Abstract
Dynamic posttranslational modification of serine and threonine residues of nucleocytoplasmic proteins by β-N-acetylglucosamine (O-GlcNAc) is a regulator of cellular processes such as transcription, signaling, and protein–protein interactions. Like phosphorylation, O-GlcNAc cycles in response to a wide variety of stimuli. Although cycling of O-GlcNAc is catalyzed by only two highly conserved enzymes, O-GlcNAc transferase (OGT), which adds the sugar, and β-N-acetylglucosaminidase (O-GlcNAcase), which hydrolyzes it, the targeting of these enzymes is highly specific and is controlled by myriad interacting subunits. Here, we demonstrate by multiple specific immunological and enzymatic approaches that histones, the proteins that package DNA within the nucleus, are O-GlcNAcylated in vivo. Histones also are substrates for OGT in vitro. We identify O-GlcNAc sites on histones H2A, H2B, and H4 using mass spectrometry. Finally, we show that histone O-GlcNAcylation changes during mitosis and with heat shock. Taken together, these data show that O-GlcNAc cycles dynamically on histones and can be considered part of the histone code.
Keywords: epigenetics, histones
Initially described more than 25 y ago (1), the dynamic and abundant modification of serine (Ser) and threonine (Thr) residues of nuclear and cytoplasmic proteins by single β-N-acetylglucosamine residues (O-GlcNAc) remains difficult to detect and functionally elusive (2, 3). O-GlcNAc is analogous to phosphorylation in that it is highly dynamic, cycling on and off sites on polypeptides in response to and eliciting specific changes in protein functions, localization, or interactions. O-GlcNAc has extensive crosstalk with phosphorylation (4). Like phosphorylation, O-GlcNAc is highly abundant with a diverse protein and functional distribution. Its donor substrate, UDP-GlcNAc, exists in high intracellular concentrations that are second only to ATP in terms of relative abundance of high-energy small molecules (3). Unlike phosphorylation, however, in mammals only one gene encodes the enzyme that catalyzes the addition of the sugar O-GlcNAc transferase (OGT), and one gene encodes the enzyme that removes the sugar moiety β-N-acetylglucosaminidase (O-GlcNAcase). Thus far, more than 1,000 different substrates have been identified as targets of these enzymes, ranging from signaling proteins such as kinases and adaptor molecules, structural proteins, and nuclear pore proteins to the transcriptional machinery including RNA polymerase II itself. UDP-GlcNAc, the direct donor substrate for O-GlcNAcylation, is at the nexus of several metabolic pathways: glucose metabolism, amino acid metabolism, nucleotide metabolism, and fatty acid metabolism. Because UDP-GlcNAc is centered around many metabolic pathways and because of the variety of substrates modified, protein O-GlcNAcylation plays a role in the etiology of metabolic diseases such as diabetes, neurodegenerative disorders, and cancer (5–7).
DNA is wound around an octamer of basic proteins called “histones.” These core histones are comprised of a tetramer of histone 3 (H3) and histone 4 (H4) and two dimers of histone 2, H2A and H2B (8). This octamer forms a core domain around which the DNA wraps. Projecting from the nucleosome are the N-terminal histone tails. Histones are modified by a wide variety of posttranslational modifications including phosphorylation, methylation, acetylation, and sumoylation, which regulate both their ability to interact with DNA and their ability to recruit enzymes to remodel local chromatin structure necessary for transcription, replication, repair, recombination, and chromosomal compaction and segregation during mitosis (9, 10). Although the majority of modification sites have been mapped to the tails, a number of modifications that mediate histone–DNA contacts and histone–histone contacts also have been identified in the core (11). It has been proposed that these modifications and their various combinations serve as a “code” interpreted by proteins to elicit certain responses.
