Abstract
The development of HIV integrase (IN) strand transfer inhibitors (INSTIs) and our understanding of viral resistance to these molecules have been hampered by a paucity of available structural data. We recently reported cocrystal structures of the prototype foamy virus (PFV) intasome with raltegravir and elvitegravir, establishing the general INSTI binding mode. We now present an expanded set of cocrystal structures containing PFV intasomes complexed with first- and second-generation INSTIs at resolutions of up to 2.5 Å. Importantly, the improved resolution allowed us to refine the complete coordination spheres of the catalytic metal cations within the INSTI-bound intasome active site. We show that like the Q148H/G140S and N155H HIV-1 IN variants, the analogous S217H and N224H PFV INs display reduced sensitivity to raltegravir in vitro. Crystal structures of the mutant PFV intasomes in INSTI-free and -bound forms revealed that the amino acid substitutions necessitate considerable conformational rearrangements within the IN active site to accommodate an INSTI, thus explaining their adverse effects on raltegravir antiviral activity. Furthermore, our structures predict physical proximity and an interaction between HIV-1 IN mutant residues His148 and Ser/Ala140, rationalizing the coevolution of Q148H and G140S/A mutations in drug-resistant viral strains.
Integrase (IN) is the essential retroviral enzyme that orchestrates insertion of a reverse-transcribed DNA replica of the viral genome into host cell chromosomal DNA (reviewed in refs. 1 and 2). In the context of viral infection, IN catalyzes two reactions acting upon the viral DNA ends. During 3′ processing, it removes di- or trinucleotides to expose 3′ hydroxyls attached to invariant CA dinucleotides. Following nuclear import, IN carries out strand transfer, using these 3′ hydroxyls for nucleophilic attacks at a pair of phosphodiester bonds on opposing strands of chromosomal DNA, resulting in transesterification and subsequent joining of the 3′ ends of viral DNA to the chromosome.
The reactions catalyzed by IN proceed in the context of the preintegration complex (PIC), a poorly characterized nucleoprotein assembly, which in addition to IN and viral DNA contains other viral as well as cellular components (3, 4). The minimal functional unit within the PIC, comprising an IN tetramer assembled on the viral DNA ends (5–7), is referred to as the stable synaptic complex or intasome. Recently, we reported a crystal structure of the functional retroviral intasome containing full-length prototype foamy virus (PFV) IN and oligonucleotide mimics of its preprocessed viral U5 DNA end (6). The structure revealed a twofold symmetric IN tetramer, with viral DNA ends coordinated to the active sites within the catalytic core domains of the inner IN subunits and held in place by cis and trans interactions with the N- and C-terminal domains of the same IN subunits. Three acidic residues composing the invariant DDX35E motif form the basis of the retroviral IN active site, through coordination of a pair of catalytically essential divalent metal cations (Mg2+ or Mn2+) (6, 8, 9).
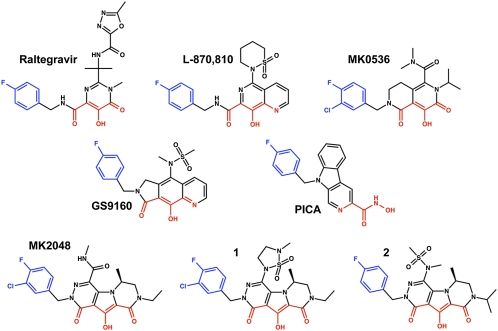
The strand transfer reaction catalyzed by HIV-1 IN is the target for a clinically validated class of antiretroviral drugs, termed IN strand transfer inhibitors (INSTIs; examples shown in Fig. 1, reviewed in refs. 10 and 11). All INSTIs described to date are structurally related to the predecessor diketo acid and naphthyridine carboxamide scaffolds (12, 13). Raltegravir, the first INSTI approved for clinical use, has demonstrated the great therapeutic promise of this class of antiretrovirals (14). Later-developed INSTIs, including elvitegravir (15) and S/GSK-1349572 (16), are in advanced clinical trials. We recently reported structures of the PFV intasome complexed with raltegravir and elvitegravir. The INSTIs bind to the catalytic metal cations, inactivating the intasome by blocking the active site and dislocating the terminal 3′ nucleotide of the viral DNA (6). An experimental structure of the HIV-1 intasome has not yet been determined. However, due to the high level of conservation between retroviral INs, in particular within their active sites (6, 8, 9, 17), PFV IN can serve as a convenient proxy for structural studies of HIV INSTIs (18, 19). Ser217 is the only PFV IN residue in the immediate vicinity of the catalytic carboxylates that differs from its HIV-1 counterpart, and this amino acid does not directly contact bound INSTIs (6).
Fig. 1.
Chemical structures of the INSTIs used in this work. The metal chelating heteroatoms are colored red and the halogenated benzene groups are blue.
