Abstract
Regulated conformational changes of proteins are critical for cellular signal transduction. The spindle checkpoint protein Mad2 is an unusual protein with two native folds: the latent open conformer (O-Mad2) and the activated closed conformer (C-Mad2). During mitosis, cytosolic O-Mad2 binds to the Mad1–Mad2 core complex at unattached kinetochores and undergoes conformational activation to become C-Mad2. C-Mad2 binds to and inhibits Cdc20, an activator of APC/C, to prevent precocious anaphase onset. Here, we show that the conformational transition of Mad2 is regulated by phosphorylation of S195 in its C-terminal region. The phospho-mimicking Mad2S195D mutant and the phospho-S195 Mad2 protein obtained using intein-mediated semisynthesis do not form C-Mad2 on their own. Mad2S195D fails to bind to Cdc20, a low-affinity ligand, but still binds to high-affinity ligands, such as Mad1 and MBP1, forming ligand-bound C-Mad2. Overexpression of Mad2S195D in human cells causes checkpoint defects. Our results indicate that Mad2 phosphorylation inhibits its function through differentially regulating its binding to Mad1 and Cdc20 and establish that the conformational change of Mad2 is regulated by posttranslational mechanisms.
Keywords: conformational change, protein structure, spindle checkpoint, NMR, mitosis
Faithful duplication and segregation of chromosomes are critical tasks during the cell cycle. To ensure accurate chromosome segregation during mitosis, the spindle checkpoint inhibits the anaphase-promoting complex or cyclosome (APC/C) to prevent the degradation of securin and cyclin B until all sister chromatids are properly attached to spindle microtubules (1, 2). Key components of the spindle checkpoint include the evolutionarily conserved Mad (mitotic arrest deficiency) and Bub (budding uninhibited by benomyl) proteins. In response to unattached or untense kinetochores, these proteins collaborate to inhibit APC/C and block anaphase onset.
Among the APC/C-inhibitory checkpoint mechanisms, Mad2 directly binds to the mitotic APC/C activator Cdc20, as part of a large complex that also contains BubR1 and Bub3, and inhibits its ability to activate APC/C (3, 4). Mad2 is a two-state protein that has two native folds: a latent N1 or open conformer (O-Mad2) and an N2 or closed conformer (C-Mad2) that is more active in Cdc20 binding and APC/C inhibition (5–7). A major difference between the two Mad2 conformers lies in the conformation of their C-terminal region. O-Mad2 is a kinetically trapped high-energy conformer and spontaneously converts to C-Mad2 with slow kinetics (t1/2 on the order of hours) in vitro. In cells, cytosolic O-Mad2 is recruited to the kinetochore-bound Mad1–Mad2 core complex through O-Mad2–C-Mad2 asymmetric dimerization (7–12). O-Mad2 bound to the Mad1–Mad2 core converts to an intermediate conformer (I-Mad2), which may directly bind to Cdc20 (12–14). I-Mad2 can also complete the O─C conformational change on its own and dissociates from the Mad1–Mad2 core as unliganded C-Mad2, which then binds to Cdc20 (14–16).
After all sister kinetochores attach properly to spindle microtubules, the spindle checkpoint is inactivated to allow APC/C activation and anaphase onset. The Mad2 inhibitor p31comet is critical for checkpoint inactivation (17, 18). It binds to the Mad1–Mad2 core complex and prevents further Mad1-assisted conformation activation of Mad2 (13, 19). It also binds to Cdc20-bound Mad2 and promotes UbcH10-dependent Cdc20 autoubiquitination and the disassembly of the Mad2–Cdc20-containing APC/C-inhibitory complexes (18, 20, 21). Consistently, depletion of p31comet by RNA interference (RNAi) from HeLa cells delays mitotic exit following the release from nocodazole-mediated mitotic arrest (18). On the other hand, p31comet depletion does not cause a mitotic arrest in the absence of spindle poisons, suggesting the existence of additional checkpoint inactivation mechanisms.
The Mad2ΔC mutant with its C-terminal 10 residues deleted only adopts the O-Mad2 conformation and cannot bind to either Mad1 or Cdc20, indicating the importance of the C-terminal region of Mad2 in its conformational transition (7). Intriguingly, multiple residues in the C-terminal region of Mad2 are phosphorylated, including S170, S178, S185, and S195 (22). Overexpression of phospho-mimicking mutants Mad2-4D (with S170, S178, S185, and S195 mutated to aspartate) and Mad2-3D (with S170, S178, and S195 mutated to aspartate) that do not bind to either Mad1 or Cdc20 in human cells causes checkpoint defects, suggesting that Mad2 phosphorylation inhibits its function and may contribute to checkpoint inactivation (22). The mechanism by which these Mad2 phospho-mimicking mutants abrogate the spindle checkpoint in a dominant-negative manner is not understood.