Mounting evidence suggests that O-GlcNAc plays a critical role in chromatin structure. Kelly et al. (12) demonstrated that a number of chromatin-associated proteins are O-GlcNAcylated. Additionally, immunostaining of Drosophila polytene chromosomes with a lectin that recognizes GlcNAc residues as well as radiolabeling O-GlcNAc moieties with a galactosyltransferase (GalT), which transfers galactose (Gal) to GlcNAc, showed that chromosomes contain many O-GlcNAcylated proteins. Interestingly, detection of O-GlcNAc was intense in the condensed chromatin, whereas the puff regions, indicative of transcriptionally active regions, had less concentrated O-GlcNAc. Moreover, RNA polymerase II and many transcription factors are O-GlcNAcylated (3, 6, 13–15). O-GlcNAcylation of transcription factors can affect protein stability as well as its ability to activate transcription (16–18). O-GlcNAc directly affects chromatin remodelers, such as mammalian Sin3a (mSin3a), histone deacetylase 1 (HDAC1), and the histone methyltransferase myeloid/lymphoid or mixed-lineage leukemia 5 (MLL5) (19, 20). Recent studies established that Drosophila OGT is encoded by the Polycomb group (PcG) gene super sex combs (sxc) and that the major sites of O-GlcNAc modification on polytene chromosomes correspond to PcG-binding sites. These studies provide a direct link of O-GlcNAcylation to PcG-mediated gene silencing, important in development, stem cell maintenance, and genomic imprinting (21, 22). Additionally, ChIP-chip whole-genome tiling arrays in Caenorhabditis elegans demonstrated that O-GlcNAc cycling occurs on chromatin, particularly in promoter regions of genes involved in metabolism, stress, and aging (23).
Here, we show that histones are modified with O-GlcNAc within the nucleosomal core. This study provides experimental evidence of this highly dynamic posttranslational modification on these evolutionarily conserved proteins.
Results
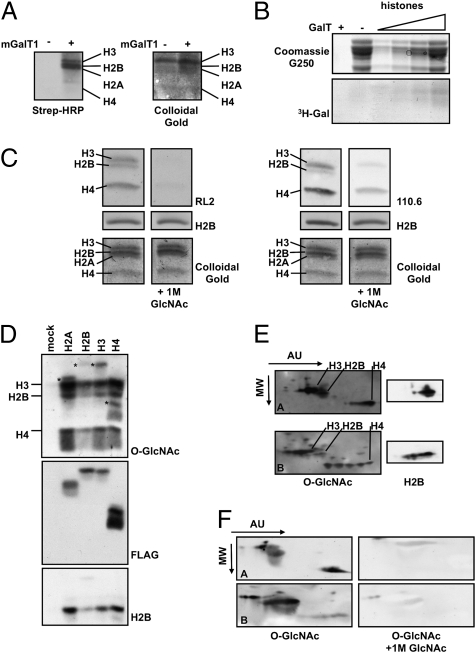
To show that histones are indeed modified with O-GlcNAc, we acid-extracted histones from HeLa cells and detected O-GlcNAc moieties using a chemoenzymatic approach implementing a mutant galactosyltransferase, mGalT1, to tag O-GlcNAc residues with an azido sugar, azide-modified galactosamine (GalNAz). The azido group is further reacted with a biotin alkyne in a Huisgen cycloaddition reaction (24, 25). O-GlcNAcylated proteins then are detected by blotting with streptavidin-conjugated HRP. This methodology is highly specific, because the mGalT recognizes terminal GlcNAcs with high affinity (26). Using this approach, we detected bands running at the molecular weight of the histones, suggesting that histones are O-GlcNAcylated (Fig. 1A). Additionally, we performed a GalT reaction using UDP-[3H]-Gal as the donor substrate and found that acid-extracted histones were enzymatically labeled with 3H-Gal (Fig. 1B).
Fig. 1.
Histones are modified with O-GlcNAc. (A) Acid-extracted histones were labeled using the Click-IT kit (Invitrogen). Briefly, GlcNAc moieties were labeled with an azido-modified Gal using mGalT1. The azido sugar was reacted further with a biotin alkyne, thereby tagging O-GlcNAc residues with biotin. Blots then were probed with streptavidin-HRP (Strep-HRP) to reveal O-GlcNAcylated proteins and stained with colloidal gold for total protein. (B) O-GlcNAc on histones was labeled with 3H-Gal using GalT and UDP-[3H]-Gal. The upper gel represents the total protein, and the lower gel is the autoradiograph. (C) Natively purified histones were probed with two separate antibodies that recognize O-GlcNAc, RL2 (Left) and 110.6 (Right). For specificity, 1 M GlcNAc competition blots were included. Blots also were probed with histone H2B and were stained with colloidal gold for protein loading. (D) FLAG-tagged histones were immunoprecipitated from transfected HeLa cells and probed for O-GlcNAc (RL2), FLAG, and H2B. Asterisks indicate the FLAG-tagged histones. (E) Acid-extracted histones from asynchronously growing HeLa cells (Upper) and HeLa cells treated with sodium butyrate (Lower) were separated in two dimensions. The first dimension was based on charge (AU), and the second dimension was based on molecular weight (MW). Blots also were probed with H2B for loading. (F) Acid-extracted histones were resolved as in E and probed for O-GlcNAc with or without 1 M GlcNAc for specificity.
