Abstract
In the Drosophila ovary, bone morphogenetic protein (BMP) signaling activated by the niche promotes germline stem cell (GSC) self-renewal and proliferation, whereas E-cadherin–mediated cell adhesion anchors GSCs in the niche for their continuous self-renewal. Here we show that Lissencephaly-1 (Lis1) regulates BMP signaling and E-cadherin–mediated adhesion between GSCs and their niche and thereby controls GSC self-renewal. Lis1 mutant GSCs are lost faster than control GSCs because of differentiation but not because of cell death, indicating that Lis1 controls GSC self-renewal. The Lis1 mutant GSCs exhibit reduced BMP signaling activity, and Lis1 interacts genetically with the BMP pathway components in the regulation of GSC maintenance. Mechanistically, Lis1 binds directly to and stabilizes the SMAD protein Mothers against decapentaplegic (Mad), facilitates its phosphorylation, and thereby regulates BMP signaling. Finally, the Lis1 mutant GSCs accumulate less E-cadherin in the stem cell–niche junction than do their wild-type counterparts. Germline-specific expression of an activated BMP receptor thickveins (Tkv) or E-cadherin can partially rescue the loss phenotype of Lis1 mutant GSCs. Therefore, this study has revealed a role of Lis1 in the control of Drosophila ovarian GSC self-renewal, at least partly by regulating niche signal transduction and niche adhesion. It has been known that Lis1 controls neural precursor/stem cell proliferation in the developing mammalian brain; this study further suggests that Lis1, which is widely expressed in adult mammalian tissues, could regulate adult tissue stem cells through modulating niche signaling and adhesion.
In adult animal tissues, stem cells normally undergo asymmetric division to generate self-renewing stem cells and differentiated cells that replace lost cells. Their behavior is tightly controlled by the concerted actions of extrinsic and intrinsic factors in a variety of systems (1, 2). Interestingly, signals from the niche often function within one cell diameter, repressing expression or functions of differentiation-promoting genes and thereby maintaining stem cell self-renewal (3). To self-renew continuously, stem cells must be anchored to the niche and constantly receive niche signals to maintain their undifferentiated state. Stem cell anchorage to the niche often is achieved through cadherin- or integrin-mediated cell adhesion (2, 4). However, how niche signaling and niche anchorage are coordinately regulated in adult stem cells remains largely unknown.
Drosophila ovarian germline stem cells (GSCs) are an attractive system for studying stem cells and the niche at the molecular and cellular level (5). Two or three GSCs are located at the tip of the germarium and directly contact cap cells and escort stem cells, which constitute a GSC niche (Fig. 1A) (6–8). In addition, they contain an intracellular spherical structure known as the “spectrosome,” which is rich in cytoskeletal proteins such as Hu li-tai shao (Hts) (Fig.1B). (9) Differentiating GSC daughters, cystoblasts, move away from the niche and further divide four times synchronously to form 16-cell cysts identified by branched fusomes. Bone morphogenic protein (BMP) signaling is necessary and sufficient for maintaining GSC self-renewal by directly repressing the expression of differentiation-promoting genes such as bag of marbles (bam) (7, 10, 11). In addition, E-cadherin–mediated cell adhesion is required to keep GSCs in the niche for continuous self-renewal (12). bam transcriptional repression by BMP signaling in the GSC is incomplete, leaving low levels of Bam expression (13). Bam-mediated E-cadherin repression in the GSC controls stem cell competition for niche occupancy, functioning as a quality control mechanism to ensure that differentiated GSCs are displaced rapidly from the niche and then are replaced by functional ones through competition (13).
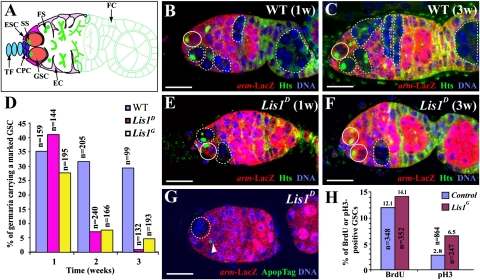
Fig. 1.