Here, we investigated the effects of Mad2 phosphorylation on the unusual two-state behavior of Mad2. Using nuclear magnetic resonance (NMR) spectroscopy, we showed that phospho-mimicking mutations blocked the O─C conformational transition of Mad2 and identified S195 as a functionally important phosphorylation site. We synthesized a Mad2 protein specifically phosphorylated on S195 using intein-mediated ligation and confirmed that S195 phosphorylation greatly hindered the conformation change of Mad2. Mad2S195D failed to bind to Cdc20 with appreciable affinity but retained its binding to Mad1 and MBP1 (an unnatural Mad2-binding peptide identified in phage display). Thus, binding of high-affinity ligands, such as Mad1 and MBP1 to Mad2S195D, compensates for the unfavorable O─C Mad2 conformational change and drives the formation of ligand-bound C-Mad2. Overexpression of the phospho-mimicking Mad2S195D mutant abrogated the spindle checkpoint in cells. Unexpectedly, Mad2S195D exhibited enhanced binding to the Mad1–Mad2 core complex through O─C Mad2 dimerization, suggesting that S195 phosphorylation might regulate the dimerization between O-Mad2 and C-Mad2. Taken together, our results show that Mad2 phosphorylation regulates its conformational transition and might contribute to checkpoint inactivation through differentially regulating Mad2 binding to Mad1 and Cdc20.
Results and Discussion
The Phospho-Mimicking Mutants of Mad2 Preferentially Adopt the O-Mad2 Conformation.
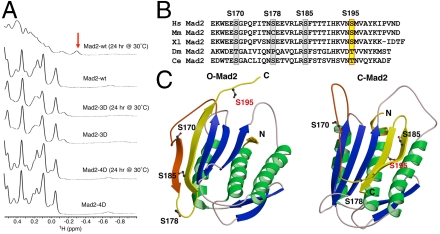
Previous studies showed that multiple residues including S170, S178, S185, and S195 in the C-terminal region of Mad2 were phosphorylated (22). Because the C-terminal region of Mad2 is critical for Mad2 structural conversion, we hypothesized that phosphorylation of this region of Mad2 regulated its conformational change. Two phospho-mimicking mutants of Mad2, Mad2-3D and Mad2-4D, blocked spindle checkpoint signaling in cells in a dominant-negative manner (22). To test the potential effect of phosphorylation on the two-state behavior of Mad2, we first monitored the conformational transition of Mad2-3D and Mad2-4D phospho-mimicking mutants in vitro using 1D 1H NMR spectroscopy. The wild-type (wt) Mad2, Mad2-3D, and Mad2-4D were purified in their O-Mad2 state with anion exchange chromatography at 4 °C and immediately analyzed with NMR. The 1D 1H NMR spectra of these proteins were almost identical in the high-field methyl region (Fig. 1A), indicating that the phospho-mimicking mutations did not grossly alter Mad2 folding and that Mad2-3D and Mad2-4D adopted O-Mad2 conformation. Upon incubation at 30 °C for 24 hrs, Mad2-wt underwent O─C conformational change and formed dimers, as evidenced by line broadening and the appearance of the -0.3 ppm peak that belonged to V197 in C-Mad2 (6, 7) (Fig. 1A, red arrow). By contrast, the spectra of Mad2-3D and Mad2-4D did not change appreciably after the 24-hr incubation at 30 °C, indicating that these mutants did not undergo the O─C conformational change during this time. Therefore, the phospho-mimicking mutations prevent Mad2 conformational transition.
Fig. 1.
The phospho-mimicking mutants of Mad2 fail to undergo the O─C conformational transition. (A) The high-field region of the 1D 1H spectra of wild-type Mad2 (Mad2-wt) or its phospho-mimicking mutants, Mad2-3D and Mad2-4D, before and after a 24-hr incubation at 30 °C. Mad2-3D contains S170D, S178D, and S195D mutations. Mad2-4D contains S170D, S178D, S185D, and S195D mutations. The -0.3 ppm peak that is unique to V197 in C-Mad2 is indicated by an arrow. (B) Sequence alignment of the C-terminal region of Mad2 proteins from Homo sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), Drosophila melanogaster (Dm), and Caenorhabditis elegans (Ce). (C) Ribbon drawing of the structures of O-Mad2 and C-Mad2. The α-helices are colored green, β-strands blue, and loops ivory. The structural elements of Mad2 that undergo major changes are in yellow and orange. S170, S178, S185, and S195 are shown as ball-and-stick.
Mad2-4D contained one additional mutation of S178, as compared to Mad2-3D. The fact that Mad2-3D and Mad2-4D behaved similarly in the cellular assays and in the NMR experiments suggested that phosphorylation of S178 might not be functionally important. Consistently, S178 was not conserved in other species (Fig. 1B). By contrast, S170, S185, and S195 were conserved in metazoans.
Phosphorylation of S195 Hinders the Conformational Transition of Mad2.
