Abstract
PII proteins control key processes of nitrogen metabolism in bacteria, archaea, and plants in response to the central metabolites ATP, ADP, and 2-oxoglutarate (2-OG), signaling cellular energy and carbon and nitrogen abundance. This metabolic information is integrated by PII and transmitted to regulatory targets (key enzymes, transporters, and transcription factors), modulating their activity. In oxygenic phototrophs, the controlling enzyme of arginine synthesis, N-acetyl-glutamate kinase (NAGK), is a major PII target, whose activity responds to 2-OG via PII. Here we show structures of the Synechococcus elongatus PII protein in complex with ATP, Mg2+, and 2-OG, which clarify how 2-OG affects PII–NAGK interaction. PII trimers with all three sites fully occupied were obtained as well as structures with one or two 2-OG molecules per PII trimer. These structures identify the site of 2-OG located in the vicinity between the subunit clefts and the base of the T loop. The 2-OG is bound to a Mg2+ ion, which is coordinated by three phosphates of ATP, and by ionic interactions with the highly conserved residues K58 and Q39 together with B- and T-loop backbone interactions. These interactions impose a unique T-loop conformation that affects the interactions with the PII target. Structures of PII trimers with one or two bound 2-OG molecules reveal the basis for anticooperative 2-OG binding and shed light on the intersubunit signaling mechanism by which PII senses effectors in a wide range of concentrations.
Keywords: metabolic signaling, nitrogen regulation, cyanobacteria, chloroplasts
The PII proteins constitute one of the largest and most widely distributed family of signal transduction proteins present in archaea, bacteria, and plants. They control key processes of nitrogen metabolism in response to central metabolites ATP, ADP, and 2-oxoglutarate (2-OG), signaling cellular energy and carbon and nitrogen abundance (1–4). These effectors bind to PII in an interdependent manner (see below), thereby transmitting metabolic information into structural states of this sensor protein (3, 5). Furthermore, PII proteins may be posttranslationally modified (1, 6). Depending on the signal input states, PII proteins bind and thereby regulate the activity of key metabolic and regulatory enzymes, transcription factors, or transport proteins (1–3). In cyanobacteria and plants, the controlling enzyme of arginine biosynthesis, N-acetyl-L-glutamate kinase (NAGK), is a major PII target (7–9). Moreover, PII affects gene expression in cyanobacteria through binding to the transcriptional coactivator of NtcA, PipX (10). In plants, acetyl-CoA carboxylase was recently shown to be regulated by PII, providing an additional link between carbon and nitrogen regulation (11). Although these PII targets share no common structural element, interaction with PII is inhibited by 2-OG.
PII proteins are homotrimers of 12- to 13-kDa subunits, built of a double ferredoxin-like fold-containing core (βαβ-βαβ), with a characteristic and highly conserved 3D structure, as revealed from numerous crystal structures (3, 12). The trimeric PII architecture resembles a flattened barrel with long and flexible T loops extending outward, from the flat side (see Fig. 1 and Fig. S1). These T loops can adopt multiple conformations and mediate the versatile protein–protein interactions (3). Each subunit further comprises two small loops (B and C loop) in the intersubunit clefts, facing each other from opposing subunits and taking part in a unique mode of ATP-ADP binding (13–15). ADP and ATP compete here for the same site. In the presence of Mg-ATP, up to three 2-OG molecules can bind per trimer (1) with ADP antagonizing 2-OG binding (16). Notably, Arabidopsis thaliana PII is an exception, because it binds 2-OG also in the presence of ADP (5, 17). Another feature characteristic for many PII proteins is also intriguing: The three ATP-binding sites and the three 2-OG–binding sites each exhibit negative cooperativity. Anticooperativity implies strong conformational coupling between the subunits, and this feature probably allows PII to sense a wide range of metabolite concentrations (5, 16, 18, 19). In contrast to the ATP-ADP–binding site, the 2-OG–binding site is controversial (3). From the crystal structure of a PII paralogue from Methanococcus jannaschii, GlnK1, one 2-OG molecule was shown to bind from outside to the distal side of the T loop in the presence of Mg-ATP (20). By contrast, a recently published structure of a PII homologue form Azospirillum brasiliense in complex with Mg-ATP and 2-OG revealed the 2-OG–binding site close to the base of the T loop and near the ATP-binding site (21). However, neither of these two structures was proved by biochemical studies nor could they explain the anticooperative binding of 2-OG.
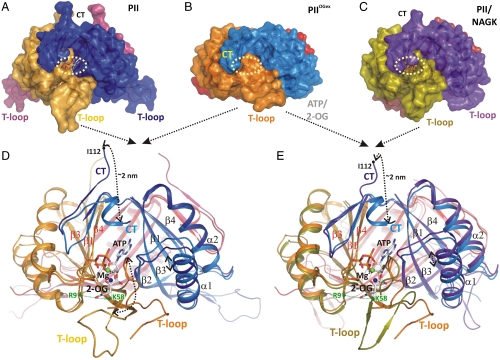
Fig. 1.
