Abstract
Proteasome inhibitors are important tools for studying the roles of the proteasome in cellular processes. In this study, we observed that the proteasome inhibitors N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal (MG132), epoxomicin, and lactacystin were ineffective and bortezomib was completely effective in inhibiting cytokine-stimulated nitric oxide production in primary cultures of human hepatocytes that had been treated with the cytochrome P450 inducer phenobarbital. The inefficacy of MG132 was due to its metabolism by CYP3A enzymes, as deduced from its rapid, ketoconazole-sensitive clearance by pooled human liver microsomes and cultured hepatocytes. The efficacy of MG132 was increased by inclusion of ketoconazole in the hepatocyte incubations and decreased by prior treatment of the cultures with the CYP3A inducers phenobarbital or rifampicin. Epoxomicin was also rapidly metabolized by CYP3A, whereas bortezomib and lactacystin were much more stable metabolically in human liver microsomes or hepatocyte cultures. Thus, bortezomib is a better choice than MG132, epoxomicin, or lactacystin in cells with high activities of CYP3A enzymes. The reason for the lack of efficacy of lactacystin in human hepatocytes has yet to be determined, but it too should not be used for studies of proteasome function in human hepatocytes.
Introduction
The proteasome is a hetero-oligomeric protein complex with diverse roles in the regulation of cellular function. One such function is the removal of damaged or misfolded proteins, which are polyubiquitinated and directed to the proteasome for subsequent processive degradation (Werner et al., 1996). Another role of the proteasome is in the rapid regulation of cellular protein function in response to extracellular signals such as inflammation. Inflammation is an adaptive response that occurs during infection and tissue injury (Medzhitov, 2008). In chronic inflammatory diseases, such as asthma, rheumatoid arthritis, or inflammatory bowel disease, nuclear factor-κB (NF-κB) is pivotal in the intracellular transmission of proinflammatory cytokine signals (Barnes and Karin, 1997). Activation of NF-κB requires proteasomal degradation of the inhibitory subunit inhibitor of NF-κB to allow the translocation of NF-κB from cytosol to nucleus (Mellits et al., 1993; Miyamoto et al., 1994), where it stimulates the transcription of many target genes (McKee et al., 1997), such as inducible nitric-oxide synthase (NOSII).
Proteasome inhibitors are important tools for studying the diverse functions of the proteasome in in vitro, cellular, and in vivo experiments (Lee and Goldberg, 1998). In addition, the proteasome inhibitor bortezomib is approved by the U.S. Food and Drug Administration for the treatment of multiple myeloma (Uttamsingh et al., 2005). Most proteasome inhibitors directly target the 20S proteasome. They can be broadly categorized into synthetic analogs and natural products (Myung et al., 2001). Synthetic inhibitors are peptide-based compounds such as peptide benzamides, peptide α-ketoamides, peptide aldehydes, peptide α-ketoaldehydes, peptide vinyl sulfones, and peptide boronic acids such as bortezomib (Adams et al., 1998). Lactacystin and its in vivo derivative clasto-lactacystin β-lactone, which is thought to be an active component of lactacystin, and epoxomicin are all natural products (Meng et al., 1999; Myung et al., 2001).
The validity of mechanistic conclusions based on studies with chemical inhibitors is always reliant on the chemical and metabolic stability of the inhibitors in the cells or tissues being studied. Yet, little is known approximately the metabolism of the proteasome inhibitors most commonly used for such studies, such as N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal (MG132), lactacystin, and epoxomicin, especially in cells with high metabolic capacity such as hepatocytes. An exception is bortezomib which, because of its use in human therapy, has been studied thoroughly with respect to its metabolism by human liver microsomes (Pekol et al., 2005). In these studies, the microsomal metabolism of bortezomib was found to be catalyzed mainly by CYP3A4, a member of the cytochrome P450 (P450) multigene family of heme-thiolate enzymes. There are 18 families of human P450 genes and 57 human P450 genes and several pseudogenes (Guengerich, 2004; http://drnelson.uthsc.edu/human.P450.table.html). Approximately one-third of human P450s are involved in metabolism of xenobiotics, and most of them are mainly expressed in liver (Guengerich, 2004). Among these, CYP3A4 contributes significantly to the metabolism of approximately one-half of clinically available drugs (Guengerich, 2004).
