Abstract
Malaria infections commonly contain multiple genetically distinct variants. Mathematical and animal models suggest that interactions among these variants have a profound impact on the emergence of drug resistance. However, methods currently used for quantifying parasite diversity in individual infections are insensitive to low-abundance variants and are not quantitative for variant population sizes. To more completely describe the in-host complexity and ecology of malaria infections, we used massively parallel pyrosequencing to characterize malaria parasite diversity in the infections of a group of patients. By individually sequencing single strands of DNA in a complex mixture, this technique can quantify uncommon variants in mixed infections. The in-host diversity revealed by this method far exceeded that described by currently recommended genotyping methods, with as many as sixfold more variants per infection. In addition, in paired pre- and posttreatment samples, we show a complex milieu of parasites, including variants likely up-selected and down-selected by drug therapy. As with all surveys of diversity, sampling limitations prevent full discovery and differences in sampling effort can confound comparisons among samples, hosts, and populations. Here, we used ecological approaches of species accumulation curves and capture-recapture to estimate the number of variants we failed to detect in the population, and show that these methods enable comparisons of diversity before and after treatment, as well as between malaria populations. The combination of ecological statistics and massively parallel pyrosequencing provides a powerful tool for studying the evolution of drug resistance and the in-host ecology of malaria infections.
Keywords: Plasmodium falciparum, next generation sequencing
The evolution of drug-resistant malaria represents an immense threat to global health. With nearly 40% of the global population at risk, 300 to 660 million cases of Plasmodium falciparum malaria occur annually, causing an estimated 1 million deaths (1). The evolutionary selection of malaria occurs both within individual hosts and within populations. Whereas the later has been extensively studied, the former has mostly been the realm of theoretical studies in humans and experimental models in animals and in vitro (2–8). An understanding of the in-host evolutionary selection of malaria parasites is critical to understanding the emergence of drug resistance (7).
Most patients with P. falciparum infections are infected with multiple genetically distinct parasite variants (also called clones, genotypes, or clonal lineages). This situation could be a result of multiple infectious mosquito bites, or bites from mosquitoes infected with multiple parasite variants (9). In areas of low transmission, such as in Asia or Latin America, patients may have infections with as few as a single variant or as many as six variants (10). In areas of high transmission, such as in sub-Saharan Africa, more than 10 variants can be routinely detected in an individual (11, 12). Thus, within-host selection among these variants, especially in areas of high transmission, is likely to play an important role in parasite evolution.
Existing methods to measure parasite heterogeneity have been shown to miss many variants within an individual (13, 14). The most common method uses nested PCR (nPCR) with gel electrophoresis to detect polymorphisms in the variable surface antigens merozoite surface protein 1 (msp1), merozoite surface protein 2 (msp2), and glutamine rich protein (glurp) (15). However, nPCR with gel electrophoresis detects only size polymorphisms and cannot detect sequence polymorphisms. Furthermore, it has been shown that nPCR methods are insensitive to low-abundance variants (3, 16). Last, the method is not quantitative for relative population sizes of different variants. Because of these limitations, it has not been possible to study within-host evolution of human malaria parasites by using these methods. To study the competition and selection between variants in a mixed malaria infection, new tools are required that are sensitive to minority populations and quantitative for relative parasite population sizes in the host (17). Next-generation sequencing techniques, such as massively parallel pyrosequencing (MPP), provide the increased resolution of in-host diversity necessary for more thorough ecological studies of in-host selection.
In this study, we investigated the use of MPP to study the diversity of P. falciparum in individuals and estimate the parasite population diversity in a malaria endemic community. More than 90 thousand partial P. falciparum msp1 block 2 and msp2 central variable repeat sequences were obtained from patients living in a region of very high transmission (Malawi) and a region of low transmission (Cambodia). These are the regions amplified by World Health Organization (WHO)-recommended genotyping protocols (15). Details about the genetic structure of these genes have been extensively reviewed (15, 18–21). Our objectives were to: (i) compare the number of variants detected (or multiplicity of infection) by deep sequencing and nPCR genotyping methods at each gene site, (ii) estimate the effects of increasing depth of sequencing on estimates of diversity, (iii) follow the changes in variant number and population sizes longitudinally in patients, and (iv) use capture-recapture methods to estimate the total genetic diversity in a community. In addition, we deep sequenced the dhfr gene from patients to show that 454 sequencing can provide information on drug resistance haplotypes, detect novel SNPs in drug resistance genes, and estimate the error-free sequencing rate of 454 sequencing in the AT-rich malaria genome.
Results
Determining the Sequencing Error Rate.
The error free rate was determined to be 99.1%, a slightly higher error rate than has previously been reported for 454 sequencing (22). This likely results in part from the highly AT-rich genome of the malaria parasite. The specific methods and types of errors are summarized in SI Results.
In-Host Parasite Diversity Revealed by MPP.