Although biotinylation of histones has been reported in the literature (27), recent evidence suggests that recognition of histones with streptavidin may be artifactual (28). The reactivity toward histones we observed with streptavidin-HRP was specific to the mGalT lanes; however, we decided to circumvent this possible issue by also using antibodies directed against O-GlcNAc. Therefore, purified core histones were resolved by SDS/PAGE, transferred to nitrocellulose, and probed with the O-GlcNAc antibodies RL2 (29) or CTD 110.6 (Fig. 1C) (30). The immunoreactivity of the histones with these antibodies was specific because 1M GlcNAc competed away the signal. Finally, we immunoprecipitated FLAG-tagged constructs of histones expressed in HeLa cells (31) and saw that the exogenously expressed histones also were recognized by an O-GlcNAc antibody (Fig. 1D). Coincidentally, endogenous histones that coimmunoprecipitated with the FLAG-tagged histones also were O-GlcNAcylated. In addition, the interaction of the endogenous histones with the FLAG-tagged histones suggests that the FLAG-tagged histones were incorporated into chromatin successfully. Moreover, histones are in vitro substrates for OGT (Fig. S1). Thus, by multiple completely independent methods, the data clearly show that histones are, in fact, O-GlcNAcylated.
Next, we determined if differentially acetylated histones are O-GlcNAcylated by resolving histones on acetic acid/urea (AU) gels. Urea, which does not affect protein charge, is used as the denaturant. Therefore, modifications that affect the net charge of histones will affect migration through the gel. Increasing acetylation of histones, for example, will retard mobility through the gel (32). Acid-extracted histones from HeLa cells or from HeLa cells treated with the histone deacetylase inhibitor sodium butyrate were separated in the first dimension by AU gel electrophoresis and in the second dimension by SDS/PAGE. Blotting with an O-GlcNAc–specific antibody showed that the O-GlcNAc modification exists in conjunction with acetylation (Fig. 1E). These blots were stripped and reprobed with histone H2B for protein loading. Specificity was confirmed using 1 M GlcNAc in competition blots (Fig. 1F).
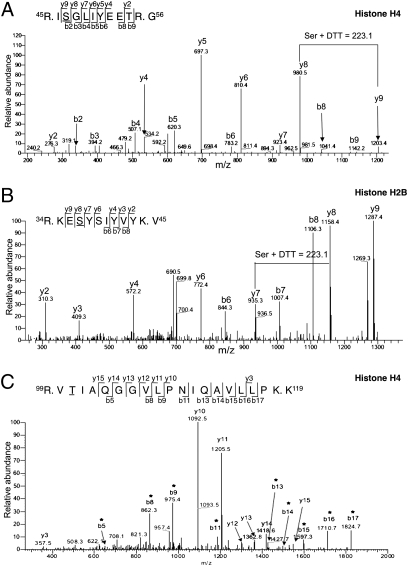
Next, we determined where histones are modified to find potential clues about the role of the sugar on histones. Therefore, O-GlcNAc sites were identified on histones isolated from HeLa cells by first enriching for O-GlcNAc–modified tryptic peptides followed by liquid chromatography (LC)-MS/MS. Tryptic peptides first were treated exhaustively with alkaline phosphatase to remove phosphates. Peptides containing O-GlcNAc residues then were labeled enzymatically with GalNAz using mGalT1. The resulting azido sugar was reacted with a biotin alkyne to tag the O-GlcNAyclated peptides with biotin. O-GlcNAcylated peptides then were highly enriched by avidin affinity chromatography and washed very stringently so that only peptides containing O-GlcNAc would bind specifically. The biotin disaccharide then was subjected to a very mild β-elimination followed by Michael addition with DTT (33), thereby releasing the biotinylated disaccharide from the Ser or Thr residues and adding a DTT marker. Unlike O-GlcNAc, DTT is stable to collision-induced dissociation (CID), and its location is readily determined by standard (i.e., CID) MS (33). This approach is highly specific for O-GlcNAc because of (i) the high specificity of the GalT used to attach GalNAz to O-GlcNAc; (ii) the exhaustive treatment of the peptides with phosphatase; (iii) the very mild β-elimination conditions, which would not release phosphate effectively; and (iv) the high selectivity of the avidin-biotin chromatography and the stringent washing of the avidin column (33–35). Any peptide that contained only a phosphate not released by the phosphatase would not remain bound to the column, and any peptide that contained both a phosphate that was β-eliminated despite the mild conditions and an O-GlcNAc would contain at least two DTT moieties.