We sought to determine which phosphorylation site was critical for regulating the conformational change of Mad2. Inspection of the O- and C-Mad2 structures revealed that S170, S178, and S185 were surface exposed in both Mad2 conformers (Fig. 1C). By contrast, S195 was located in the C-terminal tail and was surface exposed in O-Mad2, but it became buried in the interior of C-Mad2. Phosphorylation of S195 would introduce negative charges in the hydrophobic core of C-Mad2 and was expected to destabilize C-Mad2 and to favor the O-Mad2 conformation. Furthermore, the S195A mutation further reduced the phosphorylation levels of the Mad2 S170A/S178A double mutant in human cells (22), indicating S195 was phosphorylated in vivo.
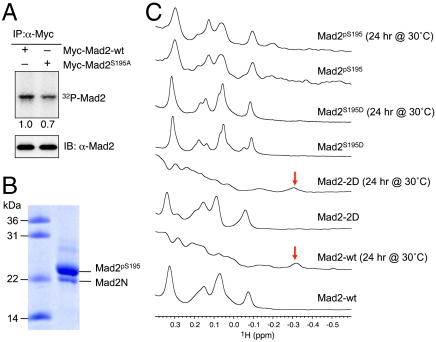
To verify that phosphorylation of Mad2 S195 occurred in vivo, we performed metabolic 32P labeling assays (Fig. 2A). Briefly, HeLa Tet-On cells were transfected with plasmids encoding Myc-tagged Mad2-wt or Mad2S195A, arrested in mitosis with nocodazole, and released into medium containing 32P-orthophosphate. The cells were lysed and immunoprecipitated with α-Myc antibodies. The levels of 32P incorporation into Myc-Mad2 were resolved by SDS/PAGE followed by autoradiography (Fig. 2A). Myc-Mad2S195A had 30% less 32P incorporation, as compared to Myc-Mad2-wt. This result confirmed that Mad2 S195 was indeed phosphorylated in vivo, although it was not the only site of phosphorylation.
Fig. 2.
Phosphorylation of S195 hinders the conformational transition of Mad2. (A) HeLa Tet-On cells were transfected with plasmids encoding Myc-tagged Mad2-wt or  , treated with nocodazole for 14 hrs, released into fresh media for 2 hrs, and labeled with 32P-orthophosphate for another 2 hrs. Myc-Mad2 proteins were immunoprecipitated with α-Myc antibody beads and blotted with α-Mad2 (Lower). The levels of 32P incorporation into Myc-Mad2 proteins were analyzed by a phosphoimager (Upper). The relative intensity of the 32P signal was quantified and normalized by the amounts of Myc-Mad2 proteins. (B) Coomassie-stained gel of phospho-S195 Mad2 (Mad2pS195) produced with intein-mediated protein ligation. A small amount of Mad2N not ligated to the C-terminal phospho-peptide was present. (C) The high-field region of the 1D 1H spectra of wild-type Mad2 (Mad2-wt), Mad2-2D (contains S170D and S185D mutations), Mad2S195D, and phospho-S195 Mad2 (Mad2pS195) before and after a 24-hr incubation at 30 °C. The -0.3 ppm peak that is unique to V197 in C-Mad2 is indicated by arrows.
, treated with nocodazole for 14 hrs, released into fresh media for 2 hrs, and labeled with 32P-orthophosphate for another 2 hrs. Myc-Mad2 proteins were immunoprecipitated with α-Myc antibody beads and blotted with α-Mad2 (Lower). The levels of 32P incorporation into Myc-Mad2 proteins were analyzed by a phosphoimager (Upper). The relative intensity of the 32P signal was quantified and normalized by the amounts of Myc-Mad2 proteins. (B) Coomassie-stained gel of phospho-S195 Mad2 (Mad2pS195) produced with intein-mediated protein ligation. A small amount of Mad2N not ligated to the C-terminal phospho-peptide was present. (C) The high-field region of the 1D 1H spectra of wild-type Mad2 (Mad2-wt), Mad2-2D (contains S170D and S185D mutations), Mad2S195D, and phospho-S195 Mad2 (Mad2pS195) before and after a 24-hr incubation at 30 °C. The -0.3 ppm peak that is unique to V197 in C-Mad2 is indicated by arrows.
We next tested whether phosphorylation of the S195 site alone affected the Mad2 conformational change. To do so, we expressed and purified the phospho-mimicking Mad2S195D mutant in the O-Mad2 state. We also generated the phospho-S195 Mad2 protein (Mad2pS195) in the O-Mad2 state using the intein-mediated protein ligation method (Fig. 2B) (23). Both Mad2S195D and Mad2pS195 proteins were incubated at 30 °C for 24 hrs and analyzed with 1D 1H NMR spectroscopy. The spectra of Mad2S195D and Mad2pS195 were very similar to that of O-Mad2-wt and remained unchanged after the 24-hr incubation at 30 °C (Fig. 2C). By contrast, the Mad2-2D mutant with S170 and S185 mutated to aspartate underwent the O─C conformational change after the incubation, similar to Mad2-wt (Fig. 2C). Therefore, phosphorylation of Mad2 S195 prevents the conformational transition of Mad2. Mad2S195D mimics this effect of S195 phosphorylation. Among the three conserved Mad2 phosphorylation sites, S195 is functionally most critical.