In total, pyrosequencing provided 115,518 sequence reads in the sections of the plate dedicated to msp1 and msp2. A total of 91,645 reads (79%) mapped to the malaria genome. Of these, 44,590 (39%) were used to build the consensus sequences for the amplicons of individual variants in the patients. The large reduction in total usable sequencing reads in part reflects the exclusion of short reads from building the consensus sequences and the exclusion of sequences containing an incomplete barcode or forward primer sequences (Materials and Methods). The average numbers of reads used per locus per patient to create the unique consensus sequences for variants in a sample were 1,371 (SD, 787) from Malawi and 2,144 (SD, 2,346) from Cambodia. The repetitive sequence alignment analysis used to create these consensus sequences served the purpose of overcoming sequencing errors. Each resulting unique consensus sequence was considered a variant. The number of variants detected by nPCR and MPP are summarized in Table 1. MPP detected as many as sixfold more variants than nPCR with gel electrophoresis. Among the Malawian samples, MPP detected on average of 5.0 more msp2 variants per individual than nPCR with gel electrophoresis [95% confidence interval (CI), 2.1–7.9] and 4.8 more msp1 variants per individual than nPCR with gel electrophoresis (95% CI, 2.4–7.1). Among the Cambodian samples, 3.3 (95% CI, 0.9–5.6) more msp2 and 2.0 (95% CI, −1.9 to 5.9) more msp1 variants were detected by MPP relative to nPCR with gel electrophoresis. Additional information about the comparison of MPP versus other genotyping methods is described in SI Results.
Table 1.
Comparison of the number of variants detected by nPCR and MPP in patients from Malawi and Cambodia
|
msp1 |
msp2 |
|||||
| 454 Sequencing |
454 Sequencing |
|||||
| Sample no. | nPCR | Total variants | Sequences | nPCR | Total variants | Sequences |
| Malawi | ||||||
| 2/041 I | 3 | 8 | 1,475 | 1 | 11 | 2,036 |
| 2/041 III | 2 | 4 | 1,333 | 2 | 3 | 3,307 |
| 1/021 I | 1 | 3 | 1,215 | ND | ND | ND |
| 2/007 I | 3 | 8 | 1,193 | 1 | 6 | 185 |
| 1/009 I | 3 | 16 | 1,191 | 3 | 12 | 1,222 |
| 1/009 II | 3 | 11 | 2,159 | 3 | 11 | 932 |
| 2/044 I | 1 | 2 | 634 | 1 | 3 | 288 |
| 1/014 I | 3 | 7 | 1,135 | 2 | 10 | 2,021 |
| 1/014 II | 1 | 3 | 339 | 2 | 2 | 2,464 |
| 1/014 III | 1 | 4 | 943 | 2 | 4 | 2,126 |
| 2/040 I | 4 | 11 | 1,238 | ND | ND | ND |
| Cambodia | ||||||
| 46 initial | 2 | 2 | 113 | 1 | 3 | 1,058 |
| 46 recurrent | 2 | 2 | 702 | 1 | 3 | 1,278 |
| 84 initial | 1 | 6 | 1,259 | 1 | 5 | 2,955 |
| 84 recurrent | 1 | 4 | 7,461 | 1 | 6 | 2,328 |
ND, not detected.
Among nine successfully sequenced Malawian patient samples at msp2, a total of 62 distinct variants were detected (GenBank accession nos. HM153086–HM153147). This represented 52 unique variants, with the remaining 10 shared between samples. BlastN analysis of these 52 variants showed five (10%) with a perfect sequence match to previously reported msp2 sequences. The average variant consensus sequence length was 245 bp (range, 153–411 bp). Similarly, among the 11 Malawian samples successfully sequenced at msp1, a total of 77 variants were detected (GenBank accession nos. HM153166–HM153242). This represented 63 unique variants (14 shared). Of these 63 variants, BlastN analysis showed nine (14%) with a perfect sequence match to previously reported msp1 sequences. The average consensus sequence length was 298 bp (range, 178–397 bp). Both patients with paired initial and recurrent parasitemias from Cambodia were successfully sequenced at both msp1 and msp2. At msp2, 17 variants were detected (GenBank accession nos. HM153080–HM153164) that contained 16 unique variants (one shared). Six of these variants (38%) had exact sequence matches to previously reported sequences by BlastN analysis. The average variant consensus sequence length was 254 bp (range, 165–365 bp). At msp1, 14 variants were detected (GenBank accession nos. HM153243–HM153256), which contained 12 unique variants (two shared). Five of these variants (42%) had exact sequence matches to previously reported sequences by BlastN analysis. The average variant consensus sequence length was 305 bp (range, 230–331 bp).
Effects of Sampling Effort on Variant Discovery.