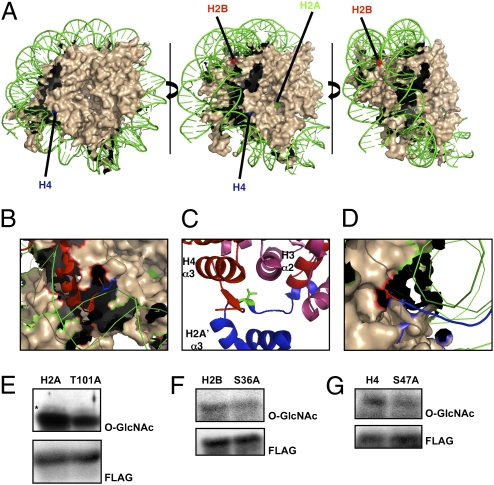
Using this approach (a schematic is shown in Fig. S2), we identified three sites, Thr101 of H2A, Ser36 of H2B, and Ser47 of H4 (Fig. 2 A–C). We were able to localize sites of O-GlcNAc modification onto the nucleosome core particle structure (Protein Data Bank code 1KX5) (Fig. 3A). Ser36 of H2B and Ser47 of H4 are located on the lateral surface of the nucleosome with the main chain of Ser36 of H2B making a direct interaction (Fig. 3D) and Ser47 of H4 making an indirect interaction with DNA (Fig. 3B) (8, 36, 37). Ser36 of H2B delineates the N-terminal tail from the nucleosomal core. Ser47 of H4, however, lies in loop 1 within the histone-fold domain. Thr101 of H2A resides in the C-terminal tail domain, which serves as a docking domain for the H3/H4 tetramer, stabilizing the histone-fold domains (Fig. 3C) (8).
Fig. 2.
Identification of O-GlcNAcylation sites on histones. (A) MS/MS spectrum of the precursor ion ([M+2H]2+ 658.62) carrying a DTT moiety identifying Ser47 of histone H4 as O-GlcNAcylated. (B) MS/MS spectrum of the precursor ion ([M+2H]2+ 708.5) carrying a DTT moiety identifying Ser36 of histone H2B as O-GlcNAcylated. (C) MS/MS spectrum of precursor ion ([M+2H]2+ 1034.2) identifying Thr101 as the O-GlcNAc site on histone H2A. Asterisks indicate ions containing the DTT tag. A, Ala; E, Glu; G, Gly; I, Ile; K, Lys; L, Leu; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; Y, Tyr.
Fig. 3.
Validation of mapped O-GlcNAc sites. (A) Core nucleosomal particle is colored in nude. (For simplicity, histone tails are not shown.) DNA is in green. H4 sites are labeled in blue, H2B sites in red, and H2A sites in green. (B) Ser47 of modified H4 is shown in blue as a stick diagram. The tail of H4 is shown in red as a ribbon. (C) Thr101 of H2A is shown in green as a stick diagram, H4 is shown in red, H3 is shown in pink, and H2A is shown in blue. The α2 and α3 domains of H3 and H4 are labeled. (D) Ser36 of H2B is shown in red in a stick diagram. The histone tail of H2B is shown in blue. To verify site-mapping data, HeLa cells were transfected with FLAG-tagged constructs for wild type and corresponding Ser/Thr-to-Ala mutations for H2A (E), H2B (F), and H4 (G). Blots were probed for O-GlcNAc (RL2) and FLAG.
To verify further the identified O-GlcNAc sites, we generated site-directed mutants [Ser/Thr- to-alanine (Ala)] for each individual site, immunoprecipitated the overexpressed site mutants from HeLa cells, and probed with an O-GlcNAc antibody. A modest decrease in O-GlcNAc levels was observed on each Ala mutant as compared with wild type, further confirming that these sites are O-GlcNAcylated in vivo (Fig. 3 E–G) and further indicating that other sites remain to be identified. There now are many known proteins in which O-GlcNAc and phosphate compete reciprocally for the same site on proteins (3). Here, all three sites also are known phosphorylation sites, although functional significance for phosphorylation has been identified only for Ser36 of H2B (Ser33 of H2B in Drosophila) (38–41). H2B is phosphorylated at this site by TATA box binding protein (TBP) associated factor 1 (TAF1). Phosphorylated H2B is found in promoters of some genes that regulate the G2 to M-phase transition, suggesting that H2B phosphorylation could regulate cell-cycle progression (39).