Structural basis of PII regulation by 2-oxoglutarate. (A–C) Side views of different S. elongatus PII structures in surface representation. The location of the ATP-binding site is indicated by a yellow dashed circle; the red boxes highlight the structure part discussed detailed in D. (A) Ligand-free PII modeled together with ATP (placed according to the superimposed 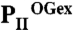 structure). The subunits are color coded in dark blue, light orange, and warm pink. The extended C termini and T loops of the proteins chains are marked CT and T-loop, respectively. (B) PII/ATP/2-OG (
structure). The subunits are color coded in dark blue, light orange, and warm pink. The extended C termini and T loops of the proteins chains are marked CT and T-loop, respectively. (B) PII/ATP/2-OG (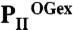 ) complex structure; subunits color coded in orange, marine blue, and red. The structure adopts a compact conformation because of the back folding of the C terminus (CT) onto the core domain structure and the T loop observed in a partially disordered conformation. (C) Structure of PII in the PII–NAGK complex (PDB ID code: 2V5H) modeled together with ATP. Subunits are color coded in magenta, deep olive, and salmon. (D) Superposition of the ligand-free PII and
) complex structure; subunits color coded in orange, marine blue, and red. The structure adopts a compact conformation because of the back folding of the C terminus (CT) onto the core domain structure and the T loop observed in a partially disordered conformation. (C) Structure of PII in the PII–NAGK complex (PDB ID code: 2V5H) modeled together with ATP. Subunits are color coded in magenta, deep olive, and salmon. (D) Superposition of the ligand-free PII and 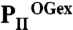 structure as ribbon models in the color codes derived from A and B. Secondary structure elements are marked with β1–β5 and α1–α2 and ATP and 2-OG are shown in stick representation, whereas the Mg ion is marked in green. The sites of two mutations, R9L and K58M, introduced to prove the binding mode of 2-OG are marked with green dots. Remarkable conformational transitions are marked with dashed lines: movement of the C terminus, a shift of helix 1, and displacement of the T-loop base by 2-OG while the tip becomes disordered. (E) Superposition of
structure as ribbon models in the color codes derived from A and B. Secondary structure elements are marked with β1–β5 and α1–α2 and ATP and 2-OG are shown in stick representation, whereas the Mg ion is marked in green. The sites of two mutations, R9L and K58M, introduced to prove the binding mode of 2-OG are marked with green dots. Remarkable conformational transitions are marked with dashed lines: movement of the C terminus, a shift of helix 1, and displacement of the T-loop base by 2-OG while the tip becomes disordered. (E) Superposition of 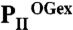 and PII–NAGK structures as ribbon models in the color codes derived from B and C. Structural details are indicated as in D.
and PII–NAGK structures as ribbon models in the color codes derived from B and C. Structural details are indicated as in D.
The structures of complexes of PII with its regulatory target NAGK from Synechococcus elongatus and A. thaliana are highly similar (22, 23), and the mode of interaction and regulation is apparently conserved in cyanobacteria and plants (24). The PII–NAGK complex involves one hexameric (trimer of dimers) NAGK toroid sandwiched between two PII trimers with the threefold axis aligned (23). Each PII subunit engages two contact surfaces in NAGK binding: A smaller surface involves the B loop and a larger is formed by the T loop, which adopts a tightly bent conformation that fits into the interdomain crevice of NAGK. Binding of PII enhances the catalytic activity of NAGK and alleviates feedback inhibition by arginine. To bind NAGK, free PII has to contract its extended T loop. Recently, a two-step process of PII–NAGK binding was proposed on the basis of the properties of a newly identified S. elongatus PII variant (I86N), which mimics the PII conformation in the NAGK-bound state (18): First, a salt bridge between PII-E85 and NAGK-R233 forms, triggering the extended T loop to fold into the tightly bent conformation that fits into NAGK.
Detailed structural information together with the well-studied biochemical features and highly sensitive complex formation assays render the PII–NAGK system ideal to study the transduction of metabolic signals into protein action. Until now, it was unclear how the 2-OG signal, perceived by PII, results in inhibition of PII–NAGK complex formation. To answer this question, we solved structures of PII with ATP, Mg2+, and 2-OG, revealing the mechanism by which the 2-OG signal is perceived by PII and controls receptor interactions.
Results
Structure of PII in Complex with ATP and 2-OG.
Crystals of recombinant PII protein from S. elongatus were obtained in the presence of excess 2-OG/Mg-ATP (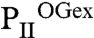 ) and at substoichiometric amounts of 2-OG (
) and at substoichiometric amounts of 2-OG (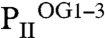 ) (Table S1). All structures were solved by molecular replacement using the free S. elongatus PII as a model [Protein Data Bank (PDB) ID code 1QY7]. The
) (Table S1). All structures were solved by molecular replacement using the free S. elongatus PII as a model [Protein Data Bank (PDB) ID code 1QY7]. The 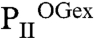 crystal diffracting to 2.2 Å contains two identical trimers in the asymmetric unit (with an rmsd of 0.2 Å2 for all C atoms superposed). Each trimer contains three Mg2+, ATP, and 2-OG molecules. In Fig. 1, this structure is compared to the structures of ligand-free PII (14) and PII in complex with NAGK (23). In the ligand-free structure, the T loop, and the C terminus adopt highly extended conformations away from the ATP-binding site (see Fig. 1 A and D). Here, PII offers an open binding cavity volume of approximately 1,000 Å3 for the ligands. Upon ATP and 2-OG binding, a significant conformational change in the T loop and the C terminus occurs: The C-terminal β-sheet of the free form moves toward the ATP-binding site by up to 20 Å (for the terminal residue I112) to change into a small helix (see Fig. 1D). The T loop contracts and the tip (from residues 44–50) becomes disordered. Part of the flexible T loop moves toward the ATP-binding site to form a kink-like structure (residues 36–41), thereby forming the scaffold for proper ATP and 2-OG assembly. As a result, the ATP/2-OG–binding cavity is completely enclosed by this unique T-loop conformation and the movement of the C terminus (see Fig. 1B, yellow circle). In the PII structure complexed with NAGK, the T loop is clamped in a different conformation (see Fig. 1 C and E). Here, access to the ligand-binding cavity is slightly reduced compared to the ATP-free form because of a bend in the T-loop conformation, thereby reducing the cavity volume to about 600 Å3.