Mad2S195D Binds to Mad1 but Not to Cdc20.
Free Mad2S195D cannot form C-Mad2 by itself. We next tested whether Mad2S195D retained binding to Cdc20 or Mad1. We performed in vitro binding assays using recombinant purified Mad2-wt, Mad2S195D, and Mad2ΔC proteins. As the negative control, Mad2ΔC did not binding to either Mad1 or Cdc20, because it could not adopt the C-Mad2 conformation (Fig. S1). Consistent with previous reports (7, 24, 25), O-Mad2-wt bound less Cdc20 as compared to C-Mad2-wt, because C-Mad2-wt had a faster on-rate in Cdc20 binding (Fig. S1). O-Mad2-wt bound more Mad1 as compared to C-Mad2-wt, presumably because O-Mad2-wt had a faster on-rate in Mad1 binding. Mad2S195D (which only adopted the O-Mad2 conformation in the absence of ligands) did not bind to Cdc20 but retained Mad1 binding, albeit to a lesser extent than O-Mad2-wt. Because all Mad2 molecules were bound to beads prior to Mad1 addition, this assay measured the formation of the Mad1–Mad2 core complex. As expected from the effect of the S195D mutation in destabilizing the C-Mad2 conformation, Mad2S195D was less efficient than O-Mad2-wt in forming the Mad1–Mad2 core complex in vitro.
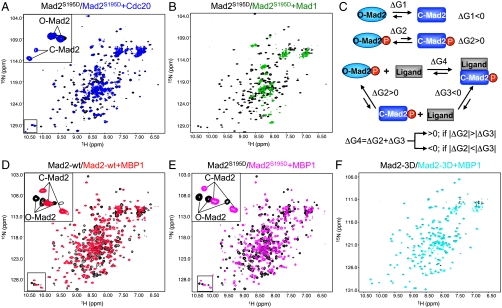
To confirm the different binding behavior of Mad2S195D toward Mad1 and Cdc20, we monitored the binding between Mad2S195D and the Mad2-binding domains of Cdc20 or Mad1 using NMR. Because C-Mad2 had a tendency to oligomerize, we used 15N-labeled monomeric mutants of Mad2 in these experiments. The 2D 1H/15N HSQC spectrum of Mad2S195D after the addition of Cdc20 contained two sets of peaks: One set belonged to free O-Mad2S195D, whereas the other set belonged to the C-Mad2S195D-Cdc20 complex (Fig. 3A). As shown in the inset of Fig. 3A, the peak intensities of C-Mad2S195D were about 30% of those of O-Mad2, indicating that less than 50% of Mad2S195D was bound to Cdc20 when both were present at about 100 μM. Therefore, Mad2S195D has minimal affinity toward Cdc20, with Kd of this interaction greater than 100 μM. By contrast, the majority of the peaks in the HSQC spectrum of Mad2S195D disappeared after the addition of Mad1 (Fig. 3B), consistent with the formation of the heterotetrameric 100 kD Mad1-Mad2S195D complex whose NMR signals were largely not observable.
Fig. 3.
Mad2S195D forms C-Mad2 when bound to high-affinity ligands. (A) Overlay of the 2D 1H/15N HSQC spectra of 15N-labeled Mad2S195D before (black contours) and after (blue contours) the addition of Cdc20. A region of the spectrum was magnified and shown in the inset, with the peaks belonging to O-Mad2 and C-Mad2 labeled. (B) Overlay of the 2D 1H/15N HSQC spectra of 15N-labeled Mad2S195D before (black contours) and after (green contours) the addition of Mad1. (C) Reaction equilibria for the conformational change and ligand binding of unmodified or phosphorylated Mad2. (D) Overlay of the 2D 1H/15N HSQC spectra of 15N-labeled Mad2-wt before (black contours) and after (red contours) the addition of MBP1. (E) Overlay of the 2D 1H/15N HSQC spectra of 15N-labeled Mad2S195D before (black contours) and after (magenta contours) the addition of MBP1. (F) Overlay of the 2D 1H/15N HSQC spectra of 15N-labeled Mad2-3D before (black contours) and after (cyan contours) the addition of MBP1.
To understand the different binding behavior of Mad2S195D toward Mad1 and Cdc20, we need to consider the thermodynamics of the following equilibrium reactions (Fig. 3C). The conversion of wild-type O-Mad2 to C-Mad2 is thermodynamically favored (ΔG1 < 0). When Mad2 is phosphorylated at S195 or contains the phospho-mimicking S195D mutation, the C-Mad2 conformer is selectively destabilized. The conversion of O-Mad2S195D to C-Mad2S195D is thermodynamically disfavored (ΔG2 > 0). The Gibbs free energy of the ligand-binding reaction of Mad2S195D (ΔG4) is the sum of the free energy of the Mad2S195D conformational change (ΔG2) and the free energy of C-Mad2S195D binding to the ligand (ΔG3). Ligand binding of C-Mad2S195D is a thermodynamically favorable reaction (ΔG3 < 0). If |ΔG3| (the absolute value of ΔG3) is greater than |ΔG2|, ΔG4 is negative, i.e., the formation of the C-Mad2S195D-ligand complex is favorable. Simply put, high-affinity Mad2 ligands can drive the formation of the C-Mad2S195D-ligand complex whereas low-affinity Mad2-binding ligands cannot. Because Mad1 binds to Mad2 more tightly than Cdc20, Mad1, but not Cdc20, binds to Mad2S195D with appreciable affinities.