Here, we studied the efficacy of various proteasome inhibitors (Fig. 1) in metabolically competent primary human hepatocytes. We found that MG132, lactacystin, and epoxomicin were each ineffective in inhibiting proteasome-dependent NOSII induction in the hepatocytes, especially when the hepatocytes were pretreated with the P450 inducers phenobarbital (PB) and rifampicin (RIF). We found that CYP3A4 was most likely responsible for the rapid metabolism of MG132 and epoxomicin by human liver microsomes. Bortezomib, on the other hand, was much more slowly metabolized and, as a result, had greater efficacy in proteasome inhibition.
Fig. 1.
Structures of the proteasome inhibitors used in this study.
Materials and Methods
Reagents.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Lactacystin, clasto-lactacystin β-lactone, MG132, and epoxomicin were purchased from Boston Biochem (Cambridge, MA), and bortezomib was purchased from LC Laboratories (Woburn, MA). Tumor necrosis factor-α (TNF-α) was from Invitrogen (Carlsbad, CA), and interferon-γ (IFN-γ) was from BD Pharmingen (San Diego, CA). Pooled human liver microsomes and baculovirus insect cell-expressed CYP3A4 Supersomes were purchased from BD Bioscience (San Jose, CA).
Human Hepatocyte Cultures and Treatments.
The human hepatocyte studies were performed in accordance with the Declaration of Helsinki and designated exempt from review by the Emory University Institutional Review Board. Human hepatocytes were obtained from Dr. Steven Strom at the University of Pittsburgh via the National Institutes of Health-sponsored Liver Tissue Cell Distribution System program. They were prepared from livers not suitable for transplantation within 24 h of procurement. Details on liver donors are provided in Table 1. Hepatocytes were isolated as described in Strom et al. (1996) by a three-step collagenase perfusion technique and cultured on collagen-coated 6- or 12-well plates. Cells were usually cultured for 24 h before delivery in hepatocyte maintenance medium (Cambrex Bio Science, Inc., Walkersville, NY). Upon receipt, cells were placed at 37°C in 5% CO2, and medium was changed 3 to 4 h later to Williams' E cell culture medium [containing 10 nM insulin, 25 nM dexamethasone, and 1× antibiotic and antimycotic solution (Invitrogen)]. Cells were cultured additionally for 3 to 4 days in the same medium, and then cells were either treated with 1 mM PB or 50 μM RIF for 2 days or cultured in the same medium.
TABLE 1.
Donor information
| Cells | Sex | Age | Race | Source or Cause of Death |
|---|---|---|---|---|
| 1390 | M | 56 yr | White | Liver resection |
| 1403 | M | 21 months | White | Liver resection |
| 1407 | F | 55 yr | Black | Liver resection |
| 1410 | F | 36 yr | White | Liver resection |
| 1422 | M | 16 yr | White | Liver resection |
| 1425 | M | 50 yr | White | Intracranial hemorrhage |
M, male; F, female.
After a 48-h induction period, cells were exposed to a mixture (ILmix) of IL-1 (2.5 ng/ml), TNF-α (5 ng/ml), and IFN-γ (5 ng/ml) in the presence or absence of proteasome inhibitors for the times indicated in the figure legends, to stimulate NOSII induction and NO production. The concentrations of proteasome inhibitors used were those documented to inhibit proteasomal activity in other cell types and that we found to inhibit proteasome activity in cultured rat hepatocytes (Lee et al., 2008, 2009). Control cells were also incubated for the same time in the absence of ILmix. At the end of the incubation period, media samples were removed and reserved for assay of the stable end products of NO production, nitrate + nitrite (NOx), using the Griess reaction (Green et al., 1982).
Inhibition of CYP3A Activity.