crystal diffracting to 2.2 Å contains two identical trimers in the asymmetric unit (with an rmsd of 0.2 Å2 for all C atoms superposed). Each trimer contains three Mg2+, ATP, and 2-OG molecules. In Fig. 1, this structure is compared to the structures of ligand-free PII (14) and PII in complex with NAGK (23). In the ligand-free structure, the T loop, and the C terminus adopt highly extended conformations away from the ATP-binding site (see Fig. 1 A and D). Here, PII offers an open binding cavity volume of approximately 1,000 Å3 for the ligands. Upon ATP and 2-OG binding, a significant conformational change in the T loop and the C terminus occurs: The C-terminal β-sheet of the free form moves toward the ATP-binding site by up to 20 Å (for the terminal residue I112) to change into a small helix (see Fig. 1D). The T loop contracts and the tip (from residues 44–50) becomes disordered. Part of the flexible T loop moves toward the ATP-binding site to form a kink-like structure (residues 36–41), thereby forming the scaffold for proper ATP and 2-OG assembly. As a result, the ATP/2-OG–binding cavity is completely enclosed by this unique T-loop conformation and the movement of the C terminus (see Fig. 1B, yellow circle). In the PII structure complexed with NAGK, the T loop is clamped in a different conformation (see Fig. 1 C and E). Here, access to the ligand-binding cavity is slightly reduced compared to the ATP-free form because of a bend in the T-loop conformation, thereby reducing the cavity volume to about 600 Å3.
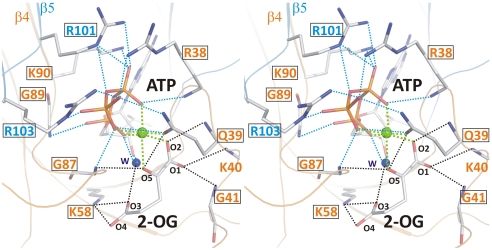
The 2-OG–binding site of PII is formed by an ATP-chelated Mg2+ ion and residues from one side of the intersubunit cleft (Fig. 2). The ATP molecule bound in the intersubunit cleft is ligated mainly by arginine residues (R38 from monomer A; R101 and R103 from monomer B) together with K90 from monomer A and a small number of hydrogen bonds. ATP fixed by these residues forms the scaffold for Mg2+-mediated binding of 2-OG. The Mg2+ ion has an almost perfect hexagonal coordination sphere of oxygen atoms, three of which are contributed by oxo groups of the α-, β-, and γ-phosphate of ATP. Two additional ligating atoms are accounted for by the O2 and O5 of 2-OG. The last coordination position is contributed by the OE1 atom of residue Q39. Notably, the 2-OG–binding sphere mainly comprises residues from the T loop. Backbone atoms from residues Q39, K40, and G41 (all contacting O1 or O2 of 2-OG) together with the Q39 side-chain ligate the O5 atom and form one area of interactions. A second interaction region is composed of the backbone nitrogen of G87 (forming a second H bridge to O5) and a strong salt bridge of K58 toward the O3 and O4 atoms (distance of 2.7 Å). Furthermore, residue R9 approaches the O4 to 3.5 Å. All these residues with the exception of K40 are highly conserved in PII proteins (4) (Fig. S1).
Fig. 2.
Stereo image of the 2-OG–binding site. Residues involved in binding of 2-OG (atoms are marked with small numbers) and the hydrophilic portion of ATP are numbered according to the sequence. Cofactors as well as side- and main-chain atoms are marked in stick representation; Mg2+ is marked as a green sphere. Colors of residue numbers (orange and blue) correspond to those of the respective subunits. Residues conserved in standard alignments are boxed. Dashed blue lines represent bonds between residues and ATP and black lines indicate bonds for the ligation of 2-OG, whereas green lines mark the hexagonal coordination of the Mg2+ ion.
To validate that 2-OG in the crystal occupies the true binding site, PII variants were constructed, in which residues K58 and R9, whose side chains according to the 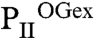 structure specifically interact with 2-OG, were replaced by similar-sized uncharged residues (K58M and R9L). Binding of 2-OG in the presence of Mg-ATP was determined by isothermal calorimetry (ITC). Indeed, the PII K58M variant was completely unable to bind 2-OG, although ATP could still be bound, showing that the loss of 2-OG binding is a specific effect of the K58 replacement. In further agreement with the structural prediction, the PII R9L variant was strongly impaired in 2-OG binding (Table 1). Moreover, both PII variants were impaired in NAGK binding, confirming that K58 is indeed pivotal for folding the T loop in the tightly bent structure. The R9 side chain is near the contact surface to NAGK and appears to stabilize the B-loop–T-loop interface (23) (Fig. S2).
structure specifically interact with 2-OG, were replaced by similar-sized uncharged residues (K58M and R9L). Binding of 2-OG in the presence of Mg-ATP was determined by isothermal calorimetry (ITC). Indeed, the PII K58M variant was completely unable to bind 2-OG, although ATP could still be bound, showing that the loss of 2-OG binding is a specific effect of the K58 replacement. In further agreement with the structural prediction, the PII R9L variant was strongly impaired in 2-OG binding (Table 1). Moreover, both PII variants were impaired in NAGK binding, confirming that K58 is indeed pivotal for folding the T loop in the tightly bent structure. The R9 side chain is near the contact surface to NAGK and appears to stabilize the B-loop–T-loop interface (23) (Fig. S2).
Table 1.