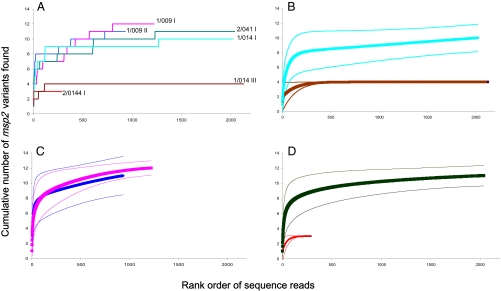
In general, the number of entities discovered is highly dependent on sampling effort. In the current context, this means that, as more sequences are analyzed, the number of variants discovered will increase. This is clear in our data (Fig. 1A), but it is also equally clear that some samples contain fewer variants than others, and the question arises whether more sequencing effort would have led to further discovery, or if we are close to a diversity asymptote. This issue also informs decisions about the depth of sequencing required in such analyses. In the current analyses, depth was determined by the amount of input DNA used to make the sequencing library. The curves in Fig. 1A are directly analogous to “species accumulation” curves in ecology (23). These can be used to estimate individual-based rarefaction curves, which estimate the number of variants that would have been observed had we made any small number of reads (24, 25). It is possible to estimate CIs for such curves, which enables comparisons of diversity even when there are differences in sampling effort (24, 25). These are shown for several of our patient samples in Fig. 1 B–D. Note that the curves plotted in Fig. 1A are single realizations of a range of variant accumulation curves that would be produced by repeated deep resequencing of the same blood samples; the smoothed rarefaction curves represent the expected variant richness values for the corresponding accumulation curves.
Fig. 1.
msp2 genotype discovery curves. These figures show genotype discovery curves and rarefaction curves for several patient samples. The genotype discovery curves are generated by plotting the number of unique sequence variants in a sample (y axis) against the rank order of sequences reads generated (x axis). Therefore, the x axis illustrates the cumulative number of sequences determined for a sample at a given site (number of sequence reads or fold coverage of the site), which are ordered by the sequential well number of the 454 picotiter plate, which reflects increasing sampling effort. (A) Number of msp2 variants discovered with increasing sequencing effort in six samples. Patient codes and sample numbers cross-reference with those in Table 1. As expected, the number of variants discovered is a function of sequencing effort, but for a given effort, some samples contain more variants than others. For six samples, expected variant richness (rarefaction), curves (thick lines), and 95% CI (thin lines) for (B) patient 1/014 before and after drug treatment [light blue (Upper) and brown (Lower) lines, respectively; samples 1 and 3], (C) patient 1/009 before and after drug treatment (pink and dark blue lines, respectively; samples 1 and 2), and (D) msp2 diversity in two different patients [2/041 I and 2/044 I, black (Upper) and red (Lower) lines, respectively].
Several conclusions emerge from these analyses. First, there is evidence that we have fully discovered all the msp2 diversity in some samples [e.g., the posttreatment sample in patient 1/014 (Fig. 1B), in which there are four clones], whereas in other samples there is little evidence that we are nearing an asymptote, implying that even deeper pyrosequencing is needed to estimate the diversity present [e.g., the pretreatment sample of patient 1/014 (Fig. 1B) and posttreatment samples of patient 1/009 (Fig. 1C)]. An implication of this is that there is no simple rule concerning the depth of pyrosequencing required to estimate the number of variants present—in effect, we went unnecessarily deep for two samples [1/014 III (Fig. 1B) and 2/044 I (Fig. 1D)], whereas even deeper sequencing would have likely been informative in other cases [1/014 I (Fig. 1B), 1/009 I and II (Fig. 1C), and possibly even sample 2/041 (Fig. 1D)]. This means that, for the latter samples, the numbers given in Table 1 for the msp2 variants are minima, whereas we conclude with high certainty that the numbers given for the former samples are sound estimates of the actual number present. These findings also show that the depth of sequencing can be varied based upon the goals of the user. As a significant proportion of diversity is detected by 250 sequencing reads in most samples, if a purpose does not require the exhaustive measurement of variants within a single sample, higher levels of multiplexing could be used to decrease the sequencing costs.
Second, the approach makes possible the comparison of diversity among samples. Such comparisons are straight forward when the depth of sequencing is similar. For patient 1/014, for example, there was similar sampling effort before and after treatment (Fig. 1B), and it is clear that diversity in the second sample was substantially lower (for clones compared with at least 10 before treatment). For patient 1/009, however, we obtained 300 more reads from the pretreatment sample, but despite this extra effort, there is no evidence that the variant richness before and after treatment differs: for any level of sampling effort, the estimates of diversity are similar, and the CIs overlap [Fig. 1C]. We can also be confident that the number of msp2 variants in patient 2/044 was substantially less than that in patient 2/041 (three vs. at least 10; Fig. 1D), even though the depth of sequencing varied sevenfold between the two samples.
To summarize, there is evidence that variant diversity differs significantly among patients, that diversity was reduced after drug treatment in one patient (Fig. 1B) but not in another (Fig. 1C), and that we can estimate with considerable confidence how many clones are present in some hosts, but not in others. The degree of confidence is poorly correlated with sampling effort.
Determining the Effect of Treatment on In-Host Variant Populations.