Requirement of Lis1 in controlling GSC self-renewal. (A) Schematic diagram of a wild-type germarium showing different cell types and organelles. CPC, cap cell; EC, escort cell; ESC, escort stem cell; FC, follicle cell. FS, fusome; GSC, germline stem cell; SS, spectrosome; TF, terminal filament. (B and C) Germaria carrying an unmarked GSC (solid circle) and a 1-wk-old (B) or 3-wk-old (C) marked wild-type GSC clone in which the marked GSC and progeny are indicated by a broken circle and broken lines, respectively. (D) Changes in percentages (y axis) of the germaria carrying a marked control and Lis1 mutant GSC clone with time (x axis). n represents the number of total germaria examined. (E) Germarium carrying an unmarked wild-type GSC (solid circle) and a marked Lis1 mutant GSC (broken circle), which produces a marked mutant cyst (broken lines). (F) Germarium carrying two unmarked wild-type GSCs (solid circles) and a marked Lis1 mutant cyst (broken line), indicative of a lost marked Lis1 mutant GSC. (G) Germarium carrying an ApopTag-negative marked Lis1-mutant GSC (broken circle) with a dying escort cell (arrowhead). (H) Lis1 mutant GSCs may divide slightly faster than control GSCs. There are almost two times more pH3-positive Lis1G mutant GSCs than control GSCs, but only slightly more Lis1G mutant GSCs than control GSCs are positive for BrdU. n represents the number of total GSCs examined. (Scale bars: 10 μm.)
Lissencephaly-1 (Lis1) first was identified as the causative gene for the human disease lissencephaly and later was shown to be involved in the regulation of neural precursor proliferation and neuronal migration in the developing brain (14, 15). Like its counterpart in other nervous systems, Lis1 is required for controlling asymmetric division of Drosophila neuronal precursors, neuroblasts, through regulating spindle orientation and for dendrite formation of differentiated neuronal cells (16, 17). Interestingly, Lis1 also regulates cyst division, oocyte formation, and oocyte nucleus migration in the Drosophila ovary (18–20). In this study, we show that it functions as an intrinsic factor to control GSC self-renewal, at least in part through regulating BMP signal transduction and E-cadherin–mediated cell adhesion.
Results and Discussion
Lis1 Is Required for GSC Self-Renewal.
To investigate the potential role of the Lis1 gene in GSC maintenance in the Drosophila ovary, we used Flippase (FLP)-mediated FLP-recognition target (FRT) mitotic recombination to generate marked armadillo (arm)-lacZ–negative Lis1 mutant GSCs and examined their maintenance and relative division rates as previously reported (7, 12). The GSCs were identified as the most anterior single germ cells containing an anteriorly anchored spectrosome and directly contacting cap cells; the marked and unmarked GSCs were identified by the absence or presence of arm-lacZ expression, respectively (Fig. 1 A and B). About 75–80% of the arm-lacZ–negative marked wild-type GSCs detected during the first week after clone induction (ACI) still remained in the niche 3 wk ACI (Fig. 1 B–D). In contrast, only 2% and 17%, respectively, of arm-lacZ-negative marked Lis1D and Lis1G10.14 (Lis1G) mutant GSCs detected 1 wk ACI remained in the niche 3 wk ACI, (Fig. 1 D–F). Lis1D and Lis1G, encoding truncated proteins, have been shown previously to represent severe or null Lis1 mutants (18).Thus, most of the germaria had already lost the marked mutant GSCs 3 wk ACI, as was evident from the presence of marked differentiated cysts in the germaria or late egg chambers (Fig. 1F). To determine further whether apoptosis contributes to the loss of the marked Lis1 GSCs, we used a TUNEL- based ApopTag labeling assay to detect the dying cells in the ovaries. Although we could detect dying somatic cells in germaria easily, we failed to detect apoptotic marked Lis1G and Lis1D mutant GSCs (n = 156), further reinforcing the idea that Lis1 mutant GSCs are lost because of differentiation rather than apoptosis (Fig. 1G). Taken together, these results demonstrate that Lis1 is required for controlling GSC self-renewal but not survival in the Drosophila ovary.