Effector molecule binding to PII variants R9L and K58M
| Kd1, μM | Kd2, μM | Kd3, μM | |
| 2-OG (+1 mM ATP) | |||
| R9L | 441 ± 40 | 123 ± 7 | 509 ± 116 |
| K58M | ND | ND | ND |
| (WT) | (5.1 ± 4.0) | (11.1 ± 1.8) | (106.7 ± 14.8) |
| ATP | |||
| R9L | NM | NM | NM |
| K58M | 10 ± 5 | 262 ± 136 | 31 ± 15 |
| (WT) | (4.0 ± 0.1) | (12.5 ± 0.9) | (47.4 ± 21.9) |
Values correspond to the mean of two independent experiments ± SEM. The raw data were fitted by using a three-site binding model for a PII trimer. For comparison, the original data for WT PII protein are given in parentheses. ND, not detectable; NM, not measured.
Structural Basis of Sequential/Anticooperative 2-OG Binding.
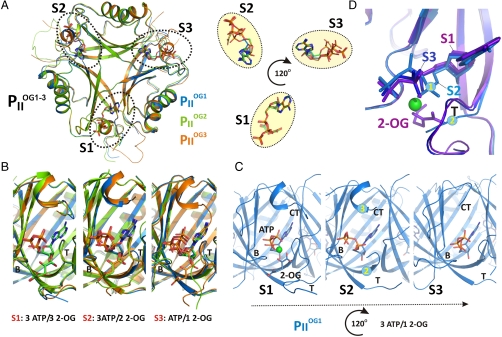
Crystallization of PII protein in the presence of low 2-OG amounts yielded PII structures with differing 2-OG content. The structure resolved at a resolution of 1.95 Å contains three PII trimers in the asymmetric unit; one trimer contained three ATP, one Mg2+, and one 2-OG (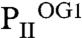 ), the second three ATP, two Mg2+, and two 2-OG (
), the second three ATP, two Mg2+, and two 2-OG (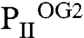 ), and the third three each ATP, Mg2+, and 2-OG (
), and the third three each ATP, Mg2+, and 2-OG (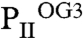 ) (Fig. 3 and Fig. S3). Additional details of structural parameters are given in SI Text (Tables S1 and S2). Binding of 2-OG does not significantly render the B-factor distribution of the three monomers significantly, unless the mobile elements (C terminus and T loop) contributing to binding are involved (Fig. S4). A superimposition of the three structurally similar trimers (Fig. 3 A and B) reveals the structure identity of the S1 site, which is occupied by 2-OG in all three
) (Fig. 3 and Fig. S3). Additional details of structural parameters are given in SI Text (Tables S1 and S2). Binding of 2-OG does not significantly render the B-factor distribution of the three monomers significantly, unless the mobile elements (C terminus and T loop) contributing to binding are involved (Fig. S4). A superimposition of the three structurally similar trimers (Fig. 3 A and B) reveals the structure identity of the S1 site, which is occupied by 2-OG in all three 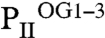 trimers, and the divergence in the S2 and S3 sites, respectively (Fig. 3B). The ligands are bound in S1 identical in topology to the mode described for the
trimers, and the divergence in the S2 and S3 sites, respectively (Fig. 3B). The ligands are bound in S1 identical in topology to the mode described for the 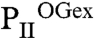 structure (Figs. 2 and 3B). The
structure (Figs. 2 and 3B). The 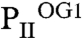 structure reveals that binding of the first 2-OG molecule to PII (S1 site) generates unequal ligand-binding sites in the adjacent monomers, and, remarkably, sites not occupied by 2-OG also lack the Mg2+ ion. The conformational differences in the nonoccupied binding sites provide a structure-based explanation for the anticooperativity observed in biochemical experiments: After occupation of the first site, the Kd for the second site increases slightly, but after occupation of the second site, the Kd for the third site increases strongly (about 20-fold higher than the Kd for the first site; see Table 1). In
structure reveals that binding of the first 2-OG molecule to PII (S1 site) generates unequal ligand-binding sites in the adjacent monomers, and, remarkably, sites not occupied by 2-OG also lack the Mg2+ ion. The conformational differences in the nonoccupied binding sites provide a structure-based explanation for the anticooperativity observed in biochemical experiments: After occupation of the first site, the Kd for the second site increases slightly, but after occupation of the second site, the Kd for the third site increases strongly (about 20-fold higher than the Kd for the first site; see Table 1). In 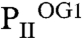 , the ATP molecule attached to the S2 site exhibits a significantly altered conformation of the phosphate moiety (Fig. 3 B–D); furthermore, the T-loop basis is displaced and the C terminus is ordered similar to the S1 site (Fig. 3 C and D). The S2 site in
, the ATP molecule attached to the S2 site exhibits a significantly altered conformation of the phosphate moiety (Fig. 3 B–D); furthermore, the T-loop basis is displaced and the C terminus is ordered similar to the S1 site (Fig. 3 C and D). The S2 site in 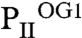 resembles the S3 site of the
resembles the S3 site of the 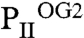 structure, which, according to the sequential 2-OG–binding mode, corresponds to the lowest affinity site (for detailed comparison of the binding sites, see Fig. S3). The S3 site of
structure, which, according to the sequential 2-OG–binding mode, corresponds to the lowest affinity site (for detailed comparison of the binding sites, see Fig. S3). The S3 site of 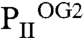 exhibits further changes, visible most significantly in the T loop, the C terminus, and a small distortion in the β4-strand. Together these changes can lead to the strongly altered affinity of 2-OG toward the stereochemically differing S2 and S3 sites.
exhibits further changes, visible most significantly in the T loop, the C terminus, and a small distortion in the β4-strand. Together these changes can lead to the strongly altered affinity of 2-OG toward the stereochemically differing S2 and S3 sites.
Fig. 3.
