Abstract
Ubiquitylation of proteins can be a signal for a variety of cellular processes beyond the classical role in proteolysis. The different signaling functions of ubiquitylation are thought to rely on ubiquitin-binding domains (UBDs). Several distinct UBD families are known, but their functions are not understood in detail, and mechanisms for interpretation and transmission of the ubiquitin signals remain to be discovered. One interesting example of the complexity of ubiquitin signaling is the Saccharomyces cerevisiae transcription factor Met4, which is regulated by a single lysine-48 linked polyubiquitin chain that can directly repress activity of Met4 or induce degradation by the proteasome. Here we show that ubiquitin signaling in Met4 is controlled by its tandem UBD regions, consisting of a previously recognized ubiquitin-interacting motif and a novel ubiquitin-binding region, which lacks homology to known UBDs. The tandem arrangement of UBDs is required to protect ubiquitylated Met4 from degradation and enables direct inactivation of Met4 by ubiquitylation. Interestingly, protection from proteasomes is a portable feature of UBDs because a fusion of the tandem UBDs to the classic proteasome substrate Sic1 stabilized Sic1 in vivo in its ubiquitylated form. Using the well-defined Sic1 in vitro ubiquitylation system we demonstrate that the tandem UBDs inhibit efficient polyubiquitin chain elongation but have no effect on initiation of ubiquitylation. Importantly, we show that the nonproteolytic regulation enabled by the tandem UBDs is critical for ensuring rapid transcriptional responses to nutritional stress, thus demonstrating an important physiological function for tandem ubiquitin-binding domains that protect ubiquitylated proteins from degradation.
Keywords: protein degradation, polyubiquitin chain protection
The ubiquitin/proteasome system governs many aspects of cellular function such as cell cycle regulation, transcription, protein localization, and vesicular trafficking. Ubiquitin is a 76-residue protein that can be covalently attached to substrates by the E1-E2-E3 cascade of enzymes. Sequential addition of ubiquitin forms polyubiquitin chains, which are best known for their role as a degradation signal, but important proteolysis-independent functions of these chains are emerging as well (1, 2). Distinct signaling functions of ubiquitylation can be determined by the type of ubiquitylation, such as monoubiquitylation, multiubiquitylation (monoubiquitin attached to several substrate lysines), and polyubiquitylation with at least 8 different chain topologies found in vivo. Importantly, the effect of these signals seems to rely on ubiquitin-binding domains (UBDs) in target proteins (3). UBDs are often found as a part of ubiquitin receptors and proteins that promote ubiquitylation or deubiquitylation of substrates. The first UBD discovered was an ubiquitin-interacting motif (UIM) in the 26S proteasome subunit S5a/Rpn10 (4). To date, more than 20 distinct UBD families are known, with ubiquitin-associated domains (UBAs) and UIMs being the most common. The various functions of UBDs are not understood in detail, but they are clearly involved in proteasome targeting, substrate ubiquitylation, and regulation of protein/protein interactions (5). Both the UBA and UIM domains have also been shown to be able to protect polyubiquitylated proteins from degradation (6–8), and recently ubiquitin-binding domains have been used as tools to purify ubiquitylated proteins (9–11). However, little is known regarding the mechanism by which UBDs are able to protect ubiquitylated proteins from degradation and spatial requirements for these effects remain unexplored. Using the S. cerevisiae Met4 transcription factor we define a portable region containing the previously identified UIM domain and a previously undescribed UBD that prevents degradation of ubiquitylated proteins. We show that this tandem arrangement of ubiquitin-binding domains is necessary for efficient stabilization of polyubiquitylated proteins in vivo, and provide in vitro evidence that ubiquitin chain elongation is affected by the presence of the tandem UBD.
Results
Met4 Contains a Portable Stabilization Domain.
