In the context of the plethora of data currently generated in molecular biology, the paper by Bourgon et al. in PNAS (1) is pivotal, because it shows that an initial data filter can appropriately increase the detection power of a high-throughput experiment. Bourgon et al. (1) showed that filtering on overall variance outperforms filtering on overall mean, but they do not address two weaknesses of the methodology. First, because filtering is done on the overall variance, it does not disentangle the biological variation (containing the potentially interesting signals) from the technical variation (i.e., the measurement noise). Second, the threshold choice when the overall variation should be considered too low is very arbitrary and makes the method subjective (2). Filtering on reliability alleviates both problems. Furthermore, filtering on reliability will typically remove more irrelevant genes compared with overall variance filtering.
Reliability can statistically be defined as the consistency among a set of items that measure the same signal. If the signal can be measured consistently (i.e., if it is larger than the measurement noise), the measurement is called reliable. In the context of filtering, a gene is called reliable or informative when the biological signal exceeds the technical noise. Otherwise, the gene is called noninformative and should be filtered out. Although reliability filtering is widely applied in disciplines such as psychology (3) and has been proposed as a filtering tool for microarray data in the form of informative/noninformative calls (I/NI calls; ref. 4), it has not yet fully penetrated the field of molecular biology (2).
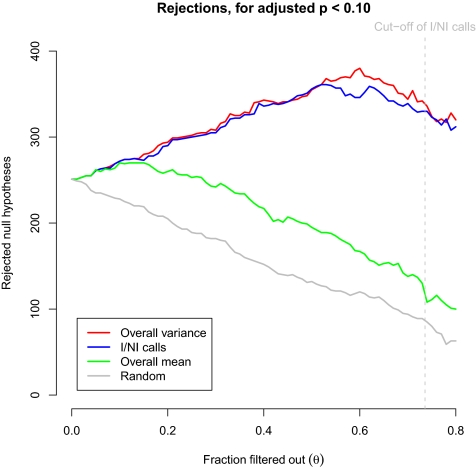
To include reliability filtering in the comparison study, we used the same B-cell acute lymphoblastic leukemia (ALL) samples as Bourgon et al. (1), reproduced their results, and superimposed the results of I/NI calls (4). Fig. 1 indicates that overall variance filtering and I/NI calls behave quite similarly, despite their strong conceptual differences, presumably because both go back to estimates of variation. It is the filtering threshold choice that makes the main difference between the two filtering methods. The threshold choice of I/NI calls is based on the point where the biological signal exceeds the technical noise. This provides a more objective foundation than an arbitrarily chosen fraction of tests passing the filter as for variance filtering. The impact of filtering threshold choice is difficult to compare, because it is arbitrary in the case of overall variance filtering. However, compared with the 50% filtering threshold used in the paper by Bourgon et al. (1), I/NI calls lead to considerably more genes being filtered out (73.6%) (Fig. 1). Because the threshold choice of I/NI calls is data-driven, it is also experiment-dependent. In experiments where many genes have a biological signal exceeding noise, fewer genes will be filtered out.
Fig. 1.
Effect of increasing filtering stringency on the number of rejected null hypotheses for the fixed adjusted P value cutoff of α = 0.1. The threshold choice of I/NI calls, the point where signal exceeds noise, is indicated and corresponds with a filtering proportion of 73.6%.
To conclude, I/NI calls filtering behaves overall quite similarly to variance filtering but has a more appealing concept and better threshold choice. More general information can be found in ref. 2, including some precautions when filtering on complex study designs. The software to run I/NI calls is available in the Farms package on Bioconductor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci USA. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Göhlmann H, Talloen W. Gene Expression Studies Using Affymetrix Microarrays. New York: Chapman & Hall; 2009. [Google Scholar]
- 3.Revelle W, Zinbarg R. Coefficients alpha, beta, omega, and the glb: Comments on Sijtsma. Psychometrika. 2009;74:145–154. [Google Scholar]
- 4.Talloen W, et al. I/NI-calls for the exclusion of non-informative genes: A highly effective filtering tool for microarray data. Bioinformatics. 2007;23:2897–2902. doi: 10.1093/bioinformatics/btm478. [DOI] [PubMed] [Google Scholar]