With the recent identification of the H2B kinase TAF1, we investigated whether knocking down TAF1 levels could elevate O-GlcNAc on this site. Therefore, we transfected HeLa cells with either H2B or H2B S36A with control or TAF1 siRNA. We immunoprecipitated with anti-FLAG-agarose and blotted for O-GlcNAc; however, there did not seem to be an increase in H2B O-GlcNAcylation in the TAF1-knockdown cells (Fig. S4A). This observation coupled with the low expression levels of FLAG-H2B S36E (Fig. S4C) suggests that phosphorylation at this site by TAF1 must be tightly regulated to prevent premature entry into mitosis and that O-GlcNAcylation at this site might add another degree of regulation; however, we could not assess this hypothesis directly without an available phospho-specific antibody.
We then determined whether we could detect changes in histone O-GlcNAcylation in vivo. We chose two different cellular states in which both O-GlcNAc and histone modifications are known to change dramatically. Global O-GlcNAcylation levels have been shown to decrease during mitosis and to return to basal levels once cells re-enter G1 (42). Moreover, recent proteomic analysis revealed that many posttranslational modifications found on histones are cell-cycle dependent (43). To determine whether histone O-GlcNAcylation also changes during mitosis, we extracted histones from HeLa cells synchronized in early M, late M, and early G1 to analyze their O-GlcNAc content by immunoblotting. O-GlcNAc levels on histones decreased during mitosis and subsequently returned to basal levels in G1 (Fig. S4C), with histone H3 being the most affected (Fig. S4C). In nucleo OGT assays also demonstrate that OGT activity toward histones decreases during mitosis (Fig. S4D).
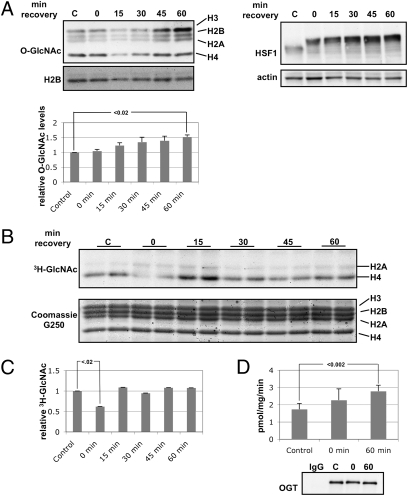
Conditions of cellular stress dramatically change global protein O-GlcNAcylation (44, 45). One of the most primitive forms of stress, heat stress, elicits an evolutionarily conserved response. We stressed HeLa cells for 1 h at 45 °C and allowed them to recover at 37 °C for the indicated times. O-GlcNAc levels on histones increased during the recovery after heat shock (Fig. 4A). As a positive control, we also probed for heat shock factor 1 (HSF1), which becomes hyperphosphorylated upon heat shock, causing a mobility shift (46). We then determined what changes occurred in OGT activity after heat shock. First, we isolated nuclei from cells treated as above and performed in nucleo OGT assays by incubating nuclei in the presence of 3H-UDP-GlcNAc (Fig. 4B). Although there was no increase in OGT activity during the recovery period, there was an ≈50% decrease in endogenous OGT activity toward the histones immediately after heat shock (Fig. 4C). Interestingly, in this assay, the major targets for OGT appeared to be histone H2A and H4. It is possible that the permeabilization technique we used prevented an accessory factor from correctly targeting OGT to recognize H3 and H2B, and, in fact, several studies have demonstrated that OGT uses interacting partners to recognize specific substrates (47, 48). We also determined OGT-specific activity by immunoprecipitating OGT from control cells, heat-shocked cells, and heat-shocked cells that had been allowed to recover for 1 h at 37 °C and performing in vitro OGT assays using a casein kinase II (CK2) peptide as substrate. We could not detect any difference in OGT-specific activity immediately after heat shock, but, as has been reported previously, we detected a slight increase in OGT-specific activity upon recovery (44).
Fig. 4.