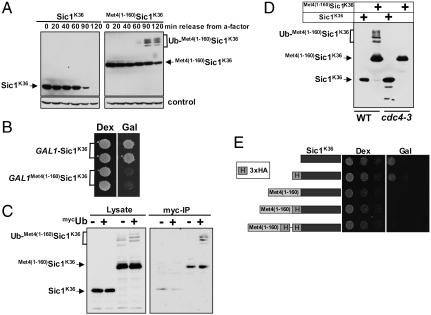
Previous studies have shown that the S. cerevisiae transcription factor Met4 contains a UIM, which is required for maintaining Met4 in a stable ubiquitylated form (6). This is remarkable because Met4 is modified with the canonical degradation signal, a K48-linked polyubiquitin chain, yet Met4 is shielded from proteasomal degradation (12). Furthermore, the function of this ubiquitin-binding region must be subject to regulation because protection is lost under some growth conditions resulting in degradation of Met4 (13, 14). Thus, Met4 presents the paradigm for regulatory functions of K-48-linked polyubiquitin chains and serves as a model system to gain detailed understanding about control of protein degradation and nonproteolytic functions for ubiquitylation. To further investigate this protective function, we tested whether the UIM is contained in a portable domain. We chose to fuse the 160 N-terminal residues comprising the UIM to the N-terminus of the well-studied proteasome substrate Sic1. Several ubiquitylation sites have been identified in Sic1, so to simplify interpretation of results we used the single ubiquitin acceptor variant SiclK36, where all lysine residues except K36 had been mutated (15). SiclK36 functions similarly to wild-type Sic1 in that it is rapidly polyubiquitylated and degraded as cells enter S-phase (15). To test whether the N-terminal region of Met4 can protect Sic1 from degradation we synchronized cells expressing SiclK36 or 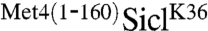 in G1, and monitored Sic1 protein levels as cells were released from the G1 cell cycle block (Fig. 1A). SiclK36 was rapidly degraded as cells entered S-phase, but
in G1, and monitored Sic1 protein levels as cells were released from the G1 cell cycle block (Fig. 1A). SiclK36 was rapidly degraded as cells entered S-phase, but 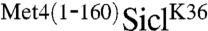 remained stable and high molecular weight derivatives of SiclK36 appeared, suggesting that the N-terminal region of Met4 contains a portable stabilization motif. Protection from degradation was not complete, and some degradation occurred despite fusion with the Met4 N-terminus. In addition to the significant stabilization, expression of
remained stable and high molecular weight derivatives of SiclK36 appeared, suggesting that the N-terminal region of Met4 contains a portable stabilization motif. Protection from degradation was not complete, and some degradation occurred despite fusion with the Met4 N-terminus. In addition to the significant stabilization, expression of 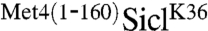 , but not SiclK36, blocked cell proliferation (Fig. 1B). This is consistent with a stabilizing activity in the N-terminus of Met4, because Sic1 degradation is required for G1/S transition, and cells unable to degrade Sic1 undergo cell cycle arrest (16).
, but not SiclK36, blocked cell proliferation (Fig. 1B). This is consistent with a stabilizing activity in the N-terminus of Met4, because Sic1 degradation is required for G1/S transition, and cells unable to degrade Sic1 undergo cell cycle arrest (16).
Fig. 1.
(A). 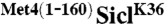 is stable and leads to cell cycle arrest. (A) Cells expressing RGS6His tagged SiclK36 or
is stable and leads to cell cycle arrest. (A) Cells expressing RGS6His tagged SiclK36 or 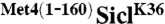 under control of the GAL1 promoter were synchronized in G1 using α-factor. Cells were released from the G1 arrest and expression from the GAL1 promoter was blocked by shifting cells to dextrose media. Protein levels were monitored at indicated time points. (B) Cells as in (A) were plated on dextrose (no expression) or galactose (expression) media. (C) Cells expressing RGS6His-SiclK36 or RGS6His-
under control of the GAL1 promoter were synchronized in G1 using α-factor. Cells were released from the G1 arrest and expression from the GAL1 promoter was blocked by shifting cells to dextrose media. Protein levels were monitored at indicated time points. (B) Cells as in (A) were plated on dextrose (no expression) or galactose (expression) media. (C) Cells expressing RGS6His-SiclK36 or RGS6His-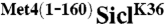 were transformed with either untagged ubiquitin or myc-ubiquitin. Ubiquitylated proteins were immunopurified using α-myc antibodies and samples were analyzed by immunoblotting using RGS6His antibodies. The signal detected for unmodified RGS6His-
were transformed with either untagged ubiquitin or myc-ubiquitin. Ubiquitylated proteins were immunopurified using α-myc antibodies and samples were analyzed by immunoblotting using RGS6His antibodies. The signal detected for unmodified RGS6His-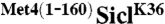 in the “myc-IP” lanes is due to low nonspecific binding to the α-myc resin. (D) WT or cdc4-3 cells expressing RGS6His-SiclK36 or RGS6His-
in the “myc-IP” lanes is due to low nonspecific binding to the α-myc resin. (D) WT or cdc4-3 cells expressing RGS6His-SiclK36 or RGS6His-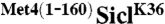 were shifted to the restrictive temperature (37 °C) and whole cell lysates were analyzed by immunoblotting with RGS6His antibodies. (E) Cells expressing the indicated proteins under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose media or galactose media. “H” refers to 3 copies of the HA epitope.
were shifted to the restrictive temperature (37 °C) and whole cell lysates were analyzed by immunoblotting with RGS6His antibodies. (E) Cells expressing the indicated proteins under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose media or galactose media. “H” refers to 3 copies of the HA epitope.