Anticooperativity of 2-OG–binding sites. (A) Top view of the 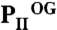 structure as a ribbon plot and superposition of the
structure as a ribbon plot and superposition of the 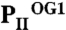 (in blue),
(in blue), 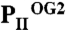 (in green), and
(in green), and 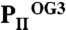 (in orange) structures. The three ATP/2-OG–binding sites are marked by dashed circles and numbered (S1, S2, and S3). The picture on the right represents the cofactors bound in the individual sites (highlighted in yellow) with three ATP and 2-OG molecules in S1, three ATP and two 2-OG molecules in S2, and three ATP and one 2-OG molecules located in S3, respectively. The clockwise consecutive 120° binding into S1 → S2 → S3 sites is shown by an arrow. (B) Zoom in (side view) of the three binding sites after superposition of the molecules. The content of the individual binding sites is marked below the picture. T and B loops are marked with T and B, respectively, for clarity. (C) Binding sites S1, S2, and S3 of the
(in orange) structures. The three ATP/2-OG–binding sites are marked by dashed circles and numbered (S1, S2, and S3). The picture on the right represents the cofactors bound in the individual sites (highlighted in yellow) with three ATP and 2-OG molecules in S1, three ATP and two 2-OG molecules in S2, and three ATP and one 2-OG molecules located in S3, respectively. The clockwise consecutive 120° binding into S1 → S2 → S3 sites is shown by an arrow. (B) Zoom in (side view) of the three binding sites after superposition of the molecules. The content of the individual binding sites is marked below the picture. T and B loops are marked with T and B, respectively, for clarity. (C) Binding sites S1, S2, and S3 of the 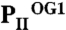 structure. In the S1 site, ATP, 2-OG, and Mg2+ (green sphere) are bound, whereas S2 and S3 contain only ATP and no Mg2+. Significant changes in the C terminus and the T loop in site S2 are marked with numbered circles. (D) Superposition of effector molecules bound to sites S1, S2, and S3 in the
structure. In the S1 site, ATP, 2-OG, and Mg2+ (green sphere) are bound, whereas S2 and S3 contain only ATP and no Mg2+. Significant changes in the C terminus and the T loop in site S2 are marked with numbered circles. (D) Superposition of effector molecules bound to sites S1, S2, and S3 in the 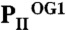 structure. The ATP molecule observed in the S2 site is significantly distorted relative to that in S1 and S3.
structure. The ATP molecule observed in the S2 site is significantly distorted relative to that in S1 and S3.
Effect of 2-OG on the Dissociation of the PII–NAGK Complex.
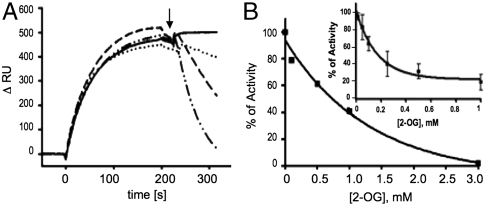
The structure of the PII Mg-ATP/2OG complex suggests that 2-OG prevents interaction of PII with NAGK by hindrance of the T loop folding into the tightly bent conformation needed for NAGK binding: The NAGK-bound PII structure involves a salt bridge between K58 and E44 (18, 23), but because K58 is an important ligand for 2-OG, formation of this salt bridge is prevented. Furthermore, binding of 2-OG introduces a significant bend in the backbone of residues 38–43 (Fig. 1E) and together with the side chain of residue 42 induces a new T-loop conformation, which is incompatible with NAGK binding. When the PII–NAGK complex has already been formed, is 2-OG still able to bind to PII and antagonize the PII–NAGK complex? Because this issue has not yet been investigated, the dissociation of the PII–NAGK complex by 2-OG was studied. First, complex dissociation was directly recorded by surface plasmon resonance (SPR) spectroscopy (Fig. 4A). The PII–NAGK complex was formed on the sensor chip, and, subsequently, 2-OG was injected (Fig. 4A, arrow) to dissociate the complex. No dissociation was observed in the presence of Mg-ATP alone; with 0.5 mM 2-OG, the complex decayed slowly with a rate of 1.8 × 10-3 s-1. With 1 mM 2-OG the decay rate increased to 9.0 × 10-3 s-1 and at 3 mM 2-OG to 28.6 × 10-3 s-1. By comparison, association of the complex was inhibited by much lower 2-OG concentrations, with an IC50 of approximately 130 μM (18). In the second assay, the catalytic activity of NAGK/PII in the presence of 50 μM arginine as an indicator of the degree of complex formation (24) was assayed (Fig. 4B). When 2-OG was added after formation of the PII–NAGK complex, the inhibitory 2-OG concentration had an IC50 of 0.9 mM. By contrast, addition of 2-OG to PII prior to the addition of NAGK inhibited the activity with an IC50 for 2-OG of approximately 120 μM (Fig. 4B, Inset) (18). Thus, 2-OG is able to dissociate the PII–NAGK complex; however, one order of magnitude higher 2-OG concentrations are required to achieve dissociation compared to those required to inhibit association.
Fig. 4.
The 2-OG effect on PII–NAGK complex dissociation. (A) SPR analysis of 2-OG-induced dissociation of NAGK–PII complex in the presence of 1 mM ATP. The response difference (ΔRU) between flow cells FC2 and FC1 (control) is shown. After binding of 100 nM PII to NAGK in FC2, a mixture of 1 mM ATP, 1 mM MgCl2, and 2-oxoglutarate [at a concentration of 0 (solid line), 0.5 (dotted line), 1 (dashed line), and 3 mM (dot-dashed line), as indicated] was injected at the point indicated by an arrow. (B) The effect of 2-OG on NAGK activity in the presence of 50 μM arginine. Increasing 2-OG concentrations were added to reaction mixtures after the formation of the PII–NAGK complex, and NAGK activity was determined as detailed in Materials and Methods. (Inset) Effect of 2-OG on NAGK activity in the presence of PII, when 2-OG was preincubated with PII.