During the analysis of the marked Lis1 mutant clones, we noticed that marked Lis1 mutant GSCs produced much fewer germline cysts than the marked control GSCs, suggesting that Lis1 is required for GSC division, cyst survival, or both (compare Fig. 1 B and E). Lis1 has been shown previously to regulate M-phase progression and spindle orientation in Drosophila and mouse neural stem cells (16, 21, 22). To determine if Lis1 is required for GSC division, we performed BrdU labeling and phosphorylated histone H3 (pH3) staining on unmarked control GSCs and marked Lis1 mutant GSCs. Surprisingly, the percentage of the BrdU-positive marked Lis1 mutant GSCs is similar to or even slightly higher than that of the unmarked control GSCs, and the percentage of the pH3-positive marked Lis1 mutant GSCs is almost two times higher than that of the control GSCs, suggesting that Lis1 mutant GSCs may cycle normally or slightly faster than control GSCs (Fig. 1H and Fig. S1). Because we did not observe TUNEL-positive Lis1 mutant cysts in the germaria carrying a Lis1 mutant GSC, this result suggests that the fewer number of mutant cysts produced by an Lis1 mutant GSC probably results from cystoblast death and the quick disappearance of the dying cystoblasts. Like Drosophila neuroblasts, Lis1-mutant GSCs exhibited a misoriented spindle more frequently than control GSCs (Fig. S2). The published studies show that the defect in spindle orientation might cause the slowdown of the M phase of the Lis1 mutant stem cells in the mouse brain and Drosophila testis (22, 23). Our finding that Lis1 mutant GSCs exhibit misoriented spindles but do not appear to proliferate more slowly than wild-type GSCs suggests that Lis1 also may be required for regulating the checkpoint control in GSCs.
To investigate if the spindle misorientation is responsible for the loss of Lis1 mutant GSCs, we generated arm-lacZ–marked hts mutant GSCs, which exhibit misoriented spindles (24), and determined their maintenance over time. Surprisingly, the marked hts mutant GSCs still were well maintained 2 wk ACI, in contrast with the dramatic loss of marked Lis1G and Lis1D GSCs 2 wk ACI (Table S1). This finding suggests that spindle misorientation could not be fully responsible for the loss of Lis1-mutant GSCs.
Lis1 Promotes BMP Signaling in GSCs and Facilitates Their Maintenance.
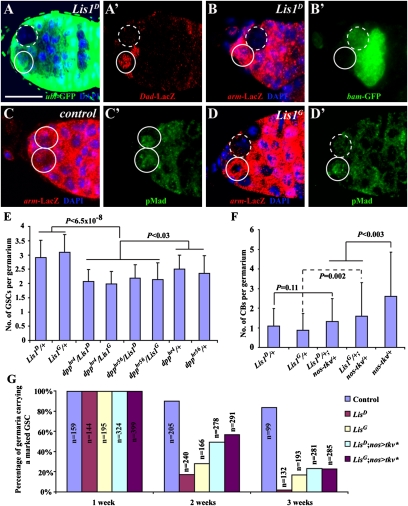
Because niche-initiated BMP signaling is required for maintaining GSC self-renewal (7, 10), we explored the possibility that Lis1 regulates BMP signal reception in the GSC. In the GSC, BMP signaling up-regulates Daughters against decapentaplegic (Dad) expression and represses bam transcription, effects that can be recapitulated by the reporters Dad-lacZ and bam-GFP, respectively (10, 11). Therefore we examined the expression of Dad-lacZ and bam-GFP in the marked Lis1 mutant GSCs. As expected, in the unmarked control GSCs, Dad-lacZ was expressed and bam-GFP was repressed (Fig. 2 A–B′). In contrast, Dad-lacZ expression was much lower in the marked Lis1D GSCs (17/34) and Lis1G GSCs (15/40) than in the neighboring unmarked control GSCs (Fig. 2 A and A′). Consistent with lower BMP signaling in the Lis1 mutant GSCs, bam-GFP expression was higher in the marked Lis1D GSCs (36/74) and Lis1G GSCs (18/59) than in the neighboring unmarked control GSCs (Fig. 2 B and B′). In Drosophila, BMP signal transduction leads to the production of phosphorylated Mothers against decapentaplegic (pMad), which works with SMAD4/Medea to regulate gene expression (25). In the control, the marked and unmarked wild-type GSCs had similar levels of pMad expression (Fig. 2 C and C′). However, the expression of pMad was down-regulated dramatically in marked Lis1D or Lis1G mutant GSCs compared with the neighboring unmarked wild-type GSCs (Fig. 2 D and D’). Taken together, these results demonstrate that Lis1 is required in the GSC to maintain BMP signaling activity.