We next confirmed that the slower migrating species of 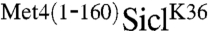 observed in Fig. 1A were ubiquitylated forms, because expression of myc-tagged ubiquitin further reduced mobility, and these low-mobility species of
observed in Fig. 1A were ubiquitylated forms, because expression of myc-tagged ubiquitin further reduced mobility, and these low-mobility species of 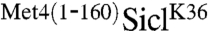 were selectively immunopurified with α-myc antibodies (Fig. 1C). Furthermore,
were selectively immunopurified with α-myc antibodies (Fig. 1C). Furthermore, 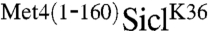 ubiquitylation was dependent on Sic1’s physiological ubiquitin ligase SCFCdc4 (17, 18) because high molecular forms were absent in cdc4 mutants (Fig. 1D). In contrast, Met4 ubiquitylation is catalyzed by SCFMet30 (19, 20) demonstrating that the protective activity contained in the Met4 N-terminus is an autonomous function, independent from a specific ubiquitin ligase.
ubiquitylation was dependent on Sic1’s physiological ubiquitin ligase SCFCdc4 (17, 18) because high molecular forms were absent in cdc4 mutants (Fig. 1D). In contrast, Met4 ubiquitylation is catalyzed by SCFMet30 (19, 20) demonstrating that the protective activity contained in the Met4 N-terminus is an autonomous function, independent from a specific ubiquitin ligase.
The Distance Between Ubiquitin Acceptor and Stabilization Domain.
We had previously identified lysine-163 as the sole ubiquitin acceptor site in Met4 (12), which is in close proximity to the UIM domain located around residue 145 (6). The spacing between ubiquitin acceptor site (lysine 36) and UIM in the 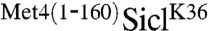 fusion was 30 residues larger than in the native Met4, suggesting that a precise distance is not essential. To further test the effect of spacing on the protective function of the Met4-UIM region, we took advantage of cell growth inhibition by the stabilized
fusion was 30 residues larger than in the native Met4, suggesting that a precise distance is not essential. To further test the effect of spacing on the protective function of the Met4-UIM region, we took advantage of cell growth inhibition by the stabilized 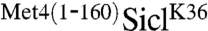 fusion and analyzed constructs with three or six copies of the HA-epitope separating the ubiquitin acceptor site and the UIM (Fig. 1E). Insertion of HA-epitopes did not significantly alter the potent antiproliferative effect of
fusion and analyzed constructs with three or six copies of the HA-epitope separating the ubiquitin acceptor site and the UIM (Fig. 1E). Insertion of HA-epitopes did not significantly alter the potent antiproliferative effect of 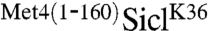 constructs, indicating that stabilization was not dependent on a precise positioning of the Met4-UIM region relative to the ubiquitylation site. This result was further confirmed by measuring the effect of the HA-epitope insertions on
constructs, indicating that stabilization was not dependent on a precise positioning of the Met4-UIM region relative to the ubiquitylation site. This result was further confirmed by measuring the effect of the HA-epitope insertions on 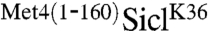 stability using Gal-shut-off experiments. Increasing the distance between the Met4-UIM and the ubiquitin acceptor site did not significantly alter the stability of
stability using Gal-shut-off experiments. Increasing the distance between the Met4-UIM and the ubiquitin acceptor site did not significantly alter the stability of 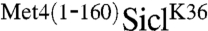 (Fig. S1). We are aware that insertion of spacer regions does not necessarily change proximity of the two regions in three dimensions. Covalent linkage of the Met4-UIM region to Sic1 was required for protection, because expression of the Met4-UIM region in trans could not protect SiclK36 (Fig. S2). Together these results suggest that stabilization is achieved in a range of different spatial configurations.
(Fig. S1). We are aware that insertion of spacer regions does not necessarily change proximity of the two regions in three dimensions. Covalent linkage of the Met4-UIM region to Sic1 was required for protection, because expression of the Met4-UIM region in trans could not protect SiclK36 (Fig. S2). Together these results suggest that stabilization is achieved in a range of different spatial configurations.
The Met4 Stabilization Domain Inhibits Efficient Polyubiquitin Chain Elongation in Vitro.