Mad2S195D Forms C-Mad2 when Bound to High-affinity Ligands.
Because the NMR signals of the Mad1-Mad2S195D complex were largely not observable, we could not be certain that Mad2S195D indeed formed C-Mad2 when bound to Mad1. To ascertain that Mad2S195D could still form C-Mad2 when bound to high-affinity ligands, we examined by NMR the binding of Mad2S195D to MBP1, a high-affinity (Kd = 87 nM) Mad2-binding peptide (6). Addition of MBP1 caused drastic changes of the HSQC spectrum of O-Mad2, indicating that Mad2 adopted the C-Mad2 conformation when bound to MBP1 (Fig. 3D). MBP1 addition also greatly altered the spectrum of Mad2S195D (Fig. 3E). No peaks belonging to O-Mad2 were observable. The HSQC spectrum of Mad2S195D bound to MBP1 was highly similar to that of Mad2 bound to MBP1 (Fig. 3D and 3E). This result confirmed that Mad2S195D could indeed adopt the C-Mad2 conformation in the presence of high-affinity ligands, such as MBP1 and possibly Mad1.
Previous studies have shown that Mad2-3D and Mad2-4D, which contain additional phospho-mimicking mutations, do not bind to Mad1 to form the Mad1–Mad2 core complex (22). Consistently, addition of MBP1 did not alter the HSQC spectrum of Mad2-3D, indicating the lack of binding between Mad2-3D and MBP1 even at high concentrations (Fig. 3F). This result suggests that, in addition to S195 phosphorylation, other phosphorylation events on Mad2 might introduce more destabilization energy to the C-Mad2 conformation (i.e. ΔG2 of Mad2-3D is greater than that of Mad2S195D), which cannot be compensated by the binding energy between Mad2 and its high-affinity ligands, such as Mad1 and MBP1.
Mad2S195D Is Ineffective to Cause Mitotic Arrest.
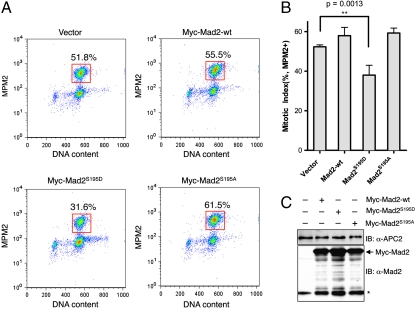
Mad2 S195 phosphorylation hinders the O─C conformational transition and diminishes Cdc20 binding in vitro. We tested whether this phosphorylation affected Mad2 function in human cells. HeLa Tet-On cells were transfected with cDNAs encoding Myc-tagged Mad2-wt, Mad2S195D, or Mad2S195A. The cells were fixed and stained with propidium iodide (PI) and the mitotic marker α-MPM2 and analyzed by flow cytometry (FACS). Mitotic cells had 4N DNA content and were MPM2-positive. Overexpression of Mad2-wt or Mad2S195A greatly increased the mitotic indices of HeLa cells (Fig. S2 A and B), indicating that high levels of Mad2 caused hyperactivation of the spindle checkpoint and blocked mitotic progression in the absence of spindle poisons. By contrast, overexpression of Mad2S195D was much less effective in triggering mitotic arrest. The three Myc-Mad2 proteins were expressed at similar levels (Fig. S2C). In addition, expression of Mad2-wt or Mad2S195A, but not Mad2S195D, increased the levels of another mitotic marker, phospho-histone H3 at S10 (Fig. S2C). Therefore, these results indicate that the Mad2S195D mutant is less active than Mad2-wt and Mad2S195A, consistent with S195 phosphorylation playing an inhibitory role.
The inhibitory effect of S195 phosphorylation could be attributed to its expected, selective destabilization of the C-Mad2 conformation. Mad2 adopts the C-Mad2 conformation when bound to Cdc20. In addition, unliganded C-Mad2 binds to Cdc20 with higher affinity than O-Mad2 does, because it binds to Cdc20 with a faster on-rate (7, 24, 25). Mad2 phosphorylation is thus expected to block the binding of Mad2 to its target Cdc20 and inhibits the function of Mad2. Indeed, as described above, the phospho-mimicking Mad2 mutants failed to bind to Cdc20 and were ineffective in eliciting mitotic arrest when overexpressed in human cells.
Mad2S195D Abrogates the Spindle Checkpoint.