Fig. 2.
Requirement of Lis1 in maintaining BMP signaling in GSCs. (A and A′) A marked Lis1 mutant GSC (broken circle) downregulating Dad-lacZ expression compared with an unmarked wild-type GSC (solid circle) in the same niche. (B and B′) A marked Lis1 mutant GSC (broken circle) up-regulating bam-GFP expression compared with an unmarked wild-type GSC (solid circle) in the same niche. (C and C′) Two unmarked wild-type GSCs (solid circles) with similar levels of nuclear pMad expression. (D and D′) A marked Lis1 mutant GSC (broken circle) with lower nuclear pMad expression than its neighboring unmarked wild-type GSC (solid circle). (E) Germaria of Lis1 and dpp heterozygous double mutants have significantly fewer GSCs than germaria of Lis1 or dpp heterozygous single mutants. Error bars represent SD. (F) Heterozygous mutation in Lis1 can suppress excess cystoblast phenotype caused by germline-specific Tkv overexpression (nos-tkv represents nos-gal4 driving UASp-tkv). (G) Germline-specific expression of an activated BMP receptor Tkv can partly slow the loss of the marked Lis1 mutant GSCs. For all genotypes, the initial percentages of the germaria carrying a marked GSC are normalized to 100%. nos > tkv*, nos-gal4 UAS-tkv* (tkv* is a constitutively active form of tkv). n represents the number of total germaria examined.
We have shown previously that GSC self-renewal is very sensitive to the dosage of decapentaplegic (dpp), which encodes a niche BMP (7, 10). To probe further the relationship between BMP signaling and Lis1 in GSC regulation, we carried out genetic interaction studies on Lis1 and dpp mutants. Interestingly, the germaria of Lis1 and dpp double heterozygotes had significantly fewer GSCs than the germaria of Lis1 or dpp heterozygotes, indicating that Lis1 and dpp interact genetically with each other in GSC regulation (Fig. 2E). GSC differentiation also is sensitive to BMP signaling dosage: More BMP signaling favors self-renewal (7, 10). Consistently, when a wild-type thickveins (tkv), encoding a BMP type I receptor (26, 27), was overexpressed in germline cells using nos-gal4 and UASp-tkv, the germaria contained an average 2.6 ± 2.3 cystoblasts per germarium, in comparison with about one cystoblast per wild-type germarium (28), indicating that boosting BMP signaling promotes GSC proliferation or slows down cystoblast differentiation (Fig. 2F). Lis1D and Lis1G heterozygous germaria had about one cystoblast, behaving like wild-type germaria (Fig. 2F). However, Lis1D or Lis1G heterozygous germaria overexpressing tkv had significantly fewer cystoblasts than wild-type germaria overexpressing tkv, indicating that reduction of Lis1 function can suppress BMP signaling in the control of GSC proliferation or differentiation (Fig. 2F). To test further that the reduced BMP signaling contributes to the loss of Lis1 mutant GSCs, we used nos-gal4–driven UAS-tkv* (a constitutively active form of tkv) to boost BMP signaling in the marked Lis1-mutant GSCs. Germline-specific tkv* expression could slow the loss of the 2-wk-old marked Lis1 mutant GSCs dramatically, but this rescue effect became less dramatic for the 3-wk-old mutant GSCs, indicating that increasing BMP signaling can, at least partially, slow the loss of the Lis1 mutant GSCs (Fig. 2G). Therefore, all the genetic data show that Lis1 interacts genetically with the dpp/BMP pathway in controlling the balance between GSC self-renewal and differentiation.
Lis1 Interacts with Mad to Regulate Its Stability and Phosphorylation.