CYP3A activity was assayed by using the P450-Glo CYP3A4 assay system with Luciferin-PPXE as a substrate of CYP3A following the manufacturer's direction (Promega, Madison, WI). In brief, 100 μM Luciferin-PPXE was mixed with either DMSO as a control or various proteasome inhibitors and then incubated with pooled human liver microsomes (1 μg/ml) or CYP3A4 Supersomes (0.5 μg/ml) without light at room temperature for 10 min. The reaction was started by addition of 100 μM NADPH and followed for 30 min. Equal volumes of the supernatant and Luciferin Detection Reagent (Promega) were mixed, and 20 min later luciferin was measured on a SpectraMax L luminometer (Molecular Devices, Sunnyvale, CA). For cellular assays, hepatocytes were incubated with substrate (1 ml of media containing 25 μM Luciferin-PPXE/well in six-well plates or 0.5 ml/well in 12-well plates) for 1 h, and then media were collected for luciferin assay as described above.
Metabolic Stability Assay.
The metabolism of epoxomicin, lactacystin, MG132, and bortezomib (1 μM) by human liver microsomes (HLM) was monitored by a substrate depletion method in 100 mM potassium phosphate buffer (pH 7.4) containing 1 mM NADPH at 37°C. Incubations with HLM contained 0.8 mg/ml protein suspended in a final volume of 1 ml. Then 50-μl samples were taken out of the incubation mixture at different time points and quenched with 150 μl of acetonitrile containing 0.2 μM tolbutamide as an internal standard. After extraction, lactacystin analytical samples were diluted by addition of water (3:1). Plates were then centrifuged at 3000 rpm for 5 min at room temperature. Midazolam (1 μM) was used as a probe substrate for CYP3A4 (95% metabolized by CYP3A4), and ketoconazole (1 μM) was used as a competitive inhibitor of CYP3A4. Intrinsic clearance for each compound was calculated using the in vitro half-life (linear regression of the ln percentage remaining of substrate as a function of incubation time). Similar experiments were conducted using human hepatocytes (1.0 × 106 cells/ml) and fresh Williams' E buffer.
Liquid Chromatography/Tandem Mass Spectral Analysis.
The LC-mass spectrometry system consisted of an API 4000 triple quadrupole mass spectrometer with an atmospheric pressure electrospray ionization source (MDS Sciex, Concord, ON, Canada) and two LC-10ADvp pumps with an SCL-10ADvp controller (Shimadzu, Columbia, MD). Analytes were separated using a reverse-phase Zorbax C-18 column (2.1 × 50 mm, 3.5 μm; Agilent Technologies, Santa Clara, CA) with a gradient elution profile and a flow rate of 0.4 ml/min. The gradient was initiated and held for the first 1 min at 100% solvent A (0.1% formic acid in water) and was then ramped linearly to 5% A:95% solvent B (0.1% formic acid in acetonitrile) over 1 min and held for 2.5 min. The profile was then immediately returned to initial conditions that were maintained for 1 min with a flow rate of 0.4 ml/min before the next injection.
Bortezomib and lactacystin were detected in negative ion mode. The deprotonated molecular ions were formed using an ion spray voltage of −4200 V, curtain gas of 25 V, collision gas of 4 eV, and source temperature of 400°C. Product ions were formed at collision energies of −29 eV (bortezomib, m/z 383.2 → 322.0), −12 eV (lactacystin, m/z 375.1 → 161.7), and −35 eV (tolbutamide, m/z 269.1 → 105.9). The positive ionization mode was used for epoxomicin and MG132. The parameters of the method are were as follows: ion spray voltage of 5500 V, curtain gas of 20 V, collision gas of 4 eV, and source temperature of 450°C. Product ions for epoxomicin (m/z 555.5 → 170.2) were formed at collision energy of 22 eV, for MG132 (m/z 476.3 → 361.2 and 91.0) at collision energy of 45 eV, and for tolbutamide (m/z 271.1 → 155.1) at collision energy 25 eV. Because MG132 was broken down into two mass fragments, both the peaks were integrated for quantitation.
Statistical Analysis.
Analysis of variance and Tukey's test were used for statistical comparisons between groups. Significance was set at p < 0.05 for all tests.
Results
NO Production by Primary Human Hepatocytes and Its Inhibition by Proteasome Inhibitors.