Discussion
The structures presented here explain the known features of PII-mediated 2-OG signaling: A Mg2+ ion, chelated by the phosphates of ATP, ligates carboxylate oxygens of 2-OG, and, therefore, Mg-ATP binding is the prerequisite for 2-OG binding to PII. Binding of 2-OG to A. thaliana PII in the presence of Mg-ADP could involve additional residues, possibly from its prolonged C-terminal segment, which contacts the effector molecule-binding site (22). The fact that all residues revealed here to be involved in 2-OG binding are highly conserved among PII proteins (see also Fig. S1) strongly suggests that the reported mode of 2-OG binding could apply to all PII proteins. The only contradiction is the previously reported structure of the PII family member GlnK1 from M. jannaschii, where 2-OG bound from outside to the apex of a bent T loop (20). Because no biochemical evidence to support this binding mode was provided, it remains to be clarified whether this 2-OG binding mode is a peculiarity of the archaeal PII protein or whether this mode of binding resulted from special crystallization conditions. In contrast, a recently described structure of the PII homologue GlnZ from the proteobacterium A. brasiliense in complex with Mg-ATP and 2-OG revealed a mode of 2-OG binding, which is highly similar to the 2-OG binding described here, in particular the involvement of the Mg2+ ion and the highly conserved key residues Q39 and K58 (21). This one and our PII structures perfectly agree with the properties of S. elongatus PII variants bearing mutations in residues R9 and K58 described in this work and with the previously described I86N variant, displaying a closed 2-OG–binding site (18). Furthermore, they agree with previously reported properties of other PII mutant variants. For instance, a Q39 mutation was shown to strongly impair 2-OG binding, whereas a deletion of the apical T-loop residues did not prevent 2-OG binding to Escherichia coli PII (25). Furthermore, a K58 substitution abolished 2-OG signaling in Rhodospirillum rubrum PII (26). Moreover, the actual structure reveals how precisely the carboxylate oxygens of 2-OG are probed by Mg2+ coordination and by interactions with protein backbone and side-chain atoms, explaining the high selectivity of PII proteins for 2-OG (1, 16, 19). All together, these evidences strongly imply that the mode of 2-OG binding described here can be generalized for PII proteins.
It has been shown that 2-OG controls PII target interactions that involve the T loop, with X-ray structural information available for the S. elongatus and A. thaliana PII–NAGK complex (22, 23), the S. elongatus PII–PipX complex (27), and the E. coli GlnK–AmtB complex (28, 29). The mechanism of 2-OG-mediated PII-target control was clarified here with the cognate PII–NAGK complex. When PII binds 2-OG, the base of the T loop (R38-G41) wraps around this metabolite, thereby adopting a unique retracted conformation. Furthermore, residues K58 and R9, which are involved in folding the T loop into the tightly bent conformation that fits into the NAGK crevice, perform ionic and H-bond interactions with the 2-OG γ-carboxylate oxygens, preventing formation of this fold. The IC50 for 2-OG to inhibit PII–NAGK association (120–130 μM, depending on the method) matches the dissociation constant of the third, low-affinity 2-OG–binding site (approximately 110 μM). This correlation implies that all three sites in PII have to be occupied by 2-OG in order to inhibit NAGK binding. Consequently, PII partially loaded with 2-OG should be able to bind NAGK, whereby 2-OG should be displaced from PII. The driving force squeezing out 2-OG could be provided by the encounter complex between PII and NAGK, which, according to a recent analysis, could be formed by an ionic interaction of the B-loop residue E85 of PII with R233 of NAGK (18).
The present study also revealed how 2-OG dissociates the PII–NAGK complex. As shown in Fig. 1C, 2-OG can access its binding site from the PII periphery, which is not shielded by NAGK in the complex. However, approximately 10-fold higher concentrations of 2-OG are required to dissociate the PII–NAGK complex than to inhibit its association. The difference could be explained by the 2-OG–binding site being closed in the PII–NAGK complex by the tightly bent T loop. The 2-OG should unlock this compact conformation to gain access to its binding site, and this process probably requires much higher concentrations than binding to the open sites, which are accessible when PII is not attached to NAGK.
The structure of the second cyanobacterial PII target complex, PII–PipX, has been determined recently (27). It reveals three PipX molecules bound on the flat bottom surface of the PII body (orientation of Fig. 1), trapped between vertically extended T loops whose tip residues grasp the PipX monomers. Notably, this extended T-loop conformation is incompatible with the T-loop fold imposed by Mg-ATP-2-OG binding (see structure overlay in Fig. S5). Binding of 2-OG to the PII–PipX complex will retract the extended T loop, releasing the bound PipX molecules, which explains the biochemical data, showing that binding of PipX to PII is antagonized by Mg-ATP/2-OG (10). A similar antagonistic mechanism of Mg-ATP/2-OG can be assumed for the complex of the PII family member GlnK with the ammonium transport channel AmtB, as deduced from the complex structure of the E. coli proteins (28, 29). In complex with AmtB, the T loop is in a vertically extended structure, resembling the T loop of S. elongatus PII in complex with PipX. In the AmtB complex, GlnK residue Q39 interacts with K58 and ADP is bound to the adenylate-binding pocket (28, 29). Given that the binding mode of Mg-ATP/2-OG to GlnK is identical as outlined above, the resulting T-loop conformation will be incompatible with formation of the GlnK–AmtB complex (21). Studies with other E. coli PII receptors such as NtrB imply that 2-OG does not always inhibit complex formation, but it may affect receptor activity at a postbinding step (16). In this case, it is conceivable that receptor binding occurs apart from the T loop (like the B-loop interaction of PII with NAGK) and the conformational changes of the T loop imposed by 2-OG binding to PII are transduced into conformational changes in the receptor, thereby altering its activity.