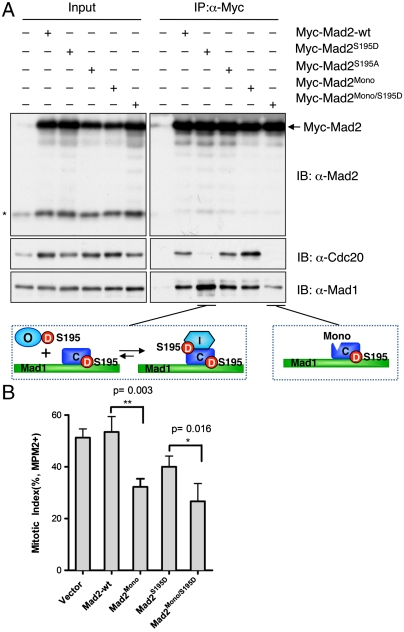
We then tested whether Mad2S195D expression affected the ability of HeLa cells to undergo mitotic arrest in the presence of spindle toxins, such as nocodazole. HeLa Tet-On cells were transfected with cDNAs encoding Myc-tagged Mad2-wt, Mad2S195D, or Mad2S195A, treated with 300 nM nocodazole for 16 hrs, stained with α-MPM2 and PI, and subjected to FACS analysis. Overexpression of Mad2S195D, but not Mad2-wt or Mad2S195A, significantly diminished the mitotic index of cells in the presence of nocodazole (Fig. 4 A and B). The expression levels of the Myc-tagged Mad2 proteins were similar (Fig. 4C). Because the endogenous Mad2 protein was not depleted from the cells in this experiment, Mad2S195D abrogated the spindle checkpoint in a dominant-negative manner.
Fig. 4.
Overexpression of Mad2S195D abrogates the spindle checkpoint. (A) The FACS analysis of HeLa Tet-On cells transfected with the indicated plasmids and treated with nocodazole. The populations of mitotic cells (with 4N DNA content and MPM2+) are boxed with the mitotic indices indicated. (B) Quantification of mitotic indices of cells described in A. The means and standard deviations of results from four experiments are shown. The p-value was calculated between Mad2S195D- and vector-transfected samples. (C) Lysates of cells described in A were blotted with the indicated antibodies. APC2 is used as the loading control. The asterisk indicates the position of the endogenous Mad2.
To determine the mechanism by which Mad2S195D blocked the function of endogenous Mad2 and abrogated checkpoint signaling, we examined the binding of Myc-Mad2S195D to the endogenous Mad1 and Cdc20 in HeLa cell lysates. HeLa Tet-On cells were transfected with control vectors or vectors encoding Myc-tagged Mad2-wt, Mad2S195D, or Mad2S195A and arrested in mitosis with nocodazole. The cell lysates were immunoprecipitated with α-Myc and blotted with α-Mad1 and α-Cdc20. Consistent with the effect of S195 phosphorylation in hindering the O─C conformational transition of Mad2, Myc-Mad2S195D failed to bind to Cdc20, whereas Myc-Mad2-wt and Myc-Mad2S195A bound to Cdc20 equally well (Fig. 5A). By contrast, Myc-Mad2S195D bound more efficiently to Mad1, as compared to Myc-Mad2-wt and Myc-Mad2S195A. The enhanced binding of Mad2S195D to Mad1 was reproducible and was observed in four independent experiments. This observation was completely unexpected. Although Mad2S195D retained binding to Mad1, it still bound with lower affinity to Mad1 as compared to Mad2-wt in vitro.
Fig. 5.
Mad2S195D binds to the Mad1–Mad2 core complex but not to Cdc20. (A) HeLa Tet-On cells were transfected with the indicated plasmids and treated with nocodazole. Lysates of these cells and the α-Myc immunoprecipitates were blotted with the indicated antibodies. The asterisk indicates the position of the endogenous Mad2. (B) Overexpression of Mad2Mono/S195D abrogates the spindle checkpoint through Mad1 sequestration. Quantification of mitotic indices of HeLa Tet-On cells transfected with the indicated plasmids and treated with nocodazole. The means and standard deviations of results from three experiments are shown. The p-values were calculated between the indicated samples.
