cellular activities in a wide range of assays. Conversely, the structurally related PIT-1i-1 and PIT-1i-2 molecules lacked activity in both in vitro PH domain binding and all cell-based assays. These results provide an important validation of the mode of action of PITs. Second, cellular activities of PITs paralleled that of the previously identified Akt kinase activity inhibitors in a number of assays, consistent with the role of Akt as an important mediator of the PIP3 signaling. Third, multiple activities of PITs can be significantly attenuated by overexpression of the PH domain-deficient activated Akt. Fourth, selectively increasing affinity of PIT-1 toward the PH domain of Akt in vitro lead to the parallel increase inhibition of PI3K/Akt signaling and induction of cell death, consistent with the central role of Akt in the regulation of cell viability (Table S1). Overall, these data suggest that PIT-1 scaffold may be a promising starting point for further optimization of the selectivity of the inhibitors toward particular PIP3-specific PH domains and, hence, PIP3-dependent cellular functions. Although initial profiling data suggest some selectivity of PIT-1 and PIT-2 toward a subset of PIP3-binding PH domains, this conclusion needs to be further verified by profiling additional PH domains.
Inhibition of the PIP3/protein binding represents a powerful approach to alter PI3K signaling, because PIP3/PH domain binding is a universal upstream step in PI3K signaling. Examples of PIP3 antagonists, using lipid scaffolds, primarily alkylphospholipids and phosphatidylinositol ether lipid analogs (PIAs), have been described (25, 26). These molecules interact with the Akt PH domain and block Akt membrane translocation and activation. However, achieving selectivity toward specific phosphoinositide targets is likely to be difficult with such scaffolds. PIA molecules, for example, use a modified nonphosphorylated inositol headgroup. Thus, cellular activities of PIAs may well require phosphorylation by endogenous lipid kinases. It may be very challenging to achieve selectivity of PIAs for a specific phosphoinositide, i.e., PIP3, versus other phosphoinositides or in the subfamily of PIP3-binding PH domains. At present, the selectivity profile of these molecules has not been established and Akt PH domain remains their only known PIP3-dependent target. Nonphospholipid/nonphosphoinositide small molecule antagonists present a good alternative starting point for the selective modulation of the PI3K signaling network. Several Akt PH domain small inhibitors have been reported (27 –29). One of these molecules, triciribine phosphate (TCN-P) binds to the PH domain of Akt with high affinity (K d = 690 nM) and blocks its recruitment to plasma membrane, resulting in the induction of apoptosis and inhibition of tumor growth in vivo (9, 29). TCN-P has been recently evaluated clinically in a tailored-personalized phase I trial where only patients whose tumors have high phosphorylated Akt levels were accrued, which showed inhibition of tumor Akt phosphorylation at the tolerated doses (30). PITs are a new structural class of directly competitive and selective nonphosphoinositide small molecule antagonists of PIP3 active in cells, in vitro and in vivo. Furthermore, our results suggest that this approach can be extended beyond inhibition of just Akt PH domains, targeting other important effectors of PIP3 signaling, such as PDK1, GRP1, and ARNO. At the same time, taking advantage of the differences in PH domain PIP3 binding pockets, selective inhibition of particular PH domains could be achieved through modification of common PIT scaffolds.
Both direct ATP-competitive inhibitors of Akt and allosteric inhibitors, targeting the interface between the PH domain and the Akt kinase domain, have been developed (25). This work describes a different approach to inhibition of cellular Akt signaling, targeting membrane localization, and activation of the kinase. Because of a different mode of action, these inhibitors can be combined with the existing Akt kinase inhibitors to achieve greater efficacy in blocking Akt signaling as shown in Fig. S4. Our results suggest a possibility of Akt-directed combination therapy. Moreover, the different mode of action of PITs may offer benefits in blocking PIP3 signaling when combined with the existing PI3K inhibitors. Importantly, efficient suppression of the PIP3 signaling by PI3K inhibitors is negatively influenced by the loss of the PIP3 phosphatase PTEN, hydrolyzing preexisting pool of PIP3. PTEN is frequently inactivated in tumors, e.g., many glioblastomas, breast, endometrium, and colon cancers (31). Thus, combining PI3K inhibitors with PITs, acting downstream from the PTEN-dependent step, may represent a promising strategy, especially against PTEN-deficient tumors. Furthermore, cells deficient in PTEN, which are adapted to growth in the presence of high PIP3 levels, display increased sensitivity to killing by PITs, compared with the matched cells expressing functional PTEN.
One important future direction of anticancer drug discovery is the targeting of specific branches of the PI3K pathway, which would limit side effects of global PI3K pathway inhibition. To this end, we show that chemical modification of PIT-1 results in profound changes in the selectivity profile of the molecule. Combining extensive data profiling PIP3/PH domain binding (4) with further chemical modification of PIT-1 or molecules with a similar mode of action may represent a promising approach to developing useful tool compounds for dissecting cellular PIP3 signaling networks. Furthermore, PIT compounds, possessing different mode of action profiles, could be attractive as leads for anticancer drug discovery.