To study the efficacy of different proteasome inhibitors in inhibiting NF-κB-dependent NOSII induction, we used a mixture of cytokines that our group found to be optimal for NOSII induction in cultured human hepatocytes (Aitken et al., 2008). ILmix (5 ng/ml IL-1β, 10 ng/ml TNF-α, and 10 ng/ml IFN-γ) was enough to produce a 10- to 50-fold induction of NOx production within 24 h (Aitken et al., 2008), NOSII mRNA levels started to rise as early as 6 h and were still elevated at 24 h (Aitken et al., 2008). We subsequently found that these concentrations of cytokines could be reduced by one-half (2.5 ng/ml IL-1β, 5 ng/ml TNF-α, and 5 ng/ml IFN-γ) and still evoke similar levels of NOx production. Therefore, we used the lower ILmix concentration to stimulate inflammatory events in the cells.
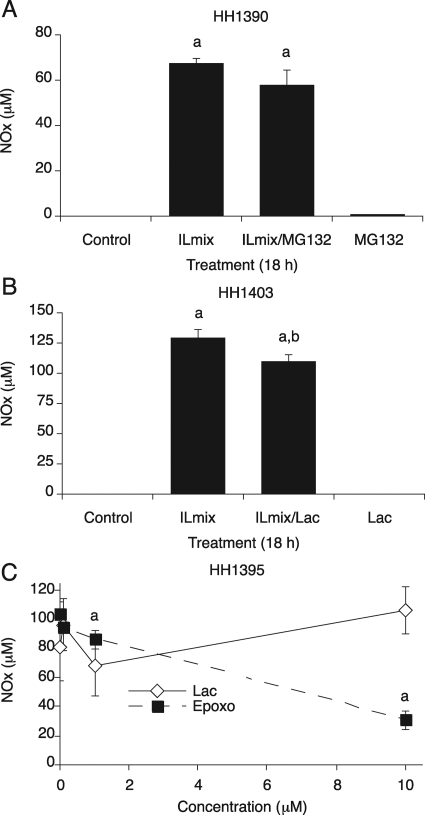
Initial experiments were conducted in hepatocytes pretreated with the P450 inducer PB. Hepatocytes (HH1390) were treated with ILmix, the proteasome inhibitor MG132 (10 μM), or both for 18 h. Because NOSII induction is dependent on the activation of NF-κB in the cells, which is in turn dependent on proteasomal activity, we expected MG132 to block the production of NOx in the media. As shown in Fig. 2A, ILmix stimulated NOx production measured as accumulation of 67 μM NOx in the media, but cotreatment of ILmix and MG132 did not inhibit this NOx production. The same results were seen in various primary hepatocyte preparations, and even 50 μM MG132 could not block ILmix-stimulated NOx production (data not shown). Likewise, lactacystin (10 μM) only minimally (by 10%) inhibited ILmix-stimulated NOx production (Fig. 2B). Next, we treated primary human hepatocytes (HH1395) with ILmix and different concentrations of epoxomicin or clasto-lactacystin for 18 h. Lactacystin, which is a prodrug, undergoes spontaneous conversion to the active proteasome inhibitor, clasto-lactacystin β-lactone, and it rapidly enters the cells (Dick et al., 1996, 1997). Therefore, we used clasto-lactacystin β-lactone instead of lactacystin. Clasto-lactacystin β-lactone also failed to inhibit ILmix-stimulated NO production at any concentration (Fig. 2C). Epoxomicin at 1 μM produced a significant but small (12%) inhibition. Although 10 μΜ epoxomicin inhibited NOx production by 65%, this effect could be due to the toxicity of this concentration to the cells (Fig. 2C; observed morphologically, data not shown). We examined inhibition of NO production by 10 μΜ epoxomicin in another primary human hepatocyte preparation (HH1407). ILmix treatment stimulated NO accumulation in media and NO production was dramatically inhibited with ILmix/epoxomicin treatment (Supplemental Fig. 1A). However, the cells were significantly stressed and detached from both 10 μΜ epoxomicin treated plates (data not shown).
Fig. 2.