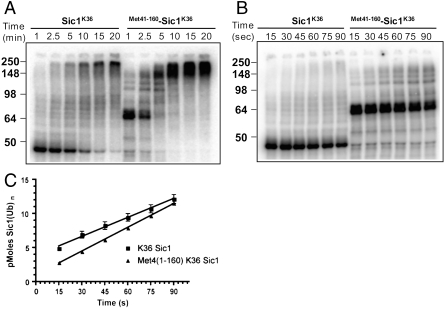
Met4 is modified by a relatively short polyubiquitin chain. We had previously suggested that the UIM region located in the Met4 N-terminus restricts polyubiquitin chain length in vivo (6). To more directly test this idea we made use of the well-defined Sic1 in vitro ubiquitylation reaction. SiclK36 and 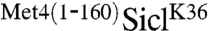 were produced and purified from Escherichia coli, phosphorylated in vitro, and incubated with an SCFCdc4 reaction mix. Both SiclK36 and
were produced and purified from Escherichia coli, phosphorylated in vitro, and incubated with an SCFCdc4 reaction mix. Both SiclK36 and 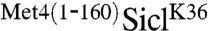 were efficiently ubiquitylated in vitro, but the presence of the N-terminal region of Met4 significantly reduced the polyubiquitin chain length attached to SiclK36 (Fig. 2A and B). A short reaction time course that captured the linear phase of ubiquitylation showed very similar rates of modification. Because the overall rate of Sic1 ubiquitylation is directly related to conjugation of the first ubiquitin—the rate limiting step of the reaction—(21), we conclude that conjugation of the first ubiquitin is not significantly affected by the Met4 N-terminus (Fig. 2B and C). We did notice slightly slower ubiquitylation for
were efficiently ubiquitylated in vitro, but the presence of the N-terminal region of Met4 significantly reduced the polyubiquitin chain length attached to SiclK36 (Fig. 2A and B). A short reaction time course that captured the linear phase of ubiquitylation showed very similar rates of modification. Because the overall rate of Sic1 ubiquitylation is directly related to conjugation of the first ubiquitin—the rate limiting step of the reaction—(21), we conclude that conjugation of the first ubiquitin is not significantly affected by the Met4 N-terminus (Fig. 2B and C). We did notice slightly slower ubiquitylation for 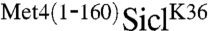 at the very early time points (Fig. 2C), indicating that the Met4 N-terminus might have a small effect on conjugation of the first ubiquitin molecule. However, overall the shorter polyubiquitin chain length on
at the very early time points (Fig. 2C), indicating that the Met4 N-terminus might have a small effect on conjugation of the first ubiquitin molecule. However, overall the shorter polyubiquitin chain length on 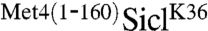 is likely predominantly due to inhibition of chain elongation, probably by stochastic capping of the terminal ubiquitin moiety. Similar inhibition of polyubiquitin chain extension has been observed after addition of Rad23 to in vitro reactions, which contains two UBA domains and can interact with polyubiquitin chains (9, 22).
is likely predominantly due to inhibition of chain elongation, probably by stochastic capping of the terminal ubiquitin moiety. Similar inhibition of polyubiquitin chain extension has been observed after addition of Rad23 to in vitro reactions, which contains two UBA domains and can interact with polyubiquitin chains (9, 22).
Fig. 2.
Fusion of Met41-160 to the N-terminus of the single lysine substrate SiclK36 is inhibitory for polyubiquitin chain elongation but not for transfer of the first ubiquitin to substrate. (A) Time-course comparing the ubiquitylation of SiclK36 (lanes 1–6) or 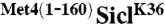 (lanes 7–12). Notice that ubiquitylated SiclK36 migrates higher in the gel than ubiquitylated
(lanes 7–12). Notice that ubiquitylated SiclK36 migrates higher in the gel than ubiquitylated 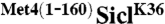 , which is indicative of a higher degree of modification. (B) same as (A), except the time-course has been shortened to capture the initial linear phase of the ubiquitylation reaction. (C) Graph showing the rate of Sic1 ubiquitylation, where substrate is defined as unmodified SiclK36 or
, which is indicative of a higher degree of modification. (B) same as (A), except the time-course has been shortened to capture the initial linear phase of the ubiquitylation reaction. (C) Graph showing the rate of Sic1 ubiquitylation, where substrate is defined as unmodified SiclK36 or 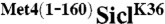 and product is substrate modified by one or more ubiquitins. Each graphical data point represents the mean of triplicate data values from three independent experiments, and the error bars are the standard deviations for those measurements.
and product is substrate modified by one or more ubiquitins. Each graphical data point represents the mean of triplicate data values from three independent experiments, and the error bars are the standard deviations for those measurements.
Two Conserved Domains are Important for Polyubiquitin Chain Protection.
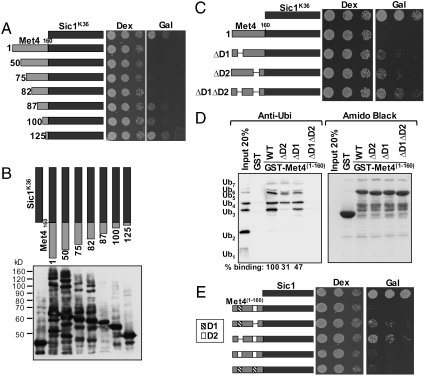
We next created a series of truncations of the Met4 N-terminus to identify the minimal region required for protection from degradation. Truncations beyond residue 82 were significantly less potent in stabilizing Sic1, as indicated by increased cell proliferation (Fig. 3A), and reduction in steady-state levels of ubiquitylated forms of Sic1 (Fig. 3B). This was unexpected because all analyzed truncations contained the UIM domain (residues 135–155). We suspected that an additional domain located between residue 83 and the previously characterized UIM was required for stabilization of polyubiquitylated proteins. Indeed, alignment of Met4 from different yeast species identified a highly conserved region between residues 86 and 96 (Fig. S3). To test the importance of this region in protecting ubiquitylated Sic1, we expressed 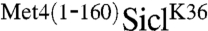 fusion proteins containing either deletions of region 86–96 (referred to as D1), deletion of the previously defined UIM (144–149) (referred to as D2 in Fig. 3C), or both domains and analyzed cell proliferation (Fig. 3C). Deletion of either D1 or D2 allowed partial cell proliferation, and mutations in both regions almost completely abolished the stabilizing activity of the Met4 N-terminal region, indicating that both D1 and D2 are important for stabilizing ubiquitylated SiclK36.