The use of raltegravir can lead to the development of viral resistance and virological failure in patients (20–22) (reviewed in ref. 23). HIV-1 mutations that give rise to raltegravir resistance map to IN and involve three primary genetic pathways: Y143H/R/C, Q148H/R/K, and N155H. Each of these changes is generally associated with secondary amino acid substitutions, some of which compensate for impaired viral fitness caused by the primary mutations (24). In the PFV intasome structure the side chain of Tyr212 makes a face-to-face π–π stacking interaction with the oxadiazole ring of bound raltegravir; this contact is likely a significant contributor to inhibitor binding and thus clearly explains why substitutions of HIV-1 IN Tyr143, the structural equivalent of PFV Tyr212, can influence drug potency. However, PFV IN residues Ser217 and Asn224, corresponding to HIV-1 IN Gln148 and Asn155, respectively, do not directly contact bound INSTIs in the cocrystal structures, leaving the molecular basis of resistance for the two major pathways unexplained. Herein, we present crystal structures of the PFV intasome complexed with an expanded set of INSTIs refined to resolutions of up to 2.5 Å. These structures allow us to describe the binding interactions between the drugs and the intasome more accurately and comprehensively. Furthermore, we present the structures of PFV intasomes containing the S217H and N224H substitutions, equivalent to HIV-1 IN Q148H/G140S and N155H, respectively, and explain the mechanism of drug resistance.
Results and Discussion
Binding Modes of INSTIs.
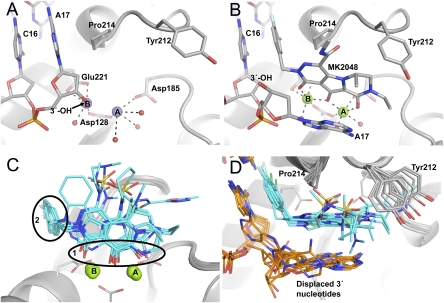
By modifying the viral DNA construct and the crystal preparation technique we were able to substantially improve the X-ray diffracting properties of the PFV intasome crystal form described previously (Table S1) (6). These modifications allowed us to collect X-ray data at higher resolution and, consequently, to build some of the previously unresolved regions of the intasome as well as place solvent molecules within the active sites and throughout the structure (Fig. 2 and Fig. S1). The structure was initially refined following crystal soaking in Mn2+ salts. Compared with Mg, Mn atoms produce much stronger electron density peaks, allowing more precise refinement of metal positions. The active site metal cations are coordinated between the side chains of Asp128 and Asp185 (metal A) or Asp128 and Glu221 (metal B) and have incomplete octahedral coordination by the active site carboxylates, the 3′ hydroxyl of viral DNA, and four bound water molecules (Fig. 2A and Fig. S1A). During the strand transfer reaction, metal B would be in position to activate the 3′ hydroxyl of the viral DNA for nucleophilic attack on a target DNA phosphodiester, with metal A stabilizing the hypothetical pentacovalent phosphorus transition state (25, 26). Soaking intasome crystals in Mg2+ salts resulted in metal binding to site A, leaving site B unoccupied (6). We speculate that binding of chromosomal DNA completes the coordination spheres for both metal binding sites, thereby increasing the affinity of site B for Mg2+.
Fig. 2.
Active site region of the PFV intasome bound to metal cofactors and INSTIs. With the exception of carbon atoms, standard atom coloring is used throughout: oxygen, red; nitrogen, blue; sulfur, yellow; fluorine, pale blue; chlorine, green; manganese, purple; magnesium, light green; phosphorus, orange. (A) The intasome active site in Mn2+-bound form. Protein is represented as a gray cartoon. Stick representations are shown for side chains mentioned in the text and DNA, colored by atom with gray used for carbon. Large purple spheres indicate Mn atoms and smaller red spheres show the water molecules coordinated to the metal atoms. (B) The Mg2+/MK2048-bound intasome active site with viewpoint and representation as in A. Here, MK2048 is also shown as a stick representation and Mg atoms as spheres. (C) Comparison of the different inhibitor-bound structures. Inhibitor molecules are shown as sticks with cyan carbon atoms. The black ovals highlight the conserved features of these drugs: 1, the metal chelating heteroatoms; and 2, the halogenated benzene ring. (D) An alternative view comparing drug binding, rotated 90° about the horizontal axis compared with C. Here, the side chain of Tyr212 is shown to highlight its involvement in binding different INSTIs; DNA is shown as sticks with carbon atoms colored orange.
To further our understanding of INSTI binding, we refined structures of PFV intasomes bound to Mg2+, the physiological cofactor, and eight small-molecule inhibitors representing six distinct INSTI scaffolds (Fig. 1): raltegravir (14), l-870,810 (13), PICA (27), GS9160 (28), MK0536 (29), MK2048 (30), 1 (30), and 2 (31). HIV-1 resistance profiles for most of these INSTIs or their close analogs have been reported (20, 32–37). Consistent with our previous results (6), the INSTIs bind to the active site, displacing the 3′-hydroxyl group (and indeed the entire 3′-terminal nucleotide of viral DNA) and thereby inactivating the intasome (Fig. 2 B–D and Fig. S1B).