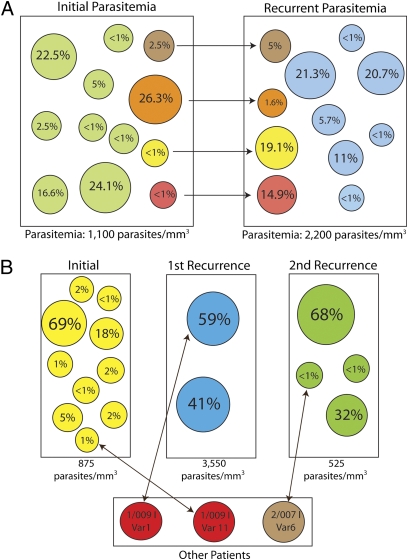
For two patients, we had MPP data before and after sulfadoxine–pyrimethamine treatment. Fig. 2A shows each of the variants present before and after treatment in a single patient (1/009, the patient also shown in Fig. 1C). The size of each variant's bubble is proportional to the population size of that variant (no. of sequencing reads used to build the consensus sequence of that variant/total no. of sequencing reads used to build the consensus sequences of all variants in the sample). In this patient, the pre- and posttreatment samples shared four variants. In addition, both the pre- and posttreatment samples have a large number of unique variants. Fig. 2B shows samples from a second patient (1/014) who had two recurrent episodes of parasitemia. In each case, the recurrent parasitemia did not contain a shared genotype with the previous infection. However, each sample did contain a shared genotype with other patients in our study. The initial treatment caused a significant decrease in complexity of infection in the second infection.
Fig. 2.
msp2 variant dynamics between longitudinal patient samples from Malawi. (A) Change in variant population in a pre- and posttreatment sample pair (1/009I and 1/009 II). The green and blue circles represent unique variants detected in the initial parasitemia and recurrent parasitemias, respectively. The brown, orange, yellow, and red circles represent population sizes of variants that are shared between the two samples. (B) Genotypes of three infections in patient 1/014. In each case, treatment appears to have cleared all of the genotypes as none of the samples contained a shared genotype within the same patient. However, all of the samples did contain a genotype identified in other patients in the study. Of note, there was a significant reduction in the complexity of infection between the first and second infection with sulfadoxine–pyrimethamine treatment.
Determination of the Community-Wide Population Diversity.
We used a capture-recapture technique to estimate the total number of variants in the parasite population at our Malawi site. Capture-recapture methods were originally developed to enumerate difficult to measure wildlife populations although have been adapted for use in epidemiological studies. When we limited the analysis to variants identified in initial parasitemias, we identified 48 unique variants of msp1 and 41 unique variants of msp2. Using estimators developed by Chao (26) that are specifically for sparse data and allow heterogeneous capture probabilities, we estimated that the total number of variants for msp1 was 356 (95% CI, 143–1,043) and that for msp2 was 421 (95% CI, 140–1,505).
Discussion
For more than 30 y, it has been clear that falciparum malaria infections are polyclonal (27, 28). However, in the past several years, it has been suggested that the amount of previous reports of diversity have described only the “tip of the iceberg” (10, 11, 16). Here we expose the extent of in-host diversity by using MPP to quantify and identify rare variants in polyclonal malaria infections in a high-throughput manner. In addition, the ability to accurately and quantitatively describe the in-host population of malaria parasite variants is critical for understanding the ecological interactions between these variants in a single infection. Mouse models suggest important ecologic interactions between variants (5, 8). However, because of the lack of appropriate tools, these issues have not been extensively studied in human populations. A body of correlational epidemiological evidence is consistent with crowding effects [“competitive interactions” (7, 29)] in human malaria infections, and some patterns of drug resistance in Africa are more readily explained by invoking differences in parasite fitness as a result of competition (5, 6, 30–33). Recrudescences consistent with competitive release, when one variant performs better after removal of its competitors by chemotherapy, were recently observed in human populations (34). If these interactions are widespread in human parasite populations, the information about interactions in mixed populations could significantly assist malaria control efforts. The experiments we report here are the first steps in developing the tools necessary to understand the dynamics of parasites within hosts and the effect of selective pressures in mixed populations.
The limitations of currently recommended WHO genotyping procedures to exhaustively detect variants in mixed infections have been extensively reviewed (13). As a result of these limitations, multiple investigators have suggested alternative methods that potentially increase the relative diversity detected (10–12, 20). MPP represents a powerful approach to detect in-host diversity of malaria infections. The key breakthrough of the MPP sequencing platform is the ability to sequence single strands of DNA from a complex mixture of DNA. This allows the identification of rare variants as well as the quantification of variant population sizes in mixed infections. In addition, compared with other next-generation sequencing methods, such as Illumina/Solexa, read length is significantly longer, allowing for larger variable regions to be studied. This is the power of 454 sequencing. HIV researchers were among the first infectious disease researchers to use this technology to describe in-host diversity, and an extensive literature is developing. Multiple reports have applied MPP to the detection and characterization of rare drug-resistant variants (35–38). This technique has also been used to detect low-frequency pretherapy chemokine (CXC motif) receptor 4 (39). In addition, MPP has been used to address questions on the dynamics of HIV quasispecies in response to selective pressure. The ability of MPP to detect minority variants and to quantitate the relative variant frequencies in an infection makes it an appropriate tool for studying the in-host ecology of malaria. These data represent an initial evaluation of in-host diversity, and additional studies to refine data analysis methods and estimate reproducibility parameters are clearly required.