NOx production of primary human hepatocytes and its inhibition by proteasome inhibitors. Primary human hepatocytes were cultured for 3 days after delivery and treated with 1 mM PB for an additional 2 days. PB was maintained in the media during treatments. NOx was measured from media after treatment. A, hepatocytes (HH1390) were treated with ILmix (2.5 ng/ml IL-1β, 5 ng/ml TNF-α, and 5 ng/ml IFN-γ), the proteasome inhibitor MG132 (10 μM), or both for 18 h. B, hepatocytes (HH1402) were treated with ILmix, the proteasome inhibitor lactacystin (10 μM), or both or ILmix/lactacystin (10 μM) for 18 h. C, concentration dependence of the effects of proteasome inhibitors on NOx production. PB-treated hepatocytes (HH1395) were cotreated with ILmix and various concentrations of lactacystin (Lac) or epoxomicin (Epoxo) for 18 h, and NOx was measured in the media. Values are the mean ± S.D. of three independent samples. a, significantly different from control; b, significantly different from ILmix alone, P < 0.05.
Effect of PB and RIF on the Efficacies of Proteasome Inhibitors.
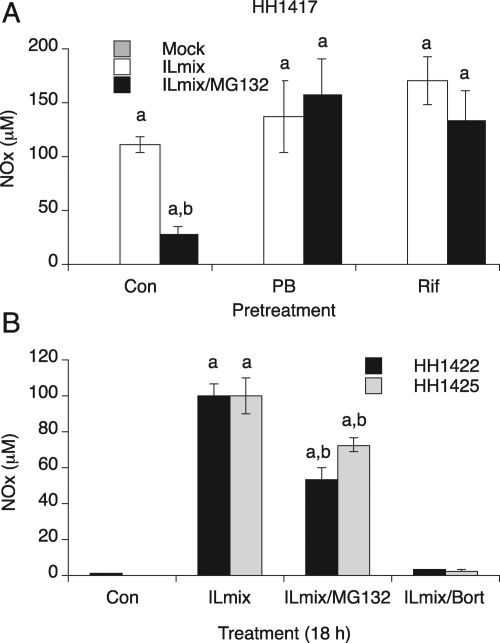
Because the cells in the previous experiments were all pretreated with PB, we were interested to determine whether induction of P450 enzymes by the barbiturate might reduce the efficacy of the proteasome inhibitors (MG132, lactacystin, and epoxomicin) via induction of their metabolism. CYP2B and CYP3A4 genes are transcriptionally activated by PB via the constitutive androstane receptor (Sueyoshi and Negishi, 2001; Tirona et al., 2003). Therefore, we next studied the efficacies of the proteasome inhibitors in the absence of PB. To differentiate the relative roles of CYP2B and CYP3A in modulating the efficacies of the inhibitors, we also studied cells pretreated with RIF, which preferentially induces CYP3A enzymes via the pregnane X receptor (Goodwin et al., 1999). HH1417 cells were pretreated with or without 1 mM PB or 50 μM RIF for 2 days and then treated with ILmix or ILmix plus MG132 for 20 h. Figure 3A shows that MG132 (20 μM) significantly suppressed NOx production by 70% in uninduced hepatocytes, whereas MG132 failed to suppress NO production in PB (1 mM) and RIF (50 μM)-induced hepatocyte samples. In contrast, the therapeutic proteasome inhibitor bortezomib (10 μM) effectively blocked ILmix-stimulated NO production in PB-induced hepatocytes from two different donors (Fig. 3B). We also found that bortezomib blocked NO production in uninduced primary cultures (data not shown).
Fig. 3.
Effect of pretreatment with PB and RIF on the efficacies of proteasome inhibitors. A, primary human hepatocytes (HH1417) were cultured for 3 days after delivery and treated with or without 1 mM PB or 50 μM rifampicin for an additional 2 days. PB or RIF were maintained in the media during treatments. Hepatocytes were treated with vehicle (mock), ILmix, or ILmix/MG132 (20 μM), and NOx was measured in the media 20 h after treatment. B, bortezomib efficacy on NOx production. Two different human hepatocyte preparations (HH1422 and HH1425) were pretreated with 1 mM PB for 2 days and then treated with vehicle ILmix, ILmix and MG132 (10 μM), or ILmix and bortezomib (Bort, 10 μM) for 18 h. NOx was measured in the media. Values are the mean ± S.D. of three independent samples. a, significantly different from control; b, significantly different from ILmix alone, P < 0.05. Con, control.