The common metal-coordinating pharmacophore of the INSTIs comprises three specifically positioned heteroatoms (oxygen or, less commonly, nitrogen) that coordinate the two metal cofactors within the active site of IN (Figs. 1 and 2C). In all INSTI-bound structures, the three heteroatoms form part of the primary coordination sphere of the catalytic metals, with the central oxygen coordinated to both metals (Fig. 2C). In contrast to the intasome with Mn2+ alone, the Mg2+/INSTI-bound structures feature complete octahedral coordination for both metal cations (Fig. 2B). The only difference in inhibitor binding orientation is with the tricyclic hydroxypyrroles (MK2048, 1 and 2). For these inhibitors the saturated nature of the piperazinone ring disrupts the coplanarity of metal A and its coordinating heteroatoms, with a consequent 15° inclination in the plane of the core ring system of the bound inhibitors (Fig. S2A).
The second conserved structural feature of INSTIs is a halogenated benzyl group (Fig. 1). In the inhibitor-bound structures, this moiety displaces the 3′ adenine of bound viral DNA, inserting between the base of the penultimate cytidine, with which it forms a face-to-face π–π interaction, and Pro214 of IN (Fig. 2 B and D). 3-Cl substituents of the phenyl ring in bound inhibitors orient “in” toward the protein, causing a small (<1 Å) outward shift of the phenyl ring and thus altering the π–π contact with the base of the viral DNA (Fig. S2B). The fluorophenyl ring of PICA is separated from the closest metal chelating heteroatom by five bonds in contrast to the four bonds in all of the other INSTIs, which results in a different angle of its projection into the binding site for this compound (Fig. S2C).
With the exception of the terminal adenosine displacement, only minor conformational adjustment of the wild-type (WT) intasome structure is required to accommodate INSTI binding (Movie S1). There are small shifts in the positions of the metal atoms to reflect the differences in spacing between the coordinating pharmacophore atoms; these are maximally 0.7 Å for metal A and 0.6 Å for metal B (Fig. 2C). The most significant change in the protein itself is the side chain of Tyr212 (discussed above for raltegravir), the hydroxyl group of which moves by 3.1 Å due to variable hydrophobic interactions between its ring and the INSTIs (Fig. 2D). The movement of Tyr212 is most pronounced for the tricyclic hydroxypyrroles (MK2048, 1 and 2) and in these cases leads to additional movement of the β1–β2 loop within the IN C-terminal domain due to an interaction with overlying Ala328. The major difference between the various INSTI-bound intasome structures is the orientation of the displaced 3′ adenosine. In all cases the adenine base stacks against either the halogenated benzyl group or the core metal-coordinating ring system of the bound INSTI (Fig. 2D). In some cases it is clear that the base can flip between multiple binding modes; thus, in cocrystals with 1, the terminal adenosine was refined in two alternative conformations. It seems likely that the energetic differences between the observed alternate conformations of the displaced 3′ adenosine are minor, but it is unclear how, if at all, this flexibility contributes to INSTI binding.
S217H and N224H PFV INs Display Partial Resistance to Raltegravir.
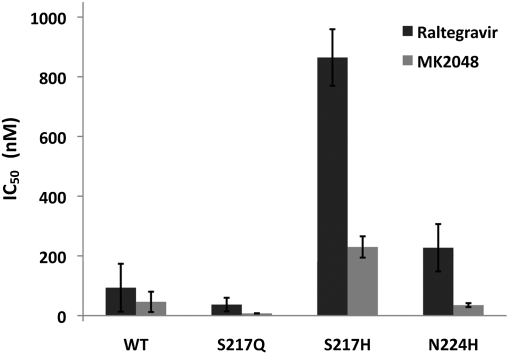
To assess the structural basis for the HIV-1 drug resistance mutations Q148H and N155H, we prepared the corresponding variant forms of PFV IN and assayed their strand transfer activities in the absence or presence of raltegravir and MK2048 (Fig. S3). PFV IN residue Ser217 corresponds to HIV-1 IN Gln148 and is the only amino acid in the direct vicinity of the INSTI-binding site that is not strictly conserved between the INs (6, 17). To evaluate the effects of amino acid substitutions at this position on INSTI binding, we evaluated S217Q and S217H PFV INs. In the absence of an INSTI, S217Q PFV IN showed ∼30% of WT PFV IN activity, whereas S217H was two- to threefold more active than WT. Raltegravir potently inhibited WT and S217Q PFV INs, with IC50 values of 90 and 40 nM, respectively (Fig. 3). Strikingly, PFV IN carrying the S217H substitution was 10-fold less susceptible to raltegravir (IC50 ∼ 900 nM) (Fig. 3). The activity of MK2048, a second-generation INSTI (30), was also affected by this substitution, although to a lesser degree, with the drug maintaining an IC50 of ∼200 nM against the S217H intasome (Fig. 3).
Fig. 3.
Bar plot showing the IC50 values of raltegravir and MK2048 against WT and mutant PFV INs. The mean values and the SDs were obtained from five independent measurements.