How could Mad2S195D bind more efficiently to Mad1 in human cells then? We reasoned that the enhanced binding seen with Mad2S195D might be owing to its increased binding to the Mad1–Mad2 core complex through O─C Mad2 dimerization in cells. It has been previously shown that the Mad2R133E/Q134A mutant (referred to as Mad2Mono hereafter) could not form O─C Mad2 dimers. We constructed the Mad2Mono/S195D mutant, which lacked the ability to form O─C dimers and was expected to lose its binding to the Mad1–Mad2 core complex. We next examined the binding of Mad2Mono and Mad2Mono/S195D to Mad1 and Cdc20 in HeLa cells. Myc-Mad2Mono bound to similar amounts of Mad1 as Myc-Mad2-wt did (Fig. 5A), confirming that the observed Mad1 binding to Mad2-wt in this assay was mostly due to the formation of the Mad1-Mad2 core complex. By contrast, Myc-Mad2Mono/S195D bound less Mad1 than Myc-Mad2S195D did, indicating that the enhanced Mad1 binding by Mad2S195D was indeed caused by the binding of a second molecule of Mad2S195D to the Mad1–Mad2 core complex through O─C Mad2 asymmetric dimerization, which eventually adopted the I-Mad2 conformation (Fig. 5A). Similar results were obtained in Mad2-RNAi cells (Fig. S3). On the other hand, because of the presence of untagged Mad2 species in the Myc-Mad2-expressing cells, we could not ascertain that the endogenous Mad2 was efficiently depleted in all samples. Therefore, our results were consistent with the notion that binding of a second molecule of Mad2S195D to the Mad1–Mad2 core complex enhanced the overall affinity of Mad2 for Mad1, independent of the Mad2 form in the core. The underlying mechanism for this enhancement is unclear at present, but one possibility is that S195 phosphorylation enhances the binding affinity between I-Mad2 and the Mad1–C-Mad2 core.
We then tested whether Mad2Mono/S195D overexpression abrogated the spindle checkpoint. HeLa Tet-On cells were transfected with plasmids encoding Myc-tagged Mad2-wt, Mad2Mono, Mad2S195D, and Mad2Mono/S195D, treated with nocodazole, and analyzed with FACS. Mad2Mono/S195D blocked the spindle checkpoint to a greater extent than Mad2S195D did (Fig. 5B). Because Mad2Mono/S195D bound to Mad1, but not to Cdc20 or C-Mad2, its dominant-negative effects on the spindle checkpoint is very likely due to the sequestration of Mad1 through the formation of an inactive Mad1-Mad2Mono/S195D core complex. Surprisingly, Mad2Mono also acted in a dominant-negative manner, despite its ability to bind to Cdc20 (Fig. 5 A and B). We speculate that Mad2 binding to Cdc20 might be insufficient to inhibit APC/C, and the Mad2 dimerization interface might be required for APC/C inhibition.
Mechanisms for the Dominant-Negative Effects of Phospho-Mimicking Mad2 Mutants.
The fact that the phospho-mimicking mutants of Mad2 are inactive is not surprising, but the observation that these phospho-mimicking mutants inhibit the function of endogenous Mad2 and block checkpoint signaling in a dominant-negative fashion is puzzling. In particular, Mad2-3D and Mad2-4D do not bind to either Mad1 or Cdc20. How can they inhibit the functions of the endogenous Mad2? Our findings with Mad2S195D reported herein now provide a solution to this puzzle. Mad2S195D binds more Mad1 than Mad2-wt and Mad2Mono/S195D (a mutant of Mad2 that cannot form O─C Mad2 dimers) do, suggesting that the apparently enhanced binding of Mad2S195D to Mad1 is due to a higher affinity of Mad2S195D toward the Mad1–Mad2 core complex. We expect that Mad2-3D and Mad2-4D also bind to the Mad1–Mad2 core complex. Because they do not directly bind to Mad1, their enhanced indirect binding to the Mad1–Mad2 core complex is negated by the loss of the direct binding to Mad1. The fact that Mad2S195D retains Mad1 binding thus allows us to uncover the effect of these phospho-mimicking mutations (and presumably phosphorylation) in enhancing binding affinity of a second Mad2 molecule to the Mad1-Mad2 core complex.
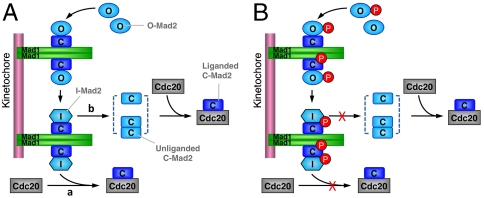
In the framework of the Mad2 conformational activation model (Fig. 6) (12, 16), these findings provide a straightforward explanation for the dominant-negative effects of phospho-mimicking mutants in cells. These mutants bind to the Mad1–Mad2 core with higher affinity than the endogenous Mad2 but cannot complete the O─C conformational transition, thereby blocking the conformational activation of the endogenous Mad2. Mad2Mono/S195D binds to Mad1, but not to Cdc20. Because Mad2Mono/S195D cannot form O─C Mad2 dimers, its binding to Mad1 creates a Mad1-Mad2Mono/S195D core complex that cannot further recruit and activate the endogenous O-Mad2 for Cdc20 binding. Therefore, Mad2Mono/S195D also blocks the spindle checkpoint in a dominant-negative manner. It has a stronger dominant-negative effect than Mad2S195D does, because the Mad1-Mad2S195D core complex can still recruit and activate the endogenous O-Mad2, albeit under the competition from free O-Mad2S195D.
Fig. 6.
Model depicting Mad1-assisted Mad2 conformational activation (A) and the proposed roles of Mad2 phosphorylation or phospho-mimicking mutations (B).
Potential Cellular Functions of Mad2 Phosphorylation.