We found that MPP, on average, detected substantially more variants per infection than the currently recommended WHO methods. More importantly, we found a highly complex milieu when looking at paired patient samples (i.e., Fig. 2). In Malawi, paired initial and recurrent parasitemia samples contained some shared variants suggestive of recrudescence. However, the majority of variants in the recurrent parasitemia were different. This suggests the potential of a significant amount of reinfection occurring in these patients. The average duration between paired samples was 28 days. Another possibility is that MPP genotyping still failed to detect all variants within the sample, as is suggested by our genotype discovery analysis (Fig. 1). An additional potential limitation is that samples from a single initial parasitemia blood spot were used and additional variants could have been sequestered out of circulation.
In the setting of a low transmission intensity in Cambodia, MPP again, on average, detected more variants than described by nested PCR. These paired patient samples had previously been genotyped by nPCR with gel electrophoresis, msp1 HTA, and msp2 HTA as part of the assessment of misclassification in antimalarial clinical trials (10, 40). In the original clinical trial, both these patients were considered to represent reinfections by nPCR with gel electrophoresis (10, 40). The MPP data confirmed the findings of our previous study: that by current practices, patient 46 should be classified as a case of recrudescence (i.e., one shared allele at msp1 and msp2) and patient 84 should be classified as a case of reinfection (i.e., one shared allele at msp1 and no shared msp2 allele).
These findings are not wholly unexpected, as previous studies that sequenced variants from mixed infections have shown more complexity than detected by nested PCR. For example, Mayengue et al. reported that three variant allele sizes detected by nested PCR actually contained 23 variants detected by sequencing (41). However, these reports required very labor-intensive methods, and newer high-throughput sequencing tools like MPP will allow for this type of analysis on a larger scale.
One concern for the use of amplicon protocols in MPP is the potential for recombination occurring during amplification creating false variants, especially for high-diversity samples requiring multiple PCR cycles (42). In fact, recombination during PCR of msp1 has been known for some time, with ratios (recombinant haplotype to parent haplotype) of 1.8% to 28.6% in an nPCR protocol reported by Tanabe et al. (18, 43). However, the amplicon they evaluated contained block 2 to 6 of msp1 and was significantly larger (approximately 1,100 bp) than the one we evaluated. The use of a much smaller fragment (approximately 300 bp) will, to a certain, extent limit the amount of recombination seen. In addition, the prolonged extension times used (1.5 min for a 300-bp fragment) will help reduce recombination. We have not directly assessed recombination rates for this project. However, a review of the consensus sequences for the variants did not show any blatant recombination events (i.e., a 3′ end sequence similar in one clone to a 5′ in another). Further mixing studies of control DNA are required to evaluate the extent of recombination and PCR amplification bias of different-sized fragments occurring during amplification of msp1 and msp2 for MPP. In addition, other approaches to minimize the effect of recombination should be evaluated, such as decreasing the number of cycles if adequate starting material is available (we were unable to do this working from dried blood spots) or more advanced techniques such as emulsion PCR or use of improved proof-reading enzymes for the initial amplification (42, 44).
Molecular techniques with improved ability to distinguish between variants have recently shown highly diverse parasite populations in areas of high transmission, in particular using the msp2 loci (45). However, the parasite variants observed in patients likely make up only a sample of circulating variants, as it is impossible to exhaustively sample the parasites circulating in a population. Capture-recapture is a method developed for wildlife ecological studies, which has since been adapted to epidemiological studies, to estimate population size in cases in which it was impossible to measure the entire population being studied. In this method, the extent of incomplete ascertainment is estimated by using information from overlapping lists of samples from distinct sources (i.e., variants shared between two independent people) (46).
Using each individual patient as a “source,” we conducted a capture-recapture evaluation to estimate the total number of variants in the population in Mepemba and Madziabango, Malawi. The accuracy of this type of assessment is dependent on several factors: (i) independence of observations, (ii) accuracy of data, (iii) equal chance of individual variants being captured, and (iv) a closed population. In an attempt to control for the first factor, only initial parasitemias were used to prevent the issue of dependence in the form of recrudescences. In addition, by including only variants identified in initial parasitemias, we simplified the analysis and reduced bias from capture probabilities possibly varying by time and no definitive way of classifying parasites found in recurrent parasitemias as reinfections or recrudescences. As MPP provides determination of variants to the sequence level, accuracy was assumed to be high. However, as there is a PCR amplification step for the procedure, this could introduce bias by the failure to amplify certain variants. Traditional capture-recapture methods assume that all individuals in the population (in this case, all parasite variants) have the same probability of being captured (i.e., “catchability”). The estimates we used were robust to violation of this assumption, which is important in this context because the underlying distribution of genetic variants in the population is not uniform. The fourth factor, a closed population, is potentially an issue for estimates of malaria diversity. This estimate of diversity is partially limited by a likely underestimate of variants from the sequencing analysis and by large CIs from the small sample sizes. However, it is a useful example of this method and demonstrates the potential for a significant unmeasured amount of diversity in a population.