Asn155 is conserved in PFV IN as Asn224; hence, the HIV-1 IN N155H substitution was reproduced as N224H. Carrying this variation, PFV IN displayed ∼10% of WT activity and was inhibited by raltegravir with an IC50 of 200 nM, indicating an ∼twofold decrease in susceptibility to the INSTI compared with WT IN (Fig. 3). By contrast, MK2048 remained fully active against the N224H intasome (IC50 ∼ 25 nM) (Fig. 3). A similar in vitro assessment of HIV-1 IN mutant resistance to raltegravir found that N155H was ninefold less susceptible and Q148H was two- to threefold less susceptible (38). Combining Q148H with the common secondary mutation G140S further decreases HIV-1 IN susceptibility to raltegravir, making it ∼40-fold resistant to the drug (38). PFV IN naturally encodes Ser at position 209, equivalent to HIV-1 IN Gly140, likely explaining the high activity and high level of resistance observed in PFV IN carrying the sole S217H substitution. Similar to our observations with PFV IN (Fig. 3), the Q148H/G140S variant of HIV-1 IN, but not N155H, displays cross-resistance to MK2048 (33, 39).
Structural Basis of Resistance to INSTIs.
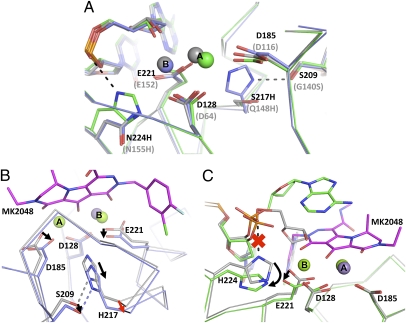
We refined structures of S217Q, S217H, and N224H mutant intasomes in the presence of Mn2+ or Mg2+ and the INSTI MK2048. In contrast to the WT PFV intasome, each of the three mutants bound only a single metal atom in the active site when soaked in the presence of Mn2+, indicating that the amino acid substitutions interfere with metal coordination. The loss of metal binding is apparently due to a combination of relatively subtle conformational adjustments in the active site region, because a single large change is not observed (Fig. 4A).
Fig. 4.
The effects of the S217H and N224H amino acid substitutions on the conformation of the PFV IN active site. (A–C) A ribbon representation is used for the protein backbone, metals are shown as spheres, and sticks are used to show relevant side chains and INSTI molecules. Standard atom coloring is used throughout except where noted. (A) Mn2+-bound active sites of WT, S217H, and N224H PFV intasomes. Carbon and manganese atoms are colored gray for WT, light blue for S217H, and green for N224H. The relevant side chains and mutations are indicated in black or gray according to the PFV or HIV-1 IN sequence, respectively. (B) Comparison of Mn2+- and Mg2+/MK2048-bound states of S217H active sites. Carbon atoms are colored gray for the Mn2+-bound structure and light blue for the MK2048 structure; carbon atoms of the MK2048 INSTI are magenta. Black arrows indicate side-chain movements necessary to accommodate INSTI binding and the red arrow indicates the required protein main-chain distortion. (C) Comparison of Mn2+- and Mg2+/MK2048-bound active sites in N224H intasome. Carbon atoms of Mn2+- and Mg2+/MK2048-bound structures are colored gray and green, respectively. Black arrows show the side-chain rearrangements required for INSTI binding, and the red “x” indicates the breaking of the His–phosphate interaction.
Ser217 is located on a 310 helix and positioned at the base of the active site, with its side chain pointing between the side chains of the catalytic residues Asp185 and Glu221. Intasome crystals containing amino acid substitutions at position 217 (both S217Q and S217H) exhibit loss of metal binding at site A due to the presence of the bulky His/Gln side chain that causes a slight shift of the Asp185 carboxylate (0.3 Å at Cγ, Fig. 4A). Because S217Q PFV IN was at least as sensitive to raltegravir as the WT enzyme was (Fig. 3), we conclude that the apparent reductions in metal binding affinity observed in our crystals are unlikely to account for the decreased raltegravir susceptibility of other PFV IN mutants.
INSTI binding to the WT PFV intasome induces very little structural change within the active site (Fig. 2 and Movie S1). Conversely, a pronounced change in protein backbone conformation is required for the S217H intasome to accommodate MK2048. Due to steric constraints, the entire His residue must give way to allow inhibitor binding, requiring a striking 1.1-Å displacement of its Cα atom (comparing WT:MK2048 and S217H:MK2048 structures) and thus weakening the hydrogen bond network of the 310 helix (Fig. 4B and Movie S2). Such an alteration in backbone conformation is likely to be energetically unfavorable, explaining why the S217H mutant is less susceptible to inhibition by INSTIs (Fig. 3). The PFV intasome encoding Gln at position 217 shows a structure intermediate between those of WT and S217H, with the main chain Cα of Gln217 shifting by 0.6 Å upon inhibitor binding. We conclude that this smaller change does not incur a significant energetic penalty. Due to the structure of the active site loop, the side chain of PFV IN Ser209, corresponding to HIV-1 IN Gly140, abuts that of Ser217 in the WT intasome. In both Mn2+- and Mg2+/MK2048-bound S217H (but not S217Q) intasomes, Ser209 is hydrogen bonded to His217. On the basis of these observations in the context of the PFV intasome, the implied proximity (and interaction) of residues 140 and 148 in HIV-1 IN likely explains the observed coevolution of resistance mutations at these positions. In a single Q148H HIV-1 IN mutant, Gly140, the smallest and most flexible amino acid, would better accommodate movement of the His side chain at position 148, thereby allowing INSTI binding with less rearrangement of the protein main chain (Fig. 4B), explaining the additional HIV-1 resistance observed with the Q148H/G140S/A double mutations over Q148H alone. Our modeling studies indicate that the bulky protonated side chains of Lys or Arg would cause an analogous active site disruption, with inhibitor binding to Q148K/R intasomes necessitating a similar main chain rearrangement.