Total histone O-GlcNAcylation changes with heat shock. (A) HeLa cells were heat shocked for 1 h at 45 °C and allowed to recover for the indicated times. Histones were probed for O-GlcNAc (RL2) and for H2B. (Left) Densitometric analysis of relative O-GlcNAc levels on histones normalized to histone H2B. (Right) Soluble extracts were probed for HSF1 and actin as controls. (B) In nucleo OGT assays were performed. The upper panel is the autoradiograph, which shows that histones H2A and H4 are major targets for OGT in this assay. The lower panel is the Coomassie G250 stain for total protein. (C) Densitometry of 3H-GlcNAc levels normalized to the amount of histones. (D) OGT was immunoprecipitated, and activity assays were performed using CK2 peptide as substrate. Activity is presented as pmol GlcNAc released per mg OGT immunoprecipitated min− assay.
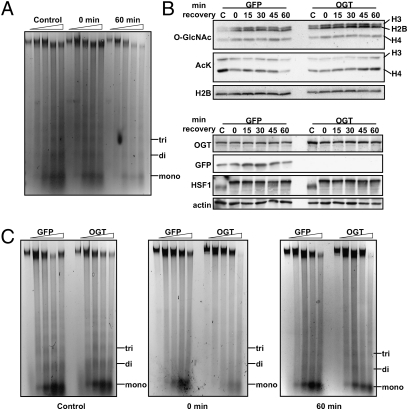
To determine a possible functional consequence for increased histone O-GlcNAcylation during recovery after heat shock, we performed micrococcal nuclease (MNase) chromatin-sensitivity assays. After 1 h heat shock, chromatin appeared more condensed, as evidenced by the generation of fewer monosomes (Fig. 5A, Control vs. 0 min recovery). This observation has been reported previously by others after staining whole cells with DAPI and visualizing areas of highly condensed chromatin in heat-shocked cells by microscopy (49). After recovery for 1 h, MNase digestion revealed that the chromatin had condensed even further (Fig. 5A), correlating with increased histone O-GlcNAcylation.
Fig. 5.
OGT overexpression affects chromatin condensation. (A) Chromatin-sensitivity assays were performed. Mono-, di-, and trinucleosomes are marked. Digestions with MNase were performed for 0, 2, 5, 10, and 15 min. (B) Histones from cells overexpressing GFP or OGT exposed to 1 h heat shock at 45 °C and allowed to recover at 37 °C as indicated were probed for O-GlcNAc and acetylated Lys (AcK). Histones H3, H2B, and H4 are indicated. Soluble extract also was probed for OGT, GFP, HSF1, and actin. (C) Chromatin-sensitivity assays were performed on cells overexpressing GFP or OGT from control cells or cells that had been heat shocked for 1 h at 45 °C and allowed to recover for 0 or 60 min at 37 °C. Digestions with MNase were performed as above.
To determine the effects of OGT overexpression on chromatin architecture, we overexpressed GFP or OGT in HeLa cells and subjected the cells to a thermal challenge of 45 °C for 1 h with recovery at 37 °C for indicated times. Although we did not observe any differences in HSF1 activation (Fig. 5B, Lower), we observed increased histone O-GlcNAcylation with OGT overexpression, as would be expected. Moreover, when probing for acetylated lysine (Lys), a histone mark that decreases during heat shock and during stress (49, 50), we saw that under control conditions histone acetylation decreased with heat shock. However, in cells overexpressing OGT, overall levels of Lys acetylation were depressed under control conditions and increased during the recovery phase after heat shock (Fig. 5B, Upper). When we compared chromatin condensation by MNase digestion, cells overexpressing OGT were consistently less sensitive to digestion, a result suggesting greater DNA compaction than in cells overexpressing GFP.
Discussion
Here we have demonstrated that histones are O-GlcNAcylated. Additionally, we have mapped three O-GlcNAc sites on histones: Thr101 of H2A, Ser36 of H2B, and Ser47 of H4. Although we did not identify any sites on histone H3, its reactivity with O-GlcNAc–specific antibodies suggests that it also bears O-GlcNAc. Also, mutation of the mapped Ser or Thr to Ala on the other three histones only partially diminished reactivity with an O-GlcNAc antibody, suggesting that more sites remain to be identified. Although we were able to map O-GlcNAc sites only to previously identified phosphorylation sites, this observation does not preclude the possibility that there are Ser or Thr residues that are O-GlcNAcylated only in vivo.