Materials and Methods
PIT-1, DM-PIT-1, inactive analogs, and PIT-2 were obtained from commercial sources. Synthesis of PIT-1 derivatives, antibodies, DNA vectors, and all other reagents are described in detail in SI Materials and Methods. FP assay was performed by using TMR-labeled PIP3 or PIP2 and bacterially expressed recombinant PH domains. For lipid overlay assay, PIP3 or PIP2 was spotted on the nitrocellulose membrane and binding of recombinant GST-PH domains was determined by using anti-GST antibody. Cell viability was measured by using commercial ATP, MTS, or Sytox assays. Membrane translocation of PH domains was evaluated by using fluorescent microscopy following transfection with GFP-PH vectors. Inhibition of tumor growth was determined following 8-d i.v. administration of DM-PIT-1 in PBS or PEG-PE mixed micelles into BALB/c mice inoculated (s.c.) with 4T1 tumors. These and all other methods are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Joan Brugge (Harvard Medical School, Boston), Dr. Or Gozani (Stanford University, Stanford, CA), Dr. Tamas Balla (National Institutes of Health, Bethesda, MD), Dr. Stanley Korsmeyer (Dana Farber Cancer Center, Boston), Dr. Philip Tsichlis (Tufts Medical Center, Boston), and Dr. Aaron J. Marshall (University of Manitoba, Winnipeg, MB, Canada) for the gifts of plasmids and cells. We thank Albert Tai for help with SPR assay. This work was supported by a Smith Family Award for Excellence in Biomedical Research and National Institute on Aging Mentored Research Scientist Career Development Award (to A.D.), an Innovator Award from the US Army (DAMD17-02-1-0403), the National Cancer Institute (RO1 CA34722/PO1-50661) (to B.S.), and the National Institute on Aging (R37 AG012859) (to J.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.N.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004522107/-/DCSupplemental.
References
- 1.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Maehama T, Dixon JE. PTEN: A tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 4.Park WS, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Várnai P, et al. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J Cell Sci. 2005;118:4879–4888. doi: 10.1242/jcs.02606. [DOI] [PubMed] [Google Scholar]
- 6.Lietzke SE, et al. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 7.Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 8.Peifer C, Alessi DR. Small-molecule inhibitors of PDK1. ChemMedChem. 2008;3:1810–1838. doi: 10.1002/cmdc.200800195. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 10.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 11.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 12.Marshall AJ, Krahn AK, Ma K, Duronio V, Hou S. TAPP1 and TAPP2 are targets of phosphatidylinositol 3-kinase signaling in B cells: Sustained plasma membrane recruitment triggered by the B-cell antigen receptor. Mol Cell Biol. 2002;22:5479–5491. doi: 10.1128/MCB.22.15.5479-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auguin D, et al. Solution structure and backbone dynamics of the pleckstrin homology domain of the human protein kinase B (PKB/Akt). Interaction with inositol phosphates. J Biomol NMR. 2004;28:137–155. doi: 10.1023/B:JNMR.0000013836.62154.c2. [DOI] [PubMed] [Google Scholar]
- 14.Pore N, Liu S, Haas-Kogan DA, O'Rourke DM, Maity A. PTEN mutation and epidermal growth factor receptor activation regulate vascular endothelial growth factor (VEGF) mRNA expression in human glioblastoma cells by transactivating the proximal VEGF promoter. Cancer Res. 2003;63:236–241. [PubMed] [Google Scholar]
- 15.Cheng CK, Fan QW, Weiss WA. PI3K signaling in glioma—animal models and therapeutic challenges. Brain Pathol. 2009;19:112–120. doi: 10.1111/j.1750-3639.2008.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaughnessy JD. Cancer: An unexpected addiction. Nature. 2008;454:172–173. doi: 10.1038/454172a. [DOI] [PubMed] [Google Scholar]
- 17.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 18.Abe K, Kurakin A, Mohseni-Maybodi M, Kay B, Khosravi-Far R. The complexity of TNF-related apoptosis-inducing ligand. Ann N Y Acad Sci. 2000;926:52–63. doi: 10.1111/j.1749-6632.2000.tb05598.x. [DOI] [PubMed] [Google Scholar]
- 19.Oka N, et al. Galectin-3 inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by activating Akt in human bladder carcinoma cells. Cancer Res. 2005;65:7546–7553. doi: 10.1158/0008-5472.CAN-05-1197. [DOI] [PubMed] [Google Scholar]
- 20.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iliopoulos D, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2:ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 23.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi H, et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 25.LoPiccolo J, Granville CA, Gills JJ, Dennis PA. Targeting Akt in cancer therapy. Anticancer Drugs. 2007;18:861–874. doi: 10.1097/CAD.0b013e3280cc2c6f. [DOI] [PubMed] [Google Scholar]
- 26.Meuillet EJ, et al. Specific inhibition of the Akt1 pleckstrin homology domain by D-3-deoxy-phosphatidyl-myo-inositol analogues. Mol Cancer Ther. 2003;2:389–399. [PubMed] [Google Scholar]
- 27.Mahadevan D, et al. Discovery of a novel class of AKT pleckstrin homology domain inhibitors. Mol Cancer Ther. 2008;7:2621–2632. doi: 10.1158/1535-7163.MCT-07-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moses SA, et al. In vitro and in vivo activity of novel small-molecule inhibitors targeting the pleckstrin homology domain of protein kinase B/AKT. Cancer Res. 2009;69:5073–5081. doi: 10.1158/0008-5472.CAN-08-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berndt N, et al. The Akt activation inhibitor TCN-P inhibits Akt phosphorylation by binding to the PH domain of Akt and blocking its recruitment to the plasma membrane. Cell Death Differ. 2010;17:1795–1804. doi: 10.1038/cdd.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrett CR, et al. Phase I pharmacokinetic and pharmacodynamic study of triciribine phosphate monohydrate, a small-molecule inhibitor of AKT phosphorylation, in adult subjects with solid tumors containing activated AKT. Invest New Drugs. 2010 doi: 10.1007/s10637-010-9479-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: Variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.