Effect of Ketoconazole on Proteasome Inhibition.
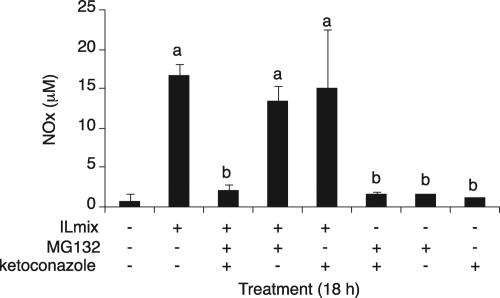
Because either PB or RIF pretreatment reduced MG132 efficacy (Fig. 3A), we reasoned that CYP3A enzymes were the main candidates to metabolize MG132 in the primary cultures. Therefore, we next investigated the effect of the competitive CYP3A4 inhibitor ketoconazole (Boxenbaum, 1999) on proteasome inhibitor action in these cells. PB-treated hepatocytes were cotreated with ketoconazole. When the HH1410 primary hepatocytes were cotreated with ketoconazole (10 μM), ILmix, and MG132 (20 μM), NO production was effectively blocked, whereas MG132 again failed to inhibit NO production in the absence of ketoconazole (Fig. 4). The effect of ketoconazole on MG132 inhibition of NO production was somewhat variable. In the experiment shown in Supplemental Fig. 1B, the inhibition caused by 20 μM MG132 in the presence of ketoconazole did not achieve statistical significance, which could be related to the much higher level of NO production in response to ILmix in the latter experiment.
Fig. 4.
Effect of ketoconazole on MG132 efficacy. Primary human hepatocytes (HH1410) were cultured for 3 days after delivery and treated with 1 mM PB for an additional 2 days. PB was maintained in the media during treatments. The cells were treated for 18 h with ILmix, ketoconazole (Ket, 10 μM), or MG132 (20 μM) in different combinations as indicated in the figure, and NOx was measured in the media. Values are the mean ± S.D. of three independent samples. a, significantly different from control; b, significantly different from ILmix alone, P < 0.05.
Proteasome Inhibitors as Inhibitors of Human Microsomal CYP3As.
The preceding data are consistent with the idea that in primary human hepatocytes, CYP3A enzymes metabolize MG132 and other proteasome inhibitors except bortezomib. We therefore hypothesized that if a proteasome inhibitor is metabolized by CYP3A, it may inhibit CYP3A metabolism of other substrates. Therefore, we tested the abilities of the proteasome inhibitors to inhibit NADPH-supported metabolism of the probe CYP3A substrate Luciferin-PPXE in pooled human liver microsomes. Bortezomib, MG132, and epoxomicin were all potent inhibitors of CYP3A activity, with >50% inhibition at 10 μM, whereas clasto-lactacystin β-lactone did not inhibit the CYP3A activity even at the 50 μM concentration (Fig. 5A). This result was confirmed by experiments in insect cell microsomes specifically expressing CYP3A4 (CYP3A4 Supersomes; BD Bioscience) and NADPH-cytochrome P450 reductase. Again, only lactacystin failed to inhibit CYP3A4 activity (Fig. 5B).
Fig. 5.