NMR studies with O-GlcNAcylated and/or phosphorylated peptides from estrogen receptor β, from the heptad repeat from the C-terminal domain of RNA polymerase II, and from the microtubule-binding protein tau indicate that although phosphorylated peptides form an extended structure, O-GlcNAcylated peptides tend to form a β-turn (51–53). With these studies in mind, we suggest that O-GlcNAcylation could induce major changes in nucleosomal structure not only by regulating peptide backbone conformation but also because of its sheer size. O-GlcNAcylation at Ser36 of H2B and at Ser47 of H4 could regulate histone tail dynamics, because these serines are located near the lateral surface, making direct (Ser36 of H2B) or indirect (Ser47 of H4) contacts with DNA. In fact, mutation of Ser47 of H4 to cysteine (Cys) in yeast displayed a switch/sucrose nonfermentable (Swi/Snf)-independent (Sin) phenotype; that is, they did not require the presence of the Swi-Snf complex in the activation of Swi-Snf genes (54). Moreover, mutation at this Ser also induced a slow-growth phenotype and sensitivity to the DNA-damaging agent hydroxyurea (54, 55).
The physiological relevance of histone O-GlcNAyclation can extend to a multitude of aspects, from normal cellular function to deviant function associated with disease states. Histone modifications, accompanied by the chromatin architecture that they encode, are responsible for the transcriptional profile that determines cell lineage, as in differentiation of pluripotent ES cells to terminally differentiated daughter cells (56), as well as cell response to extracellular stimuli such as growth factors or stress (57). In fact, the emerging role of O-GlcNAc and OGT in differentiation and development (21, 58, 59) is supported further by the absolute requirement of mouse ES cells for OGT (60). Additionally, because of the various types of modifications that histones can undergo, the nucleosome itself serves as a metabolic sensor of cellular state that can be related directly to gene expression (61). Histone acetyltransferases, for example, use acetyl-CoA, which is sensitive to fatty acid oxidation, as a donor substrate, whereas the histone deacetylase sirtuin 1 (SIRT1) is activated by an increase in the NAD+/NADH ratio (62). It has been proposed that O-GlcNAc is a total-cell metabolic sensor because UDP-GlcNAc, the donor substrate for OGT, is at the center of many metabolic pathways (63) and directly affects the levels of protein O-GlcNAcylation.
Of note, our studies show that histone O-GlcNAcylation increases with heat shock and that this increase is concomitant with DNA condensation. Interestingly, hyperthermia has been shown to sensitize tumor cells to radiotherapy. Although the mechanism for this sensitization has not been elucidated, it has been suggested that prior treatment with heat affects the cellular response to DNA damage induced by ionizing radiation (64). Our observations suggest that changes in histone O-GlcNAcylation might be another potential mechanism for radiosensitization.
Even with the numerous studies attempting to elucidate the histone code, we still are deciphering the many nuances associated with each posttranslational modification, singly and in combination. With the addition of O-GlcNAc to this repertoire of modifications, the combinatorial array of histone modifications has increased greatly.
Materials and Methods
A detailed discussion of materials and methods is given in SI Materials and Methods.
Purification of Histones.
Histones were either acid extracted or purified by hydroxylapatite chromatography as outlined in SI Materials and Methods.
MS.
A detailed protocol for preparation of samples for MS and subsequent data analysis is provided in SI Materials and Methods.
GalT Assays, OGT Assays, and MNase Chromain Accessibility Assays.
These assays were performed as described in SI Materials and Methods.
Western Blotting and Immunoprecipiation.
A detailed protocol is provided in SI Materials and Methods.
Statistical Analysis.
All experiments were performed on at least three separate occasions (n ≥ 3). In all figures, error bars represent SE. P values were calculated using the paired two-tailed Student's t test.
Supplementary Material
Acknowledgments
We thank Barbara Smith of the Johns Hopkins Core Microscopy Facility (Baltimore, MD). We also thank Drs. Win D. Cheung, Wagner B. Dias, Chad Slawson, Ronald Copeland, and other members of the G.W.H. laboratory for help and critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01 DK61671 and R01 CA42486 (to G.W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009023107/-/DCSupplemental.