A high degree of genetic variability in msp2 and msp1 has been previously reported in Africa. For example, Schoepflin et al. reported 76 msp2 variants and 29 msp1 variants in the Kilombero district of Tanzania by using a sensitive capillary electrophoresis method (45). However, these estimates are dependent on the ability of the survey to capture all the circulating variants. Similar to these previous reports, we found a high level of genetic diversity at msp2 [measured, 41 variants; estimated total, 421 (95% CI, 140–1,505)]. We also found equally high genetic diversity at msp1 [measured, 48 variants; estimated total, 356 (95% CI, 143–1,043)]. Because of the small number of patients, the estimates of population size have wide CIs and, as with any analysis that requires currently untestable assumptions, one must appreciate that there is uncertainty about the estimate. Still, our findings suggest that genetic diversity can be greatly underestimated without accounting for additional unobserved variants.
In this study, we expose the complexity of malaria infections by using MPP. Falciparum malaria infections contain more diversity than has previously been suggested, raising concerns about how we have studied parasite diversity in the past. In addition, we propose the use of classic ecological tools to improve our estimates of malaria diversity in patients as well as in a community despite limited sampling. These new tools will empower the study of the evolutionary and ecological factors involved in the spread of drug resistance.
Materials and Methods
Human Subjects.
We studied parasites from the filter paper blood spots of nine patients that were collected during two previously completed studies. Seven women who took part in a trial of intermittent preventive malaria treatment in Mpemba and Madziabango, Malawi, provided 11 samples during parasitemic episodes (47). Two patients from an in vivo efficacy trial of artesunate–mefloquine in Chumkiri, Cambodia, provided initial and recurrent parasitemia samples (40). The institutional review boards of the University of North Carolina, the Malawi College of Medicine, the Cambodian Ministry of Health, and Naval Medical Research Unit No. 2 approved these studies (40, 47).
Nested PCR Genotyping of msp1 and msp2.
Nested PCR followed previously published protocols as described in SI Materials and Methods.
Overview of Variant Genotyping by MPP.
Full details of the MPP procedures and data analysis are given in SI Materials and Methods. In brief, PCR products for 454 sequencing were prepared using primers specific for three regions of the of the malaria genome: (i) the block 2 region of msp1, (ii) the central variable region of msp2, and (iii) the segment of dhfr containing aa 51, 59, and 108. For each region, and for each patient sample, this generates a library of amplicons thought to be representative of the parasite population [based on previous reports in the HIV literature (48)]. Thus, we might have 2,000 amplicons from a patient sample for the defined region of msp2. These 2,000 amplicons are then individually sequenced using the 454, giving msp2 sequences for as many as 2,000 individual strands of amplicon DNA in that sample. The issue then is to determine from these sequences how many msp2 variants are present among those 2,000 parasites.
To ensure our measured variation was not a result of sequencing error, we developed consensus sequences for each variant. When identifying unique variants, we elected to prioritize large (>6 bp) insertions and deletions and allelic families over SNP differences. This was mediated by: (i) a desire to conservatively estimate diversity by not over-calling variants created as a result of recombination or PCR error, (ii) the results of a coincident analysis of error showing high rates of single nucleotide indels and occasional SNPs (given the average read length of just less than 300 bp and an error-free sequencing rate of 99.1%, we would expect fewer than three errors in that read length), and (iii) the unique features of pyrosequencing a highly rich AT genome, which requires the modification and development of new bioinformatic tools for analysis that are still under development. In total, our adopted approach would predictably underestimate the diversity of samples.
Briefly, the analysis occurred in a two-step format: (i) evaluation for length polymorphisms at least 6 bp followed by (ii) an evaluation of sequence homology. First, for a given patient, all sequences for a single allele were aligned using ClustalW. These initial alignments were visually divided into groups of sequences based on the allelic family of the repeat and the presence of indels of at least 6 bp. Sequences within each group were then separately realigned. Occasionally, these realignments revealed the presence of two groups within the initial group based on the presence of an additional indel of at least 6 bp, typically at the 3′ end of the sequence. In this case, the group was again subdivided and the sequences within the subgroups realigned. After that, the alignments based on length polymorphism were then evaluated for sequence homology. Subgroups were created again if sequences varied by approximately 3% (approximately 9 bp in 300 bp). These final alignments were then used to build the consensus sequence, which defined each variant. The generated consensus sequence was then evaluated by the investigator for the presence of ambiguous nucleotides (either SNPs or single BP indels), which were called by a simple approach of “majority rule” to determine the final consensus sequence that defined a variant. The consensus sequences defining each variant were then checked for correct translation and then aligned to confirm that all variants within a patient sample were unique and differed from each other by at least 5 bp as SNPs or at least 3 bp for indels. The number of variants detected did not significantly change if the number of unique SNPs required to define a variant was increased to eight or the size of the required insertion/deletion was increased to at least 6 bp. These consensus sequences were then compared with other previously reported sequences at the allele using BlastN. Multiplicity of infection for an given blood sample was then calculated separately for each genomic region, based on the number of unique variants at each region detected in the patient sample.
Variant Accumulation Curves.
Rarefaction curves were calculated using EstimateS (http://viceroy.eeb.uconn.edu/estimates) using individual-based curves (sampling without replacement) (24) based on the Michaelis–Menten models (49). We note that the impact on the shape of species-accumulation curves of different community structures is an area of active research (50); with the data at hand (six samples), we are unable to satisfactorily explore this issue.