fusion proteins containing either deletions of region 86–96 (referred to as D1), deletion of the previously defined UIM (144–149) (referred to as D2 in Fig. 3C), or both domains and analyzed cell proliferation (Fig. 3C). Deletion of either D1 or D2 allowed partial cell proliferation, and mutations in both regions almost completely abolished the stabilizing activity of the Met4 N-terminal region, indicating that both D1 and D2 are important for stabilizing ubiquitylated SiclK36.
Fig. 3.
Met41-160 contains two ubiquitin-binding domains. (A) Cells expressing SiclK36 or 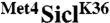 fusions under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose or galactose media. (B) Whole cell lysates from cells expressing RGS6His tagged SiclK36 or versions of
fusions under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose or galactose media. (B) Whole cell lysates from cells expressing RGS6His tagged SiclK36 or versions of 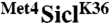 fusions were analyzed by immunoblotting with RGS6His antibodies. (C) Cells expressing the indicated proteins under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose media or galactose media. Region D1 represents residues 86–96, D2 represents previously defined UIM residues 144–149. (D) GST-tagged Met41-160 and deletion mutants were expressed in E. coli, purified on glutathione beads, and incubated with K48-linked polyubiquitin chains. (Left) Purified fractions were separated by SDS-PAGE and probed with α-ubiquitin antibodies. Quantitation of signal intensities are shown at the bottom. (Right) Amido black staining of purified GST-tagged proteins. (E) Cells expressing the indicated proteins under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose media or galactose media.
fusions were analyzed by immunoblotting with RGS6His antibodies. (C) Cells expressing the indicated proteins under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose media or galactose media. Region D1 represents residues 86–96, D2 represents previously defined UIM residues 144–149. (D) GST-tagged Met41-160 and deletion mutants were expressed in E. coli, purified on glutathione beads, and incubated with K48-linked polyubiquitin chains. (Left) Purified fractions were separated by SDS-PAGE and probed with α-ubiquitin antibodies. Quantitation of signal intensities are shown at the bottom. (Right) Amido black staining of purified GST-tagged proteins. (E) Cells expressing the indicated proteins under control of the inducible GAL1 promoter were plated in serial dilutions on either dextrose media or galactose media.
Met4 Residues 86–96 Define a Ubiquitin-Binding Domain.
We next assessed the importance of the Met4 region 86–96 (D1) for binding to ubiquitin in vitro. GST-tagged versions of the N-terminal 160 residues of Met4 with small deletions of domains D2 (UIM), D1 (∆86–96), or both D1 and D2 were expressed in E. coli, immobilized on glutathione beads, and incubated with K-48 linked polyubiquitin chains ranging from monoubiquitin to seven ubiquitins. Bound fractions were analyzed by immunoblotting with α-ubiquitin (Fig. 3D). Met4(1–160) did not bind monoubiquitin but efficiently bound polyubiquitin chains ranging from 3 to 7 ubiquitin residues. Deletion of D1 or D2 (UIM) significantly reduced binding, and combined mutations in the two regions completely abolished binding to polyubiquitin chains. Previous experiments with the N-terminal 270 residues of Met4 and radiolabeled K48-tetra ubiquitin showed a more dramatic effect of the UIM mutation (6). It is unclear whether this is due to the use of a larger Met4 fragment or because of differences in the polyubiquitin chains used. The previous study relied on polyubiquitin chains formed with ubiquitin in which all lysine residues except K48 had been mutated to alanine, whereas this study used commercial chains, which have a K29R mutation but are otherwise unmodified. Interestingly, none of the N-terminal Met4 fragments bound mono- or diubiquitin.