Asn224 is located on the PFV IN α4 helix, approximately one helical turn from the catalytic Glu221, with its side chain projecting between the side chains of Glu221 and Asp128. The side chain of Asn224 is hydrogen-bonded to the carboxylate of Asp128, contributing to the outer coordination sphere of metal B. In contrast, in the N224H PFV intasome the imidazole moiety of the mutant His residue makes an electrostatic interaction with the phosphate group of the 3′ adenosine (Fig. 4A). This interaction causes a 0.5-Å shift of the DNA backbone as well as slight shifts of the catalytic Asp128 and Glu221 carboxylates, likely accounting for the loss of metal B in the mutant intasome (Fig. 4A). INSTI binding to the N224H intasome induces and stabilizes an ordered active site conformation and, necessarily, the His side chain moves out of the way, losing its interaction with the phosphate group (Fig. 4C and Movie S3). Although the protein backbone around the variant site shifts slightly upon inhibitor binding, it does not affect the α4 helix appreciably. The most likely cause of resistance of N224H PFV IN and, by proxy, HIV-1 IN N155H, to the inhibitors is the dependence of INSTI binding on disruption of the His–phosphate interaction.
Due to the consistency of the observed INSTI binding modes (Fig. 2C), the active site structural rearrangements required to accommodate any of the INSTIs studied here are expected to be similar to those seen in our MK2048-bound structures (Fig. 4 B and C). Thus, our results provide straightforward explanations as to why the S217H and N224H amino acid substitutions in PFV IN (and the corresponding N155H and Q148H/G140S substitutions in HIV-1 IN) lead to a reduction in INSTI potency. Less clear is why some INSTIs (MK2048, MK0536, and PICA) are less susceptible than raltegravir to the N155H and Q148H/G140S substitutions in HIV-1 IN (16, 30, 33). Recent data indicate that second-generation INSTIs such as MK2048 display substantially lower dissociation rates compared with raltegravir (40). Presumably, tighter binding INSTIs would better compensate for the energetic penalty associated with reordering the active sites of mutant INs. We note that some INSTI scaffolds offer better metal coordination geometry than others. For example, the structures containing the tricyclic hydroxypyrrole (MK2048, 1 and 2) or pyrrolidinone hydroxyquinoline scaffolds (GS9160) have INSTI(O)-Mg-(O)INSTI angles closest to the ideal 90° (Fig. S4). Furthermore, our initial quantum mechanical studies suggest that the tricyclic hydroxypyrroles may be more electronically adaptable than other INSTIs to subtle changes in active site geometry and electrostatics (compare polarizability values in Table S2). In addition, the electronic character of the metal-coordinating heteroatoms themselves may also be relevant (Table S2). Work in this area, based on both small molecule and complex simulations, is ongoing.
The INSTIs target the long-lived intermediate of the retroviral integration machinery, containing the product of 3′ processing, before engagement of chromosomal DNA. However, INSTI binding involves a fully ordered active site, coordinating a pair of metal cofactors. Our results highlight the ability of IN to de-tune its active site in this state, while retaining substantial enzymatic activity. This result is not entirely unexpected. Indeed, the presence of a scissile phosphodiester bond in the active site of the intasome (before 3′ processing or upon engagement of target DNA) would provide additional ligands to the metal cofactors, helping to order the active site for catalysis. Because binding of viral and target DNA relies on extensive networks of protein–DNA interactions that are distal to the active site (6, 26), IN can tolerate amino acid substitutions in the vicinity of the active site. One challenge for INSTI discovery is to design inhibitors that can accommodate this active site variability at lower cost to inhibitor potency.
Although the current study focused on the primary HIV-1 raltegravir resistance pathways involving IN Asn155 and Gln148, we note that our cocrystal structures will help elucidate a plethora of other INSTI resistance mutations. Thus, a change of HIV-1 IN Pro145 is associated with elvitegravir resistance (21). This residue corresponds to Pro214 of PFV IN, which is in direct contact with the halobenzyl groups of bound INSTIs. A change of HIV-1 IN Gly118 was observed in an MK2048-resistant HIV-1 strain (41); this residue corresponds to Gly187 in PFV IN, which is in a Van der Waals contact with the bound inhibitor in our structures. HIV-1 mutations causing amino acid substitutions farther away from the active site, where the level of sequence identity between PFV and HIV-1 INs becomes far less significant, can be studied using molecular dynamics approaches based on updated homology models of the HIV-1 intasome (18, 19).