Estimating Population Sizes of msp1 and msp2 Variants.
We treated each individual patient as a source able to capture individual parasite variants. We calculated estimates of the total number of msp1 and msp2 variants using estimators developed by Chao (26).
Supplementary Material
Acknowledgments
We thank Xiaojun Guan for cluster computing support. We are also grateful to Dr. Socheat Duong and Dr. Sinuon Muth of the Cambodian National Malaria Control Program for their support. We thank Steve Taylor, Ian Hastings, and our anonymous reviewers for their thoughtful reviews. This project was funded by National Institute of Allergy and Infectious Diseases (NIAID) Grant 1R21AI076785, National Center for Research Resources Award KL2RR025746 (to J.J.J.), and National Research Service Award T32-AI070114-01 from NIAID (to K.P.). A.R. has benefited from discussion with members of the Research and Policy in Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HM153086–HM153256).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007068107/-/DCSupplemental.
References
- 1.World Health Organization . World Malaria Report 2008. Geneva: World Health Organization; 2008. [Google Scholar]
- 2.Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: Parasite virulence and competitive success. Evolution. 2006;60:1358–1371. [PubMed] [Google Scholar]
- 3.Liu S, Mu J, Jiang H, Su XZ. Effects of Plasmodium falciparum mixed infections on in vitro antimalarial drug tests and genotyping. Am J Trop Med Hyg. 2008;79:178–184. [PMC free article] [PubMed] [Google Scholar]
- 4.Mideo N, et al. Understanding and predicting strain-specific patterns of pathogenesis in the rodent malaria Plasmodium chabaudi. Am Nat. 2008;172:214–238. doi: 10.1086/591684. [DOI] [PubMed] [Google Scholar]
- 5.Wargo AR, Huijben S, de Roode JC, Shepherd J, Read AF. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc Natl Acad Sci USA. 2007;104:19914–19919. doi: 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hastings IM. Malaria control and the evolution of drug resistance: an intriguing link. Trends Parasitol. 2003;19:70–73. doi: 10.1016/s1471-4922(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 7.Hastings IM, D'Alessandro U. Modelling a predictable disaster: The rise and spread of drug-resistantmalaria. Parasitol Today. 2000;16:340–347. doi: 10.1016/s0169-4758(00)01707-5. [DOI] [PubMed] [Google Scholar]
- 8.Huijben S, et al. Chemotherapy, within-host ecology and fitness of drug resistant malaria parasites. Evolution. 2010 doi: 10.1111/j.1558-5646.2010.01068.x. 10.1111/j.1558-5646.2010.01068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talisuna AO, Okello PE, Erhart A, Coosemans M, D'Alessandro U. Intensity of malaria transmission and the spread of Plasmodium falciparum resistant malaria: A review of epidemiologic field evidence. Am J Trop Med Hyg. 2007;77(6 suppl):170–180. [PubMed] [Google Scholar]
- 10.Juliano JJ, et al. Misclassification of drug failure in Plasmodium falciparum clinical trials in southeast Asia. J Infect Dis. 2009;200:624–628. doi: 10.1086/600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenhouse B, et al. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am J Trop Med Hyg. 2006;75:836–842. [PMC free article] [PubMed] [Google Scholar]
- 12.Kwiek JJ, et al. Estimating true antimalarial efficacy by heteroduplex tracking assay in patients with complex Plasmodium falciparum infections. Antimicrob Agents Chemother. 2007;51:521–527. doi: 10.1128/AAC.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juliano JJ, Gadalla N, Sutherland CJ, Meshnick SR. The perils of PCR: Can we accurately ‘correct’ antimalarial trials? Trends Parasitol. 2010;26:119–124. doi: 10.1016/j.pt.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juliano JJ, Taylor SM, Meshnick SR. Polymerase chain reaction adjustment in antimalarial trials: Molecular malarkey? J Infect Dis. 2009;200:5–7. doi: 10.1086/599379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization . Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy: Genotyping to Identify Parasite Populations. Geneva: World Health Organization; 2008. [Google Scholar]
- 16.Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13:872–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 17.Hastings IM, Nsanzabana C, Smith TA. A comparison of methods to detect and quantify the markers of antimalarial drug resistance. Am J Trop Med Hyg. 2010;83:489–495. doi: 10.4269/ajtmh.2010.10-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanabe K, et al. In vitro recombination during PCR of Plasmodium falciparum DNA: A potential pitfall in molecular population genetic analysis. Mol Biochem Parasitol. 2002;122:211–216. doi: 10.1016/s0166-6851(02)00095-6. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira MU, et al. Allelic diversity at the merozoite surface protein-1 locus of Plasmodium falciparum in clinical isolates from the southwestern Brazilian Amazon. Am J Trop Med Hyg. 1998;59:474–480. doi: 10.4269/ajtmh.1998.59.474. [DOI] [PubMed] [Google Scholar]
- 20.Ngrenngarmlert W, et al. Measuring allelic heterogeneity in Plasmodium falciparum by a heteroduplex tracking assay. Am J Trop Med Hyg. 2005;72:694–701. [PubMed] [Google Scholar]