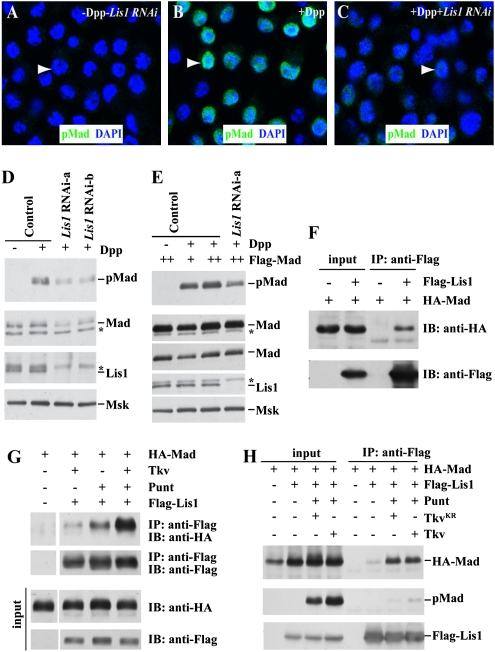
Because of the difficulty in obtaining sufficient GSCs, we sought to use S2 cells to carry out biochemistry studies to obtain further mechanistic insights into how Lis1 regulates BMP signaling. Dpp stimulation led to the nuclear accumulation of pMad in S2 cells in comparison with no detectable pMad in the S2 cells without Dpp, confirming the previous finding that S2 cells are capable of responding to Dpp/BMP (Fig. 3 A and B) (29). Interestingly, knockdown of Lis1 expression using dsRNAs also resulted in the reduction of nuclear pMad expression, just as in Lis1-mutant GSCs, indicating that Lis1 regulates BMP signaling in S2 cells (Fig. 3C). Two independent dsRNAs could knock down Lis1 protein expression efficiently, as was verified by Western blotting (Fig. 3D). In the Lis1 knockdown S2 cells, both Mad and pMad expression levels were significantly reduced, as compared with controls, indicating that Lis1 is required for maintaining Mad protein expression and/or phosphorylation (Fig. 3D and Fig. S3). Interestingly, pMad levels still remained lower in the Lis1 knockdown cells in which total Mad levels were brought back nearly to normal by forced expression than in the controls, suggesting that Lis1 also is involved in the regulation of Mad phosphorylation (Fig. 3E). Therefore, Lis1 is required for maintaining Mad protein expression levels and facilitating Mad phosphorylation.
Fig. 3.
Regulation of Mad stabilization and phosphorylation by Lis1. (A–C), Suppression of Dpp-stimulated nuclear pMad expression by Lis1 RNAi knockdown Untreated S2 cells have low pMad expression (A). Dpp treatment promotes nuclear accumulation of pMad in S2 cells (B), which can be suppressed by RNAi-mediated Lis1 knockdown (C). Arrowheads point to nuclei of representative cells. (D) Knockdown of Lis1 expression by two independent dsRNAs causing the reduction of both Mad and pMad protein expression. Moleskin (Msk) a nuclear import factor for Mad, acts as a loading control. (E) Knockdown of Lis1 expression still exhibits the reduction of pMad expression even in the presence of normal Mad levels. The normal Mad level is achieved through using a higher concentration of CuSO4. The two lanes showing Mad protein expression represent two different exposures. Asterisks in D and E denote nonspecific protein ban. (F) Flag-tagged Lis1 can specifically bring down HA-tagged Mad in S2 cells. IB, immunoblot. (G) The presence of Tkv or Punt can strengthen the physical interaction between Lis1 and Mad. IP, immunoprecipitation. (H) The physical interaction between Lis1 and Mad is enhanced by the presence of Punt and wild-type Tkv and even by kinase-dead Tkv.