What are the physiological functions of Mad2 phosphorylation? Is Mad2 phosphorylation regulated during the cell cycle? What are the kinases responsible for these phosphorylation events? Addressing these questions awaits the development of phospho-specific antibodies that can readily detect the phosphorylated forms of endogenous Mad2 in human cells. Nevertheless, it is clear that phosphorylation of Mad2 negatively regulates its activity. We envision two nonexclusive functions for Mad2 phosphorylation. First, Mad2 phosphorylation may attenuate spontaneous, unregulated conversion of O-Mad2 to C-Mad2 (which is more thermodynamically stable in vitro), thereby preventing unscheduled activation of Mad2. Second, Mad2 phosphorylation may contribute to checkpoint inactivation by capping the Mad1–Mad2 complex and blocking further conformational activation of Mad2, a function that is analogous to that of p31comet (16, 19).
Conclusion
In this study, we have further explored the functions of Mad2 phosphorylation and defined S195 as a critical phosphorylation site for regulating the conformational transition of Mad2. Our studies establish a posttranslational regulatory mechanism for the conformational change of Mad2. Furthermore, taking advantage of the unique binding properties of the phospho-mimicking Mad2S195D mutant, we have unexpectedly obtained evidence to suggest that S195 phosphorylation regulates O─C Mad2 dimerization. These results provide a straightforward explanation for the dominant-negative effects of Mad2 phospho-mimicking mutants and further support the in vivo relevance of conformational activation of Mad2 in the spindle checkpoint.
Materials and Methods
Cell Culture, Transfection, and Flow Cytometry (FACS).
HeLa Tet-On (Clontech) cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum. pCS2-Myc Mad2 mutant vectors were generated using the QuikChange XL mutagenesis kit (Stratagene). Plasmid and siRNA transfections were performed with the Effectene reagent (Qiagen) and Lipofectamine RNAiMAX (Invitrogen), respectively. When indicated, cells were arrested at metaphase with 300 nM nocodazole (Sigma) for 16–18 hrs prior to harvesting. Details of the FACS analysis are included in SI Text.
Metabolic 32P Labeling.
Metabolic 32P labeling was performed essentially as described previously (22). Details are included in SI Text.
Protein Purification and in Vitro Binding Assay.
Purification of Mad2 proteins and in vitro binding assays were performed as described (7, 14). Intein-mediated protein ligation is described in SI Text.
NMR Spectroscopy.
NMR experiments were performed at 30 °C on Varian Inova 600 or 800 MHz spectrometers equipped with four channels and pulsed-field gradients. 2D 1H/15N HSQC spectra were acquired on samples containing 0.8–1.2 mM 15N-labeled Mad2Mono (Mad2R133A), Mad2Mono/S195D (Mad2R133A/Q134E/S195D), and Mad2-3D in the NMR buffer (50 mM phosphate, pH 6.8, 300 mM KCl, and 1 mM DTT), in the absence or presence of 1∶1.2 molar ratio of unlabeled MBP1 or the Mad2-binding fragments of Cdc20 (residues 124–137 of human Cdc20) or Mad1 (residues 495–718 of human Mad1). To monitor the conformational change of Mad2, a series of 1D 1H NMR spectra, each lasting 30 min, was acquired on samples containing 0.1 mM Mad2-wt, Mad2-2D, Mad2-3D, Mad2-4D, Mad2S195D, and Mad2pS195 initially in the O-Mad2 state in the NMR buffer.
Supplementary Material
Acknowledgments.
We thank Anthony Pham for technical assistance with intein-mediated protein ligation. This research was supported by grants from the National Institutes of Health (GM61542 to H.Y. and GM085004 to X.L.) and the Welch Foundation (I-1441). H.Y. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009000107/-/DCSupplemental.
References
- 1.Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 2.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 3.Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- 4.Yu H. Cdc20: A WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Luo X, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 6.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 7.Luo X, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 8.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah JV, et al. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 10.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 11.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: Structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 12.Mapelli M, Musacchio A. MAD contortions: Conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol. 2007;17:716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Mapelli M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, et al. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:e50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: The two-state Mad2 model versus the Mad2 template model. J Cell Biol. 2006;173:153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo X, Yu H. Protein metamorphosis: The two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habu T, Kim SH, Weinstein J, Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21:6419–6428. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia G, et al. Conformation-specific binding of p31comet antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M, et al. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 21.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 22.Wassmann K, Liberal V, Benezra R. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J. 2003;22:797–806. doi: 10.1093/emboj/cdg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muralidharan V, Muir TW. Protein ligation: An enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 24.Simonetta M, et al. The influence of catalysis on Mad2 activation dynamics. PLoS Biol. 2009;7:e10. doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lad L, Lichtsteiner S, Hartman JJ, Wood KW, Sakowicz R. Kinetic analysis of Mad2-Cdc20 formation: conformational changes in Mad2 are catalyzed by a C-Mad2-ligand complex. Biochemistry. 2009;48:9503–9515. doi: 10.1021/bi900718e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.