- 21.Felger I, et al. Sequence diversity and molecular evolution of the merozoite surface antigen 2 of Plasmodium falciparum. J Mol Evol. 1997;45:154–160. doi: 10.1007/pl00006215. [DOI] [PubMed] [Google Scholar]
- 22.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magurran AE. Measuring Biological Diversity. Malden, MA: Blackwell Science; 2004. [Google Scholar]
- 24.Gotelli NJ, Colwell RK. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett. 2001;4:379–391. [Google Scholar]
- 25.Colwell RK, Mao CX, Chang J. Interpolating, extrapolating and comparing incidence based species accumulation curves. Ecology. 2004;85:2717–2727. [Google Scholar]
- 26.Chao A. Estimating population size for sparse data in capture-recapture experiments. Biometrics. 1989;45:427–438. [Google Scholar]
- 27.Snounou G, Beck HP. The use of PCR genotyping in the assessment of recrudescence or reinfection after antimalarial drug treatment. Parasitol Today. 1998;14:462–467. doi: 10.1016/s0169-4758(98)01340-4. [DOI] [PubMed] [Google Scholar]
- 28.Day KP, Koella JC, Nee S, Gupta S, Read AF. Population genetics and dynamics of Plasmodium falciparum: An ecological view. Parasitology. 1992;104(suppl):S35–S52. doi: 10.1017/s0031182000075235. [DOI] [PubMed] [Google Scholar]
- 29.Read AF, Taylor LH. The ecology of genetically diverse infections. Science. 2001;292:1099–1102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- 30.Bruce MC, et al. Cross-species interactions between malaria parasites in humans. Science. 2000;287:845–848. doi: 10.1126/science.287.5454.845. [DOI] [PubMed] [Google Scholar]
- 31.Daubersies P, et al. Rapid turnover of Plasmodium falciparum populations in asymptomatic individuals living in a high transmission area. Am J Trop Med Hyg. 1996;54:18–26. doi: 10.4269/ajtmh.1996.54.18. [DOI] [PubMed] [Google Scholar]
- 32.Mercereau-Puijalon O. Revisiting host/parasite interactions: molecular analysis of parasites collected during longitudinal and cross-sectional surveys in humans. Parasite Immunol. 1996;18:173–180. doi: 10.1046/j.1365-3024.1996.d01-79.x. [DOI] [PubMed] [Google Scholar]
- 33.Talisuna AO, et al. Two mutations in dihydrofolate reductase combined with one in the dihydropteroate synthase gene predict sulphadoxine-pyrimethamine parasitological failure in Ugandan children with uncomplicated falciparum malaria. Infect Genet Evol. 2004;4:321–327. doi: 10.1016/j.meegid.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Harrington WE, et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Natl Acad Sci USA. 2009;106:9027–9032. doi: 10.1073/pnas.0901415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann C, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozera G, et al. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology. 2009;6:15. doi: 10.1186/1742-4690-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsuya Y, et al. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J Virol. 2008;82:10747–10755. doi: 10.1128/JVI.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Archer J, et al. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. Aids. 2009;23:1209–1218. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers WO, et al. Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar J. 2009;8:10. doi: 10.1186/1475-2875-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayengue PI, et al. Submicroscopic Plasmodium falciparum infections and multiplicity of infection in matched peripheral, placental and umbilical cord blood samples from Gabonese women. Trop Med Int Health. 2004;9:949–958. doi: 10.1111/j.1365-3156.2004.01294.x. [DOI] [PubMed] [Google Scholar]
- 42.Lahr DJ, Katz LA. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques. 2009;47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- 43.Kaneko O, Kimura M, Kawamoto F, Ferreira MU, Tanabe K. Plasmodium falciparum: Allelic variation in the merozoite surface protein 1 gene in wild isolates from southern Vietnam. Exp Parasitol. 1997;86:45–57. doi: 10.1006/expr.1997.4147. [DOI] [PubMed] [Google Scholar]
- 44.Williams R, et al. Amplification of complex gene libraries by emulsion PCR. Nat Methods. 2006;3:545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- 45.Schoepflin S, et al. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar J. 2009;8:250. doi: 10.1186/1475-2875-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hook EB, Regal RR. Capture-recapture methods in epidemiology: methods and limitations. Epidemiol Rev. 1995;17:243–264. doi: 10.1093/oxfordjournals.epirev.a036192. [DOI] [PubMed] [Google Scholar]
- 47.Kalilani L, et al. A randomized controlled pilot trial of azithromycin or artesunate added to sulfadoxine-pyrimethamine as treatment for malaria in pregnant women. PLoS ONE. 2007;2:e1166. doi: 10.1371/journal.pone.0001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsibris AM, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS ONE. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keating KA, Quinn JF. Estimating species richness: The Michaelis-Menten model revisited. Oikos. 1998;81:411–416. [Google Scholar]
- 50.Wei SG, et al. Comparative performance of species-richness estimators using data from a subtropical forest tree community. Ecol Res. 2010;25:93–101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.