To investigate how Lis1 regulates Mad protein expression and phosphorylation, we tested whether Lis1 and Mad proteins could interact physically in S2 cells. Interestingly, Flag-tagged Lis1 and HA-tagged Mad immunoprecipitated each other down in the S2 cells (Fig. 3F). Furthermore, Flag-tagged Lis1 could bring down HA-tagged Mad more efficiently in the presence of Tkv than in its absence, indicating that Tkv can enhance the Lis1–Mad interaction (Fig. 3G). The interaction between Lis1 and Mad became even stronger in the presence of both Punt and Tkv than in the presence of Tkv alone, indicating that the BMP receptor complex can help strengthen the Lis1–Mad interaction (Fig. 3 G and H and Fig. S3C). In Drosophila, Dpp requires the kinase functions of both the type I receptor Tkv and the type II receptor Punt for transducing its signal (26, 27, 30, 31). Interestingly, wild-type Tkv and kinase-dead TkvKR could equally promote the Lis1–Mad interaction, but the presence of TkvKR caused the reduction of pMad expression, suggesting that BMP receptors, not BMP signaling per se, facilitate the Lis1–Mad interaction (Fig. 3H). A comparison of the amount of Mad and pMad brought down by Lis1 indicated that Lis1 had stronger interaction with the unphosphorylated form of Mad than with pMad, suggesting that Lis1 releases Mad following its phosphorylation (Fig. 3H). Taken together, these results suggest two distinct roles of Lis1 in the regulation of BMP signal transduction: stabilizing Mad protein through physical interaction and promoting Mad phosphorylation, possibly by facilitating interaction between Mad and the receptor complex.
Lis1 Maintains Normal E-Cadherin Accumulation in the GSC–Niche Junction.
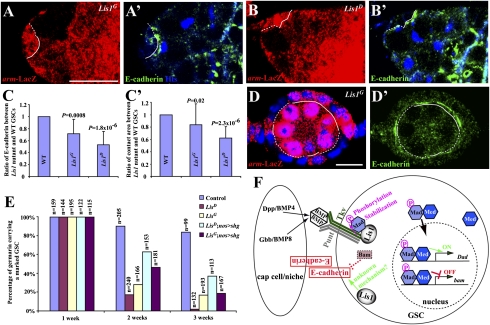
E-cadherin is important for anchoring GSCs in the niche and thus for continuous GSC self-renewal (12). To investigate if Lis1 also is required to maintain the accumulation of E-cadherin in the stem cell–niche junction, we measured and compared E-cadherin levels in marked Lis1 mutant GSCs and their wild-type neighboring GSCs by reconstructing 3D images based on thin confocal sections, according to our published experimental procedures (13). Interestingly, the marked Lis1D and Lis1G mutant GSCs had significantly less E-cadherin accumulation in the stem cell–niche junction than the unmarked control GSCs in the same niches (Fig. 4 A–C). Recently, we showed that a GSC expressing more E-cadherin gradually gains more contact area with the niche than a GSC expressing less E-cadherin and, competing for niche occupancy, gradually pushes the latter out of the niche (13). Indeed, the Lis1G and Lis1D mutant GSCs had less contact area than the unmarked control GSCs in the same niches, indicating that E-cadherin–mediated competition might be responsible in part for loss of Lis1 mutant GSCs (Fig. 4C′). Our previous finding that Bam up-regulation in GSCs results in E-cadherin down-regulation raises the interesting possibility that the decrease of E-cadherin expression in Lis1 mutant GSCs could result from the up-regulation of bam shown earlier (28). To investigate the possibility that Lis1 regulates E-cadherin accumulation independently of bam, we examined E-cadherin expression in Lis1-mutant follicle cells, in which bam is not expressed. Lis1 mutant follicle cells had less E-cadherin accumulation in the germ cell–follicle interface than neighboring wild-type follicle cells in the same egg chambers, suggesting that Lis1 can regulate E-cadherin levels independently of bam (Fig. 4 D and D′). To test further whether the reduced E-cadherin accumulation in the GSC–niche junction contributes to the loss of Lis1 mutant GSCs, we used nos-gal4–driven UAS-shotgun (shg, which encodes Drosophila E-cadherin) to increase E-cadherin expression in the marked Lis1-mutant GSCs. Germline-specific E-cadherin expression could slow the loss of the 2-wk-old marked Lis1 mutant GSCs dramatically, and the rescue effect also decreased for the 3-wk-old mutant GSCs, indicating that increasing E-cadherin accumulation can, at least in part, slow the loss of Lis1 mutant GSCs (Fig. 4E). Together, these results argue strongly that Lis1 maintains E-cadherin accumulation in the stem cell–niche junction, contributing to GSC maintenance.