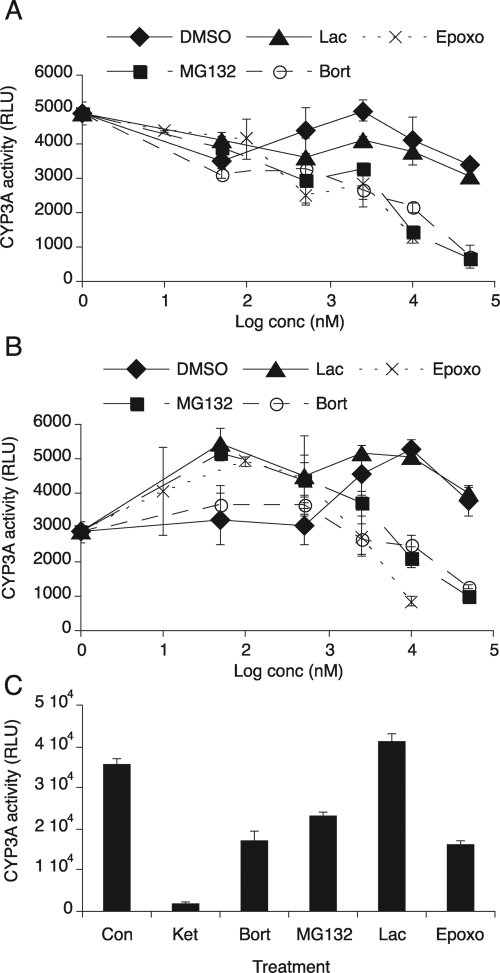
Effect of proteasome inhibitors on CYP3A activity. CYP3A activity was measured by using the P450-Glo CYP3A4 assay system with Luciferin-PPXE as a substrate of CYP3A. A, CYP3A activity in pooled human liver microsomes. Luciferin-PPXE (100 μM) was mixed with either DMSO as a control or various concentrations (0.05–50 μM) of MG132, lactacystin (Lac), or bortezomib (Bort) or 0.01 μM to 10 μM epoxomicin (Epoxo) and then incubated with microsomes (1 μg/ml) for 10 min. The reaction was started by addition of NADPH. After 30 min, luminescence was measured with a luminometer. B, CYP3A4 activity in CYP3A4 Supersomes. Luciferin-PPXE was mixed with either DMSO as a control or various concentrations of proteasome inhibitors as described above and then incubated with CYP3A4 Supersomes (0.5 μg/ml) for 10 min. Thirty minutes after addition of NADPH to the reaction, luminescence was measured with a luminometer. C, hepatocellular CYP3A activity. Two-day PB-treated primary human hepatocytes (HH1425) were incubated with various proteasome inhibitors [20 μM MG132, 20 μM bortezomib, 10 μM clasto-lactacystin β-lactone (Lac), or 2.5 μM epoxomicin] or 20 μM ketoconazole (Ket) as a positive control (Con). Four hours later, hepatocytes were incubated with 25 μM Luciferin-PPXE for 1 h, and the media were collected for luminescence assay as described above. Values are the mean ± S.D. of three independent samples. RLU, relative light units.
We also examined CYP3A inhibition in cell cultures. Luciferin-PPXE (50 μM) was added to PB-treated cultures of primary human hepatocytes (HH1425) in the presence or absence of the proteasome inhibitors. After 4 h of incubation, media were harvested for luciferin assay. Ketoconazole (20 μM) as a positive control effectively inhibited CYP3A activity, whereas bortezomib (20 μM), MG132 (20 μM), and epoxomicin (2.5 μM) each inhibited CYP3A activity by approximately 50% (Fig. 5C). Clasto-lactacystin β-lactone (the cell-permeable, active form of lactacystin, 10 μM) had no effect. These results are similar to those of the in vitro inhibition assays.
In Vitro and Cellular Clearance of Proteasome Inhibitors.