Materials and Methods
Small-Molecule Inhibitors.
The INSTIs studied here (Fig. 1) were synthesized as described: l-870,810 (42), raltegravir (14), MK0536 (43), GS9160 (44), PICA (27), MK2048 (30), 1 (45) and 2 (31).
Protein Purification.
PFV IN was produced essentially as previously described (6). Briefly, PFV IN was expressed from pSSH6P-PFV-INFL (17) in Escherichia coli PC2 cells (46) for 4 h at 24 °C. Following cell lysis and lysate clarification, hexahistidine-tagged PFV IN was purified by NiNTA (Qiagen) affinity chromatography, and the tag removed by digestion with human rhinovirus 14 (HRV14) 3C protease. Subsequent affinity chromatography using a 5-mL HiTrap Heparin HP column (GE Healthcare) and size exclusion chromatography on a HiLoad 16/60 Superdex-200 column (GE Healthcare) in 500 mM NaCl, 50 mM Tris-HCl, pH 7.4, yielded pure protein. For storage, PFV IN was concentrated to 10–20 mg·mL−1, supplemented with 10% glycerol, flash frozen in liquid nitrogen, and kept at −80 °C. Mutations were introduced into pSSH6P-PFV-INFL, using a quick-change site-directed mutagenesis procedure (Stratagene). Expression and purification of PFV IN mutants followed the same protocol as that for WT PFV IN.
Intasome Preparation and Crystallization.
For PFV intasome assembly, donor DNA was prepared by annealing HPLC-grade oligonucleotides 5′-TGCGAAATTCCATGACA and 5′-ATTGTCATGGAATTTCGCA. The resulting construct had two A:T-to-G:C base pair substitutions (mutant nucleotides are underlined), introduced to improve stability of the crystal lattice. These changes involve nucleotides not specifically recognized by IN (6) and, consequently, do not affect the efficiency of intasome assembly. PFV intasome crystals were grown by hanging drop vapor diffusion at 18 °C against reservoirs containing 1.2–1.4 M ammonium sulfate, 25% (vol/vol) glycerol, 4.8% (wt/vol) 1,6-hexanediol, 1 mM EDTA, 50 mM 2-(N-morpholino) ethanesulfonic acid (Mes)-NaOH, pH 6.5. Whereas WT and N224H intasomes crystallized readily, S217Q and S217H intasomes required microseeding with WT intasome crystals. Before freezing, crystals were soaked for 2–9 d in 1.0 M ammonium sulfate, 25% (vol/vol) glycerol, 4.8% (wt/vol) 1,6-hexandiol, 50 mM Mes-NaOH, pH 6.5, supplemented with 25 mM MnCl2 or 25 mM MgCl2 and 1 mM INSTI (compounds not readily soluble under these conditions were used at above saturation concentration). The combined use of the stabilized viral DNA construct and the lower ammonium sulfate concentration during crystal soaking improved the resolution of X-ray diffraction from the original 2.9–2.8 Å (6) to 2.6–2.5 Å (Table S1). The highest resolution was obtained with crystals of N224H intasome soaked in the presence of Mn2+, which diffracted X-rays to 2.0 Å (Table S1), due to a fortuitous introduction of an additional crystal contact involving a Mn2+ ion and His224 from the outer IN subunit of the intasome.
Data Collection and Structure Refinement.
Data were collected on the following synchrotron beamlines: Proxima1 of Soleil, Paris, France (WT:MK0518, WT:MK2048, WT:l-870,810, WT:MK0536, WT:GS9160, WT:1, WT:2, S217H:Mn, and S217H:MK2048); X06DA of Swiss Light Source, Villigen PSI, Switzerland (WT:PICA); and I02 and I04 of Diamond Light Source, Oxfordshire, UK (WT:Mn, N224H:Mn, N224H:MK2048, S217Q:Mn, and S217Q:MK2048). Diffraction data were processed using Mosflm and Scala programs of the CCP4 suite (47). All crystals were near isomorphous and belonged to space group P41212 with unit cell dimensions of a = b ≈ 160 Å and c ≈ 123.5 Å (Table S1). The structure was first refined against data collected from WT crystals soaked in Mn2+, using PDB ID 3L2S coordinates (6) as a starting model. Following several rounds of manual editing in Coot (48), including adding solvent molecules, simulated annealing using Phenix (49), and restrained refinement in Refmac (50), the final model had Rwork/Rfree values of 0.197/0.222 with good geometry (Table S1). The final WT:Mn structure was used as a starting point for building the Mn2+-bound mutant and WT:MK2048 structures, which were all refined using a combination of manual editing in Coot and restrained refinement in Refmac. The WT:MK2048 structure was subsequently used as a starting point for building all remaining drug-bound structures. Final structures were validated with MolProbity (51). Data collection and structure refinement statistics are given in Table S1. Coordinates and structure factors were deposited with the Protein Data Bank; the corresponding accession codes are listed in Table S1. Morphing of the inhibitor-free and -bound structures was done using LSQMAN (52); figures and movies were generated with PyMol (http://www.pymol.org).