Fig. 4.
Regulation of E-cadherin accumulation in the stem cell–niche interface by Lis1. (A–B′) The interface (broken lines) between a marked Lis1G mutant GSC (A and A′) or a Lis1D mutant GSC (B and B′) and the niche accumulates much less E-cadherin than the interface (solid line) between an unmarked wild-type GSC and the niche. (C and C′) Marked Lis1 mutant GSCs have significantly less E-cadherin accumulation in the GSC–niche junction (C) and have a significantly smaller contact area with the niche (C′) than unmarked control GSCs. (D and D′) Marked Lis1 mutant follicle cells (broken lines) have much less E-cadherin in their interface with germ cells than unmarked wild-type follicle cells (solid lines). (Scale bars in A and D: 10 μm.) (E) Germline-specific E-cadherin expression can partly slow the loss of the marked Lis1 mutant GSCs. For all genotypes, the initial percentages of the germaria carrying a marked GSC are normalized to 100%. nos > shg = nos-gal4 UAS-shg. (F) A working model explaining how Lis1 might regulate BMP signaling in the GSC and E-cadherin accumulation in the GSC–niche junction. Gbb, glass-bottom boat; Med, Medea.
In this study, we show that Lis1 controls GSC self-renewal by maintaining BMP signaling in GSCs and E-cadherin accumulation in the stem cell–niche interface. To our knowledge, this is the first time that Lis1 has been shown to regulate signaling and adhesion in any cell type, including stem cells. We also show that Lis1 regulates BMP signaling by modulating Mad stability and phosphorylation, probably through physical interaction. Based on our findings, we propose that Lis1 can stabilize Mad protein and facilitate its phosphorylation by the BMP receptor complex, likely through direct association, and also can promote E-cadherin accumulation in the stem cell–niche junction, possibly through bam-dependent and independent mechanisms (Fig. 4F). Because BMP signaling is known to regulate self-renewal of different stem cell types, and E-cadherin is known to regulate stem cell anchorage and cell migration, our findings may provide insight into how Lis1 controls neural stem cell self-renewal and neuronal migration in mammalian systems. In addition, this study also demonstrates the function of Lis1 in regulating an adult stem cell type other than neural stem cells or precursors. Because Lis1 is widely expressed in mammalian adult tissues, this study raises the possibility that Lis1 also regulates other adult stem cell types in mammals, including humans.
Materials and Methods
Drosophila Stocks and Genetic Clonal Analysis.
The information about the different Drosophila strains (bam-GFP, Dad-lacZ, dpphr4, dpphr56, Lis1D, Lis1G10.14, nos-gal4, UASp-tkv, UASp-tkv*, UASp-shg, FRTG13, armadillo-lacZ, and ubiquitin-GFP) is described in FlyBase (http://flybase.org). All of the Drosophila crosses and cultures were done at 25 °C. The experimental procedures for using FLP-mediated FRT recombination to generate the marked control and Lis1 mutant GSCs were described previously (7, 12). For statistical analyses, the student's t test was used.
Immunohistochemistry.
The following antisera were used: monoclonal mouse anti-Hts antibody 1B1, 1:4 [Developmental Studies Hybridoma Bank (DSHB)]; monoclonal rat anti-E-cadherin DCAD2, 1:6 (DSHB); polyclonal rabbit anti-β-galatosidase antibody, 1:300 (Cappel); monoclonal mouse anti-β-galatosidase antibody, 1:100 (Promega); polyclonal rabbit anti-GFP antibody, 1:200 (Molecular Probes); D-PLP, 1:1,000 (a gift from J. Raff University of Oxford, Oxford), pMad, 1:200 (a gift from P. ten Dijke, Leiden University Medical Center, Leiden, the Netherlands); pH3, 1:500 (Upstate); and Alexa 488- and Alexa 568-conjugated goat anti-mouse, anti-rabbit, and anti-rat, 1:300 (Molecular Probes). The immunostaining protocol and the TUNEL assay using the ApopTag kit from Chemicon have been described previously (7, 12, 32). All micrographs were taken using a Leica TCS SP2 confocal microscope.