These results suggested that both the UIM and D1 define domains that bind polyubiquitin chains. However, it was formally possible that both regions are part of one larger domain, although this was unlikely because the Met4-UIM region is clearly a member of the well-defined UIM family (6, 23). Nevertheless, to address this question in vivo, we generated 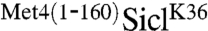 fusions with tandem copies of either the UIM (residues 131–160 duplicated) or D1 (residues 86–112 duplicated). Both tandem configurations were significantly more potent in stabilizing Sic1 as compared to the single domain mutations (Fig. 3E). Most importantly, the construct containing duplication of region D1 lacked the UIM but was nevertheless potent in protecting Sic1 from degradation. Together the in vitro binding assay (Fig. 3D) and in vivo protection results (Fig. 3E) strongly support that domains D1 and D2 are independent ubiquitin-binding domains that act in concert to protect polyubiquitylated proteins from degradation. Whereas the UIM is well-defined, the region between residues 86–96 lacks key residues characteristic of the canonical UIM (23). However, this unique ubiquitin-binding motif clearly shares several features with UIMs such as the small size and a predicted alpha-helical structure. We termed this motif a UIM-like domain.
fusions with tandem copies of either the UIM (residues 131–160 duplicated) or D1 (residues 86–112 duplicated). Both tandem configurations were significantly more potent in stabilizing Sic1 as compared to the single domain mutations (Fig. 3E). Most importantly, the construct containing duplication of region D1 lacked the UIM but was nevertheless potent in protecting Sic1 from degradation. Together the in vitro binding assay (Fig. 3D) and in vivo protection results (Fig. 3E) strongly support that domains D1 and D2 are independent ubiquitin-binding domains that act in concert to protect polyubiquitylated proteins from degradation. Whereas the UIM is well-defined, the region between residues 86–96 lacks key residues characteristic of the canonical UIM (23). However, this unique ubiquitin-binding motif clearly shares several features with UIMs such as the small size and a predicted alpha-helical structure. We termed this motif a UIM-like domain.
Met4 Is Regulated by Two Ubiquitin-Binding Domains.
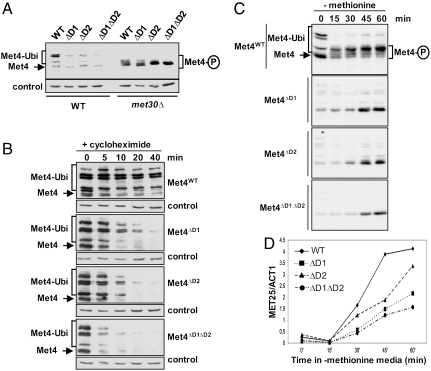
During normal growth conditions Met4 is usually sequestered in an inactive, polyubiquitylated form, which is protected from the proteasome (12–14, 19). To evaluate the contribution of the individual ubiquitin-binding domains to Met4 stabilization, we expressed various Met4 D1 and D2 mutants under control of the MET4 promoter and analyzed steady-state levels (Fig. 4A). Mutations in either region or their combination led to a significant decrease in Met4 steady-state levels. Reduced Met4 levels were due to SCFMet30-dependent degradation because deletion of MET30 not only abolished Met4 ubiquitylation but also restored normal steady-state levels of Met4 (Fig. 4A). We noted that mutations in the UIM region also changed the phosphorylation pattern, but the significance of this observation is still unknown (Fig. 4A). To further characterize the importance of the UIM-like domain, we measured degradation of Met4 mutants (Fig. 4B). Wild-type Met4 was very stable and mutations in the UIM or the UIM-like region significantly destabilized Met4. Importantly, Met4 lacking both UIM and UIM-like functions was degraded faster than either single domain mutant (Fig. 4B). Thus, the presence of the UIM and UIM-like domains convert a highly unstable protein with a half-life of less than 5 min to an almost completely stable, yet K48-polyubiquitylated protein.
Fig. 4.
Both Met4 ubiquitin-binding domains are required to maintain stably polyubiquitylated Met4. (A) Steady-state levels of Met4 or Met4 ubiquitin-binding domain mutants expressed from the MET4 promoter in wild-type cells or met30Δ mutants. (B) Cells expressing Met4 or Met4 ubiquitin-binding domain mutants under the endogenous promoter were treated with cycloheximide to block protein expression, and samples as indicated were analyzed by immunoblotting. (C) Cells expressing the indicated proteins under control of the MET4 promoter were grown to midlog phase in medium containing methionine. Cells were then shifted to medium lacking methionine to activate Met4 and samples as indicated were analyzed by immunoblotting. (D) Experiment as in (C), but RNA levels of the Met4 target gene MET25 were analyzed by RT-qPCR and plotted normalized to actin (ACT1).
To address the biological importance of the UIM and UIM-like domains we tested the transcriptional response of Met4 D1 and D2 mutants to methionine stress. As previously suggested, maintaining a pool of inactive, polyubiquitylated Met4 rather than inactivation by degradation should allow cells to react more rapidly to environmental and nutrient stress (6). To test this hypothesis we compared wild-type Met4 and mutants lacking individual or both UBDs. We first confirmed that in contrast to wild-type Met4, whose transcriptional activity is controlled by polyubiquitylation independently of proteolysis, polyubiquitylation regulates the mutant versions of Met4 at the level of protein abundance (Fig. 4C). To this end, cells expressing endogenous levels of Met4 were shifted to methionine-lacking medium to activate Met4. As expected, the levels of wild-type Met4 remained constant but a dramatic change in modification from the transcriptionally inactive polyubiquitylated species to phosphorylated active forms was evident. In contrast, Met4 mutants lacking UBDs responded to methionine depletion by accumulation of Met4 protein levels. We next followed induction kinetics for the Met4-dependent gene MET25 in response to methionine depletion (Fig. 4D). Cells expressing wild-type Met4 responded significantly faster to nutritional stress than either single UBD or double UBD mutants. These results support the notion that the UBD regions in Met4 allow proteolysis-independent regulation by polyubiquitylation, which permits a more rapid response to nutrient stress than regulation by ubiquitin-mediated degradation.