PFV Integration Assay and Quantification.
For quantitative strand transfer assays, donor DNA substrate was formed by annealing HPLC grade oligonucleotides 5′-GACTCACTATAGGGCACGCGTCAAAATTCCATGACA and 5′-ATTGTCATG GAATTTTGACGCGTGCCCTATAGTGAGTC. Reactions (40 μL) contained 0.75 μM PFV IN, 0.75 μM donor DNA, 4 nM (300 ng) supercoiled pGEM9-Zf(−) (Promega) target DNA, 125 mM NaCl, 5 mM MgSO4, 4 μM ZnCl2, 10 mM DTT, 0.8% (vol/vol) DMSO, and 25 mM BisTris propane–HCl, pH 7.45. INSTIs were added at indicated concentrations. Reactions were initiated by addition of 2 μL PFV IN diluted in 150 mM NaCl, 2 mM DTT, and 10 mM Tris-HCl, pH 7.4, and stopped after 1 h at 37 °C by addition of 25 mM EDTA and 0.5% (wt/vol) SDS. Reaction products, deproteinized by digestion with 20 μg proteinase K (Roche) for 30 min at 37 °C followed by ethanol precipitation, were separated in 1.5% agarose gels and visualized by staining with ethidium bromide.
Integration products were quantified by quantitative real-time PCR, using Platinum SYBR Green qPCR SuperMix (Invitrogen) and three primers: 5′-CTACTTACTCTAGCTTCCCGGCAAC, 5′-TTCGCCAGTTAATAGTTTGCGCAAC, and 5′-GACTCACTATAGGGCACGCGT. PCR reactions (20 μL) contained 0.5 μM of each primer and 1 μL diluted integration reaction product. Following a 5-min denaturation step at 95 °C, 35 cycles were carried out in a CFX96 PCR instrument (Bio-Rad), using 10 s denaturation at 95 °C, 30 s annealing at 56 °C and 1 min extension at 68 °C. Standard curves were generated using serial dilutions of WT PFV IN reaction in the absence of INSTI.
Computational Methods.
Inhibitor conformations were taken from the intasome-bound complexes reported in the present study. Hydrogen atoms were added and the hydroxyls that coordinate to Mg2+ were deprotonated. Single-point B3LYP/6–31G**+ calculations were performed with Jaguar (Maestro 9.1; Schrödinger) to calculate various electronic properties (Table S2).
Supplementary Material
Acknowledgments
We thank Alan Engelman for critical reading of the manuscript and helpful discussions, and the staff of Proxima1 (Soleil, Paris, France), X06DA (Swiss Light Source, Villigen PSI, Switzerland), and I02 and I04 (Diamond Light Source, Oxfordshire, United Kingdom) beamlines for assistance with data collection. This work was supported by United Kingdom Medical Research Council Grant G0900116.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3OY9, 3OYA, 3OYB, 3OYC, 3OYD, 3OYE, 3OYF, 3OYG, 3OYH, 3OYI, 3OYJ, 3OYK, 3OYL, 3OYM, and 3OYN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010246107/-/DCSupplemental.
References
- 1.Craigie R. Retroviral DNA integration. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 613–630. [Google Scholar]
- 2.Lewinski MK, Bushman FD. Retroviral DNA integration—mechanism and consequences. Adv Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 3.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: Studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: Reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J Mol Biol. 2010;399:491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McColl DJ, Chen X. Strand transfer inhibitors of HIV-1 integrase: Bringing IN a new era of antiretroviral therapy. Antiviral Res. 2010;85:101–118. doi: 10.1016/j.antiviral.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Marchand C, Maddali K, Métifiot M, Pommier Y. HIV-1 IN inhibitors: 2010 update and perspectives. Curr Top Med Chem. 2009;9:1016–1037. doi: 10.2174/156802609789630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazuda DJ, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 13.Egbertson MS, et al. A potent and orally active HIV-1 integrase inhibitor. Bioorg Med Chem Lett. 2007;17:1392–1398. doi: 10.1016/j.bmcl.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 14.Summa V, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 15.Sato M, et al. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem. 2006;49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 16.Johns B, et al. 17th Conference on Retroviruses and Opportunistic Infections. San Francisco: 2010. The discovery of S/GSK1349572: A once-daily next generation integrase inhibitor with a superior resistance profile. [Google Scholar]
- 17.Valkov E, et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang J, Maddali K, Pommier Y, Sham YY, Wang Z. Scaffold rearrangement of dihydroxypyrimidine inhibitors of HIV integrase: Docking model revisited. Bioorg Med Chem Lett. 2010;20:3275–3279. doi: 10.1016/j.bmcl.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnan L, et al. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci USA. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper DA, et al. BENCHMRK Study Teams Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, et al. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 2008;80:213–222. doi: 10.1016/j.antiviral.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Charpentier C, et al. Drug resistance profiles for the HIV integrase gene in patients failing raltegravir salvage therapy. HIV Med. 2008;9:765–770. doi: 10.1111/j.1468-1293.2008.00628.x. [DOI] [PubMed] [Google Scholar]