Experimental details for RNAi in S2 cells, constructs, immunoprecipitation, and immunostaining of S2 cells are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Hays (University of Minnesota, Minneapolis), D. McKearin (University of Texas Southwestern, Dallas), R. Padgett (Rutgers, Piscataway, NJ), J. Raff (University of Oxford, Oxford), R. Steward (Rutgers, Piscataway, NJ), and P. ten Dijke (Leiden University, Leiden, the Netherlands), the Developmental Studies Hybridoma Bank (Iowa City, IA), and Bloomington Drosophila Stock Center (Bloomington, IN) for reagents; the Xie laboratory members for stimulating discussions, D. Chao, W. Neaves, and A. Spradling for comments; and C. Lee for administrative assistance and proofreading. This work was supported by National Institutes of Health Grants R01GM64428 (to T.X.) and R01CA108509 (to L.X.), by the Stowers Institute for Medical Research (T.X.), and by the Worcester Foundation of Biomedical Research (L.X.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008606107/-/DCSupplemental.
References
- 1.Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Xie T. Stem cell niche: Structure and function. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 3.Xi R, Kirilly D, Xie T. Molecular mechanisms controlling germline and somatic stem cells: Similarities and differences. Curr Opin Genet Dev. 2005;15:381–387. doi: 10.1016/j.gde.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: Stem cells and their niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 5.Kirilly D, Xie T. The Drosophila ovary: An active stem cell community. Cell Res. 2007;17:15–25. doi: 10.1038/sj.cr.7310123. [DOI] [PubMed] [Google Scholar]
- 6.Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- 7.Xie T, Spradling AC. Decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- 8.Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: Similar somatic stem cells and signals. Dev Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- 10.Song X, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–1857. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- 13.Jin Z, et al. Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell. 2008;2:39–49. doi: 10.1016/j.stem.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynshaw-Boris A, Gambello MJ. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Walsh CA. Protein-protein interactions, cytoskeletal regulation and neuronal migration. Nat Rev Neurosci. 2001;2:408–416. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- 16.Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev Biol. 2008;319:1–9. doi: 10.1016/j.ydbio.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat Cell Biol. 2000;2:776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Xie T, Steward R. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development. 1999;126:4477–4488. doi: 10.1242/dev.126.20.4477. [DOI] [PubMed] [Google Scholar]
- 19.Lei Y, Warrior R. The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev Biol. 2000;226:57–72. doi: 10.1006/dbio.2000.9848. [DOI] [PubMed] [Google Scholar]
- 20.Swan A, Nguyen T, Suter B. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat Cell Biol. 1999;1:444–449. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- 21.Hebbar S, et al. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–1071. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yingling J, et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng J, et al. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng W, Lin H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- 25.Affolter M, Basler K. The Decapentaplegic morphogen gradient: From pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 26.Penton A, et al. Identification of two bone morphogenetic protein type I receptors in Drosophila and evidence that Brk25D is a decapentaplegic receptor. Cell. 1994;78:239–250. doi: 10.1016/0092-8674(94)90294-1. [DOI] [PubMed] [Google Scholar]
- 27.Nellen D, Affolter M, Basler K. Receptor serine/threonine kinases implicated in the control of Drosophila body pattern by decapentaplegic. Cell. 1994;78:225–237. doi: 10.1016/0092-8674(94)90293-3. [DOI] [PubMed] [Google Scholar]
- 28.Shen R, Weng C, Yu J, Xie T. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci USA. 2009;106:11623–11628. doi: 10.1073/pnas.0903325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimmi O, Umulis D, Othmer H, O'Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letsou A, et al. Drosophila Dpp signaling is mediated by the punt gene product: A dual ligand-binding type II receptor of the TGF beta receptor family. Cell. 1995;80:899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 31.Ruberte E, Marty T, Nellen D, Affolter M, Basler K. An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell. 1995;80:889–897. doi: 10.1016/0092-8674(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhu CH, Xie T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development. 2003;130:2579–2588. doi: 10.1242/dev.00499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.