Discussion
Our study defines a tandem ubiquitin-binding domain in the N-terminus of the transcription factor Met4 that protects polyubiquitylated proteins from degradation by the proteasome. Importantly, the tandem UBDs of Met4 function autonomously in that they can be functionally fused to other proteins. Ubiquitin-associated domains (UBAs), which also bind ubiquitin, have previously been reported to protect proteins in cis from degradation by the proteasome (7, 24). However, the mechanism of stabilization seems to be different from that of the N-terminal region in Met4. A recent report suggests that protein stabilization by UBA domains does not correlate with the ability to bind ubiquitin, but rather with the structural integrity of the domain (24). UBA domains might thus interfere with processes at the proteasome and not with recognition of ubiquitylated substrates by the proteasome. Consistent with this idea, fusion of the UBA2 domain of Rad23 to SiclK36 completely stabilized Sic1, but the vast majority of 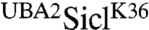 was in its deubiquitylated form (Fig. S4). This is in stark contrast to the function of the Met4 tandem UBDs, which stabilized Sic1 mainly as ubiquitylated species. The mechanism of stabilization by the Met4 tandem UBDs is therefore distinct from UBA-mediated stabilization.
was in its deubiquitylated form (Fig. S4). This is in stark contrast to the function of the Met4 tandem UBDs, which stabilized Sic1 mainly as ubiquitylated species. The mechanism of stabilization by the Met4 tandem UBDs is therefore distinct from UBA-mediated stabilization.
To the best of our knowledge, we report here a unique example of a physiologically relevant domain that protects proteins in their ubiquitylated form from degradation by the proteasome. By protecting the ubiquitylated form of a protein, the tandem UBDs of Met4 enable the polyubiquitin chain to function as an activity switch that is independent of proteolysis. We demonstrate that this ubiquitylation-based regulatory mechanism uncouples the cellular response to nutrient stress from new protein synthesis and therefore makes possible a faster adaptation to stress. How and why a canonical degradation signal (K48-polyubiquitin chain) evolved to be a functionally different regulatory device is not completely clear, in part because cells could simply use monoubiquitylation or regulatory K63-polyubiquitin chains instead. However, under certain physiological growth conditions Met4 is degraded by the proteasome (13, 14, 25), and a conditional degradation signal that is masked by the tandem UBDs under normal growth conditions might be an effective way to integrate several regulatory pathways. Interestingly, such a conditional degradation function implies that the protective function of the Met4 tandem UBDs is regulated either by posttranslational modifications or specific binding proteins that neutralize UBD activity.
Tandem ubiquitin-binding domains are frequently observed in proteins (23), and it has recently been shown that polyubiquitin-chain topology-specific binding can be achieved by such tandem UBDs (26). Our results demonstrate that the tandem arrangement of ubiquitin-binding domains can have important physiological functions by protecting ubiquitylated forms of proteins from degradation by the 26S proteasome. Tandem ubiquitin-binding domains might thus be critical mediators of the functions of ubiquitylation that do not involve proteolysis.
Materials and Methods
Protein Analysis.
Lysis conditions for immunoblotting (denaturing conditions in urea-buffer) and immunopurification (Triton-buffer) were as described (6). Antibodies used in this study were as follows: α-myc 9E10 (1∶2000; Covance), α-RGS4H (1∶2000; Qiagen), and α-ubiquitin P4G7 (1∶2000; Santa Cruz Biotechnology) for immunoblotting. α-myc antibodies (SC-789-G; Santa Cruz Biotechnology, CA) for immunopurification.
Ubiquitin Binding.
E. coli expressing GST or GST-tagged Met4 variants were lysed in Triton-buffer (6). Proteins were bound to glutathione beads (50 μL bead slurry/1 mg protein) for 4 h at 4 °C, beads were washed, and incubated with 5 μg K48-linked polyubiquitin chains Ub2-7 (Enzo Life Sciences International, Inc.) in 50 μL Ub binding buffer (0.05% NP-40, 20 mM Tris, pH 7.5, 10 mM NaCl, 25 μg/mL BSA) for 1 h at room temperature. Ubiquitin binding was analyzed by immunoblotting.
Yeast Methods and Protein Half-Life.