- 23.Métifiot M, Marchand C, Maddali K, Pommier Y. Resistance to integrase inhibitors. Viruses. 2010;2:1347–1366. doi: 10.3390/v2071347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delelis O, et al. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 2009;37:1193–1201. doi: 10.1093/nar/gkn1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X ray structures of its key intermediates. Nature. 2010 doi: 10.1038/nature09517. 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuki A, Li X, Plewe MB, Wang H, Zhang J. HIV-integrase inhibitors, pharmaceutical compositions, and methods for their use. 2006 US Patent 7,001,912 B2. [Google Scholar]
- 28.Jones GS, et al. Preclinical evaluation of GS-9160, a novel inhibitor of human immunodeficiency virus type 1 integrase. Antimicrob Agents Chemother. 2009;53:1194–1203. doi: 10.1128/AAC.00984-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perlow DS, et al. 235th ACS National Meeting. New Orleans: 2008. Discovery and synthesis of a potent long-acting inhibitor of HIV integrase. [Google Scholar]
- 30.Vacca J, et al. Fourth International AIDS Socienty Conference. Sydney: 2007. Discovery of MK-2048—subtle changes confer unique resistance properties to a series of tricyclic hydroxypyrrole integrase strand transfer inhibitors. [Google Scholar]
- 31.Wai JS, et al. Pyrazinopyrrolopyridazines as HIV integrase inhibitors, their preparation, pharmaceutical compositions, and use to prevent or treat HIV infection. 2005 PCT Appl WO2005/110415. [Google Scholar]
- 32.Goethals O, et al. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol. 2008;82:10366–10374. doi: 10.1128/JVI.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goethals O, et al. Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology. 2010;402:338–346. doi: 10.1016/j.virol.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 34.Hazuda D, Miller M, Nguyen B, Zhao J. XVI International HIV Drug Resistance Workshop. St Michael, Barbados: 2007. Resistance to the HIV-integrase inhibitor raltegravir: Analysis of protocol 005, a phase II study in patients with triple-class resistant HIV-1 infection. [Google Scholar]
- 35.Johnson VA, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 36.Paredes R, Clotet B. Clinical management of HIV-1 resistance. Antiviral Res. 2010;85:245–265. doi: 10.1016/j.antiviral.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 37.Hombrouck A, et al. Mutations in human immunodeficiency virus type 1 integrase confer resistance to the naphthyridine L-870,810 and cross-resistance to the clinical trial drug GS-9137. Antimicrob Agents Chemother. 2008;52:2069–2078. doi: 10.1128/AAC.00911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Métifiot M, et al. Biochemical and pharmacological analyses of HIV-1 integrase flexible loop mutants resistant to raltegravir. Biochemistry. 2010;49:3715–3722. doi: 10.1021/bi100130f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey KK, Bera S, Vora AC, Grandgenett DP. Physical trapping of HIV-1 synaptic complex by different structural classes of integrase strand transfer inhibitors. Biochemistry. 2010;49:8376–8387. doi: 10.1021/bi100514s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grobler JA, Stillmock K, Miller MD, Hazuda DJ. Mechanism by which the HIV integrase active-site mutation N155H confers resistance to raltegravir. Antivir Ther. 2008;13(Suppl 3):A41. [Google Scholar]
- 41.Bar-Magen T, et al. Identification of novel mutations responsible for resistance to MK-2048, a second-generation HIV-1 integrase inhibitor. J Virol. 2010;84:9210–9216. doi: 10.1128/JVI.01164-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anthony NJ, Xu W, Lepore JV, Mahajan AV. Process for the preparation of a Na salt of a 5-(dioxidothiazinanyl)naphthyridine-7-carboxamide HIV integrase inhibitor. 2003 PCT Appl WO2003/016315. [Google Scholar]
- 43.Matty LJ, Wang Y. Preparation of crystalline 6-(3-chloro-4-fluorobenzyl)-4-hydroxy-2-isopropyl-N,N-dimethyl-3,5-dioxo-2,3,5,6,7,8-hexahydro-2,6-naphthyridine-1-carboxamide sodium salt as an HIV integrase inhibitor. 2006 PCT Appl WO2006/107478. [Google Scholar]
- 44.Evans JW, et al. Processes and intermediates useful for preparing integrase inhibitor compounds. 2008 US Patent Appl US2008/0039487. [Google Scholar]
- 45.Wai JS, et al. 14th Conference on Retroviruses and Opportunistic Infections. Los Angeles: 2007. Next generation of inhibitors of HIV-1 integrase strand transfer inhibitor: Structural diversity and resistance profiles. [Google Scholar]
- 46.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.