PII proteins are highly sophisticated devices for measuring the concentration of central metabolites ATP, ADP, and 2-OG in an interdependent manner. This study reveals the mechanisms underlying this process. Binding of one, two, or three 2-OG molecules generates, via intersubunit communication, distinct structural states of PII. Intermolecular signaling is based on the highly conserved trimeric architecture of the PII proteins. The β2-strands, which directly connect the three binding sites, could play an important role. Binding of 2-OG to one site affects the two neighboring sites asymmetrically, generating the anticooperativity that allows metabolite sensing in a wide concentration range. Moreover, the free site in clockwise orientation displays a characteristic T-loop structure. PII receptors perceive the signal via intimate T-loop interactions, which affect binding or influence the receptor at a postbinding stage (16, 18). This mode of signal transduction by PII is unique, and the complexity of interactions explains the remarkably high conservation of PII proteins.
Materials and Methods
Full protocols are available in SI Materials and Methods.
Overexpression and Purification of Recombinant PII and NAGK.
The R9L and K58M variants were created with artificial glnB genes carrying the respective mutations and cloned into the Strep-tag fusion vector pASK-IBA3 (IBA) after restriction with BsaI as described previously (7). Overexpression of wild-type and mutant S. elongatus glnB in E. coli RB9060 (30) and purification of recombinant PII proteins with a C-terminal-fused Strep-tag II peptide were performed according to Heinrich et al. (7). His6-tagged recombinant NAGK from S. elongatus was overexpressed in E. coli strain BL21(DE3) (31) and purified as reported previously (8).
SPR Detection.
SPR experiments were performed by using a BIAcore X biosensor system (GE Healthcare) at 25 °C in Hepes-buffered saline-Mg buffer as described previously (8). In order to analyze the effect of 2-OG on the dissociation of the PII–NAGK complex, 100 nM PII was first bound to immobilized His6–NAGK in flow cell 2 (FC2) (ascending curves). Subsequently, 50 μL buffer containing 1 mM ATP and different concentrations of 2-OG was injected (start of injection indicated by the arrow). Binding and dissociation of PII to NAGK was recorded as the response signal difference (ΔRU) of FC2-FC1; FC1, reference cell without His6–NAGK.
ITC.
ITC experiments were performed on a VP-ITC microcalorimeter (MicroCal, LCC) in ITC buffer containing 10 mM Hepes-NaOH, pH 7.4, 50 mM KCl, 50 mM NaCl, and 1 mM MgCl2 at 20 °C as described previously (18). For determination of ATP- and 2-OG–binding isotherms for PII variants R9L and K58M, solutions with different protein concentration were titrated with 1 mM ATP or 4 mM 2-OG (in the presence of 1 mM ATP). The binding isotherms were calculated from received data and fitted to a three-site binding model using the MicroCal ORIGIN software (Northampton) as indicated.
Coupled NAGK Activity Assay.
Activity of NAGK was assayed by a coupled assay (32) with modifications as described previously (24), in the buffer consisting of 50 mM imidazole, pH 7.5, 50 mM KCl, 20 mM MgCl2, 0.4 mM NADH, 1 mM phosphoenolpyruvate, 10 mM ATP, 0.5 mM DTT, 11 U lactate dehydrogenase, 15 U pyruvate kinase, 50 μM arginine, 1.2 μg PII, and 3 μg NAGK. The mixture was preincubated for 3 min to allow PII–NAGK complex formation. Then the reaction was started by the addition of 50 mM NAG and 2-OG (to determine the effect of increasing 2-OG concentrations on disruption of PII–NAGK complex in the presence of NAGK-inhibiting concentrations of arginine). Then, 20 s after addition of substrate, the change in absorbance at 340 nm was recorded for 10 min. Linear kinetics were observed over that period of time.
Crystallization of Recombinant S. elongatus PII Protein.
Crystallization was performed with the sitting-drop technique by mixing 400 nL of the protein solution with equal amounts of the reservoir solution by using the honeybee robot (Genomic Solutions Ltd). Drops were incubated at 20 °C, and pictures were recorded by the RockImager system (Formulatrix). The crystallization buffer was composed of 10 mM Tris (pH 7.4), 0.5 mM EDTA, 100 mM NaCl, 1% glycerol, and 2 mM ATP-Mg, and also 2 mM 2-OG was added; crystals appeared in a precipitant condition containing PEG 4000. Glycerol was used as the cryoprotectant, and the crystals were flash-frozen in liquid nitrogen. Diffraction data were collected at the Swiss Light Source. Diffraction images were recorded on a MarCCD camera 225 (Marresearch), and images were processed by using the XDS/XSCALE software (33). The structure was solved by molecular replacement using the program Molrep (34). Rebuilding of the structure and structure refinement was performed by using the programs Coot and Refmac (35, 36). The quality of the structure was analyzed by the Procheck program (37). Figures were generated by using PyMOL (www.pymol.org).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Gary Sawers for critically reading the manuscript. The help of Reinhard Albrecht in crystallization of PII proteins and Christina Herrmann for technical assistance is gratefully acknowledged. This work was strongly supported by the Max Planck Society and the PXII Beamline personnel and grants from the Deutsche Forschungsgemeinschaft (Fo195).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.L.B. is a guest editor invited by the Editorial Board.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2XUL and 2XUN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007653107/-/DCSupplemental.