References
- 1.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 2.Wang Z, et al. Enrichment and site-mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation (ETD) mass spectrometry. Mol Cell Proteomics. 2009;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci USA. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer's disease. Mol Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 6.Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 7.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2009;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 11.Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83:468–476. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- 12.Kelly WG, Hart GW. Glycosylation of chromosomal proteins: Localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell. 1989;57:243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- 13.Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: Implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 14.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 15.Jackson SP, Tjian R. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc Natl Acad Sci USA. 1989;86:1781–1785. doi: 10.1073/pnas.86.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang WH, et al. NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc Natl Acad Sci U S A. 2008;105:17345–17350. doi: 10.1073/pnas.0806198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Housley MP, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: Post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 20.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 21.Sinclair DA, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 23.Love DC, et al. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci USA. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khidekel N, et al. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 25.Gurcel C, et al. Identification of new O-GlcNAc modified proteins using a click-chemistry-based tagging. Anal Bioanal Chem. 2008;390:2089–2097. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- 26.Ramakrishnan B, Qasba PK. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: Point mutation broadens beta 4Gal-T1 donor specificity. J Biol Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 27.Hassan YI, Zempleni J. A novel, enigmatic histone modification: Biotinylation of histones by holocarboxylase synthetase. Nutr Rev. 2008;66:721–725. doi: 10.1111/j.1753-4887.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 28.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal Biochem. 2008;373:71–77. doi: 10.1016/j.ab.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Holt GD, et al. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 1987;104:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 31.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 33.Wells L, et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Vosseller K, et al. Quantitative analysis of both protein expression and serine / threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics. 2005;5:388–398. doi: 10.1002/pmic.200401066. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Hart GW. Glycomic approaches to study GlcNAcylation: Protein identification, site-mapping, and site-specific O-GlcNAc quantitation. Clin Proteomics. 2008;4:5–13. [Google Scholar]
- 36.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Richmond TJ. DNA binding within the nucleosome core. Curr Opin Struct Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- 38.Imaoka T, Imazu M, Ishida N, Takeda M. Comparison of two forms of pig heart phosphoprotein phosphatase. Biochim Biophys Acta. 1978;523:109–120. doi: 10.1016/0005-2744(78)90014-1. [DOI] [PubMed] [Google Scholar]
- 39.Maile T, Kwoczynski S, Katzenberger RJ, Wassarman DA, Sauer F. TAF1 activates transcription by phosphorylation of serine 33 in histone H2B. Science. 2004;304:1010–1014. doi: 10.1126/science.1095001. [DOI] [PubMed] [Google Scholar]
- 40.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 41.Mayya V, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 42.Slawson C, et al. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 43.Bonenfant D, et al. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Zachara NE, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 45.Cheung WD, Hart GW. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J Biol Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyer SP, Hart GW. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 48.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-GlcNAc transferase substrate specificity is regulated by MYPT1 and other interacting proteins. J Biol Chem. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fritah S, et al. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol Biol Cell. 2009;20:4976–4984. doi: 10.1091/mbc.E09-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YX, et al. Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta. Chem Biol. 2006;13:937–944. doi: 10.1016/j.chembiol.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Simanek EE, et al. Glycosylation of threonine of the repeating unit of RNA polymerase II with b-linked N-acetylglucosamine leads to a turnlike structure. J Am Chem Soc. 1998;120:11567–11575. [Google Scholar]
- 53.Daly NL, Hoffmann R, Otvos L, Jr, Craik DJ. Role of phosphorylation in the conformation of tau peptides implicated in Alzheimer's disease. Biochemistry. 2000;39:9039–9046. doi: 10.1021/bi0004807. [DOI] [PubMed] [Google Scholar]
- 54.Fleming AB, Pennings S. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hyland EM, et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 57.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 58.Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007;26:4368–4379. doi: 10.1038/sj.emboj.7601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim HS, et al. Excessive O-GlcNAcylation of proteins suppresses spontaneous cardiogenesis in ES cells. FEBS Lett. 2009;583:2474–2478. doi: 10.1016/j.febslet.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 60.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorne JL, Campbell MJ, Turner BM. Transcription factors, chromatin and cancer. Int J Biochem Cell Biol. 2009;41:164–175. doi: 10.1016/j.biocel.2008.08.029. [DOI] [PubMed] [Google Scholar]
- 62.Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23:1849–1869. doi: 10.1101/gad.1807009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 64.Laszlo A, Fleischer I. Heat-induced perturbations of DNA damage signaling pathways are modulated by molecular chaperones. Cancer Res. 2009;69:2042–2049. doi: 10.1158/0008-5472.CAN-08-1639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.