Standard yeast methods and growth conditions were used (27). For the spot assays, cells were grown to A600 = 0.5 and serially diluted in 5-fold increments starting with 5000 cells. Cells were spotted onto the indicated agar plates. To measure Sic1 degradation cells expressing Sic1 fusions as indicated under control of the GAL1 promoter were grown in galactose containing medium to an A600 = 0.3 and synchronized in G1 with 0.1 μg/mL α-factor for 3 h. Cells were released from the pheromone arrest in dextrose containing media to block production of Sic1 fusion constructs and samples were analyzed at the time intervals as indicated by immunoblotting. To measure Met4 protein stability, cells expressing Met4 under control of the native promoter were cultured in rich medium (1% yeast extract, 2% peptone, 2% dextrose) to A600 = 0.6. Cycloheximide was added to a final concentration of 0.2 mg/mL to block protein synthesis, and cell lysates were analyzed by immunoblotting.
In Vitro Ubiquitylation Aassay.
G1-CDK and SCF were expressed and purified from insect cells as previously described (28). Both SiclK36 and 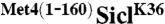 were phosphorylated and labeled with 32P at a final concentration of ∼12 μM (28). Ubiquitylation reactions were carried out in 30 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 2 mM ATP, and 2 mM dithiothreitol. In a 1.5 mL eppendorf tube, 200 nM SCF, 0.8 μM E1, 10 μM Cdc34, and 60 μM ubiquitin were briefly incubated at 22 °C and the reactions were initiated by the addition of 1.2 μM SiclK36 or
were phosphorylated and labeled with 32P at a final concentration of ∼12 μM (28). Ubiquitylation reactions were carried out in 30 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 2 mM ATP, and 2 mM dithiothreitol. In a 1.5 mL eppendorf tube, 200 nM SCF, 0.8 μM E1, 10 μM Cdc34, and 60 μM ubiquitin were briefly incubated at 22 °C and the reactions were initiated by the addition of 1.2 μM SiclK36 or 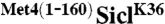 . Time-points were extracted from the reaction mixture (30 μL) and quenched with reducing SDS-PAGE buffer. The samples were then separated on a 10% Tris-glycine SDS-PAGE gel, which was dried and exposed to a phosphor screen for analysis. Quantification was performed with ImageQuant (GE Healthcare) and plotted using Prism. The rate of ubiquitylated Sic1 product formation was calculated by dividing the quantity of all ubiquitylated Sic1 species (product) by the sum of unmodified substrate and product and then multiplying by 36 pmol.
. Time-points were extracted from the reaction mixture (30 μL) and quenched with reducing SDS-PAGE buffer. The samples were then separated on a 10% Tris-glycine SDS-PAGE gel, which was dried and exposed to a phosphor screen for analysis. Quantification was performed with ImageQuant (GE Healthcare) and plotted using Prism. The rate of ubiquitylated Sic1 product formation was calculated by dividing the quantity of all ubiquitylated Sic1 species (product) by the sum of unmodified substrate and product and then multiplying by 36 pmol.
Supplementary Material
Acknowledgments.
We thank N. Dantuma and C. Heinen for discussion of unpublished data. This work was supported by National Institutes of Health Grants GM66164 and GM66164AS1 (P.K.) and A.T. is supported by an NCI Institutional Training Grant Award (5 T32 CA009054-32). R.J.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010648107/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67(425):425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Winget JM, Mayor T. The diversity of ubiquitin recognition: Hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 5.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains—From structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flick K, Raasi S, Zhang H, Yen JL, Kaiser P. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat Cell Biol. 2006;8:509–515. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]
- 7.Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Ortolan TG, et al. The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjerpe R, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Flick K, et al. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat Cell Biol. 2004;6:634–641. doi: 10.1038/ncb1143. [DOI] [PubMed] [Google Scholar]
- 13.Kuras L, et al. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol Cell. 2002;10:69–80. doi: 10.1016/s1097-2765(02)00561-0. [DOI] [PubMed] [Google Scholar]
- 14.Chandrasekaran S, et al. Destabilization of binding to cofactors and SCFMet30 is the rate-limiting regulatory step in degradation of polyubiquitinated Met4. Mol Cell. 2006;24:689–699. doi: 10.1016/j.molcel.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Petroski MD, Deshaies RJ. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell. 2003;11:1435–1444. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 16.Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 17.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 18.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 20.Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30 )complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 22.Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 24.Heinen C, et al. Mutant p62/SQSTM1 UBA domains linked to Paget’s disease of bone differ in their abilities to function as stabilization signals. FEBS Lett. 2010;584:1585–1590. doi: 10.1016/j.febslet.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Wu CY, et al. Repression of sulfate assimilation is an adaptive response of yeast to the oxidative stress of zinc deficiency. J Biol Chem. 2009;284:27544–27556. doi: 10.1074/jbc.M109.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 28.Petroski MD, Deshaies RJ. In vitro reconstitution of SCF substrate ubiquitination with purified proteins. Methods Enzymol. 2005;398:143–158. doi: 10.1016/S0076-6879(05)98013-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.