References
- 1.Ninfa AJ, Jiang P. PII signal transduction proteins: Sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–377. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- 3.Forchhammer K. P(II) signal transducers: Novel functional and structural insights. Trends Microbiol. 2008;16:65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Sant’Anna FH, et al. The PII superfamily revised: A novel group and evolutionary insights. J Mol Evol. 2009;68:322–336. doi: 10.1007/s00239-009-9209-6. [DOI] [PubMed] [Google Scholar]
- 5.Jiang P, Ninfa AJ. Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in vitro. Biochemistry. 2007;46:12979–12996. doi: 10.1021/bi701062t. [DOI] [PubMed] [Google Scholar]
- 6.Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: From signals to targets. FEMS Microbiol Rev. 2004;28:319–333. doi: 10.1016/j.femsre.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Heinrich A, Maheswaran M, Ruppert U, Forchhammer K. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol Microbiol. 2004;52:1303–1314. doi: 10.1111/j.1365-2958.2004.04058.x. [DOI] [PubMed] [Google Scholar]
- 8.Maheswaran M, Urbanke C, Forchhammer K. Complex formation and catalytic activation by the PII signaling protein of N-acetyl-L-glutamate kinase from Synechococcus elongatus strain PCC 7942. J Biol Chem. 2004;279:55202–55210. doi: 10.1074/jbc.M410971200. [DOI] [PubMed] [Google Scholar]
- 9.Burillo S, Luque I, Fuentes I, Contreras A. Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J Bacteriol. 2004;186:3346–3354. doi: 10.1128/JB.186.11.3346-3354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinosa J, Forchhammer K, Burillo S, Contreras A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol. 2006;61:457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 11.Feria Bourrellier AB, et al. Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc Natl Acad Sci USA. 2010;107:502–507. doi: 10.1073/pnas.0910097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helfmann S, Lu W, Litz C, Andrade SL. Cooperative binding of MgATP and MgADP in the trimeric PII protein GlnK2 from Archaeoglobus fulgidus. J Mol Biol. 2010;402:165–177. doi: 10.1016/j.jmb.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, et al. GlnK, a PII-homologue: Structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J Mol Biol. 1998;282:149–165. doi: 10.1006/jmbi.1998.1979. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, et al. The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803. Acta Crystallogr, Sect D: Biol Crystallogr. 2003;59:2183–2190. doi: 10.1107/s0907444903019589. [DOI] [PubMed] [Google Scholar]
- 15.Sakai H, et al. Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8. J Struct Biol. 2005;149:99–110. doi: 10.1016/j.jsb.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Jiang P, Ninfa AJ. Alpha-ketoglutarate controls the ability of the Escherichia coli PII signal transduction protein to regulate the activities of NRII (NtrB) but does not control the binding of PII to NRII. Biochemistry. 2009;48:11514–11521. doi: 10.1021/bi901158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CS, Weljie AM, Moorhead GB. Molecular properties of the putative nitrogen sensor PII from Arabidopsis thaliana. Plant J. 2003;33:353–360. doi: 10.1046/j.1365-313x.2003.01634.x. [DOI] [PubMed] [Google Scholar]
- 18.Fokina O, Chellamuthu VR, Zeth K, Forchhammer K. A novel signal transduction protein P(II) variant from Synechococcus elongatus PCC 7942 indicates a two-step process for NAGK-P(II) complex formation. J Mol Biol. 2010;399:410–421. doi: 10.1016/j.jmb.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Forchhammer K, Hedler A. Phosphoprotein PII from cyanobacteria—analysis of functional conservation with the PII signal-transduction protein from Escherichia coli. Eur J Biochem. 1997;244:869–875. doi: 10.1111/j.1432-1033.1997.00869.x. [DOI] [PubMed] [Google Scholar]
- 20.Yidiz Ö, Kalthoff C, Raunser S, Kühlbrandt W. Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake. EMBO J. 2007;26:589–599. doi: 10.1038/sj.emboj.7601492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truan D, et al. A new PII protein structure identifies the 2-oxoglutarate binding site. J Mol Biol. 2010;400:531–539. doi: 10.1016/j.jmb.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno Y, Moorhead GB, Ng KK. Structural basis for the regulation of N-acetylglutamate kinase by PII in Arabidopsis thaliana. J Biol Chem. 2007;282:35733–35740. doi: 10.1074/jbc.M707127200. [DOI] [PubMed] [Google Scholar]
- 23.Llacer JL, et al. The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc Natl Acad Sci USA. 2007;104:17644–17649. doi: 10.1073/pnas.0705987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beez S, Fokina O, Herrmann C, Forchhammer K. N-acetyl-L-glutamate kinase (NAGK) from oxygenic phototrophs: PII signal transduction across domains of life reveals novel insights in NAGK control. J Mol Biol. 2009;389:748–758. doi: 10.1016/j.jmb.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 25.Jiang P, et al. Structure/function analysis of the PII signal transduction protein of Escherichia coli: Genetic separation of interactions with protein receptors. J Bacteriol. 1997;179:4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson A, Nordlund S. In vitro studies of the uridylylation of the three PII protein paralogs from Rhodospirillum rubrum: The transferase activity of R. rubrum GlnD is regulated by alpha-ketoglutarate and divalent cations but not by glutamine. J Bacteriol. 2007;189:3471–3478. doi: 10.1128/JB.01704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llacer JL, et al. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc Natl Acad Sci USA. 2010;107:15397–15402. doi: 10.1073/pnas.1007015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conroy MJ, et al. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruswitz F, O’Connell J, 3rd, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc Natl Acad Sci USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bueno R, Pahel G, Magasanik B. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P, Ninfa AJ. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J Bacteriol. 1999;181:1906–1911. doi: 10.1128/jb.181.6.1906-1911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.