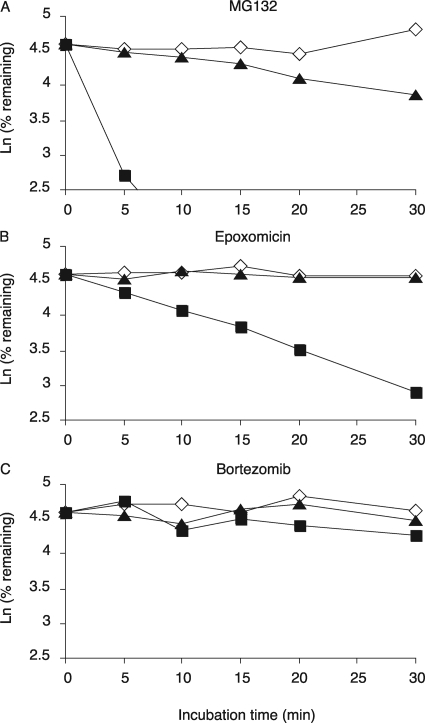
Finally, we studied the metabolic clearance of the proteasome inhibitors by pooled human liver microsomes and cultured primary human hepatocytes using LC-MS-MS. Midazolam, which is metabolized and cleared more than 95% by CYP3A4, was used as a positive control (Ghosal et al., 1996), and ketoconazole was used as a specific competitive inhibitor of CYP3A4 (Boxenbaum, 1999). In human liver microsomes, midazolam was rapidly cleared [intrinsic clearance (Clint) = 284 ml/(min · kg)], and this clearance was blocked by ketoconazole (data not shown). MG132 was also cleared rapidly [Clint = 306 ml/(min · kg)] (Fig. 6A). Whereas epoxomicin was cleared more slowly (Fig. 6B) [Clint = 82 ml/(min · kg)], metabolism of both MG132 and epoxomicin was inhibited effectively by ketoconazole. In sharp contrast, bortezomib was cleared much more slowly [Clint = 18.2 ml/(min · kg)] (Fig. 6C), and lactacystin was not significantly cleared by pooled human liver microsomes (data not shown).
Fig. 6.
Metabolic stability of proteasome inhibitors and midazolam using pooled human liver microsomes. The metabolism of MG-132 (A), epoxomicin (B), and bortezomib (C) by HLM were monitored by a substrate depletion method. Aliquots were taken from the incubation mixtures at 0, 5, 10, 15, 20, and 30 min, and the quenched samples were analyzed by LC-MS/MS. Ketoconazole (1 μM) was used as a competitive inhibitor of CYP3A4.
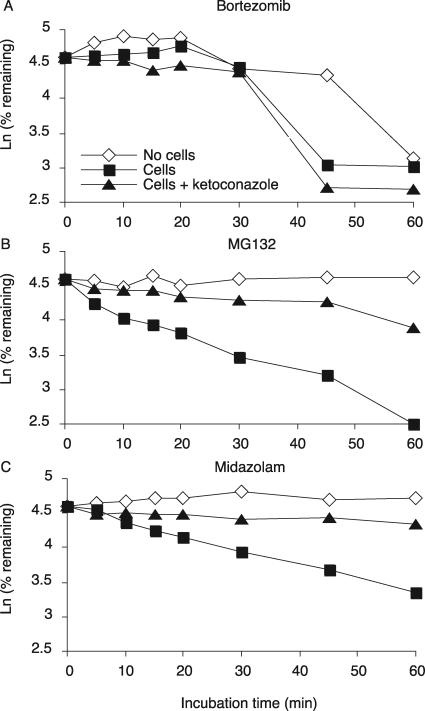
The results from the above experiments in human liver microsomes were confirmed by studies in cryopreserved primary human hepatocyte cultures. In cultures incubated with midazolam, 50% of midazolam was cleared within 30 min (Fig. 7A). MG132 was more rapidly cleared than midazolam, in that only 32.4% of the total MG132 remained after 30 min of incubation, and the metabolism of both drugs was blocked by coincubation of ketoconazole (Fig. 7B). However, bortezomib was not cleared until 30 min of incubation, and then concentrations rapidly declined by 80% in the next 15 min. However, its clearance was not affected by ketoconazole coincubation, suggesting that bortezomib disappearance was not due to CYP3A but may be due to instability of bortezomib over the 45 min of this experiment. In a second experiment, bortezomib was not significantly cleared within 45 min by cryopreserved hepatocytes (Supplemental Fig. 2B), either in the presence or absence of ketoconazole. However, this preparation of hepatocytes was even more active in midazolam clearance (Supplemental Fig. 2A) than the preparation used in Fig. 7. Taken together, our data clearly indicate that MG132 is much more rapidly cleared than bortezomib in cryopreserved primary human hepatocytes in the first 30 min and that this clearance is mediated by one or more CYP3A enzymes.
Fig. 7.
Metabolic stability of proteasome inhibitors and midazolam using cryopreserved primary human hepatocyte cultures. The metabolism of bortezomib (A), MG132 (B), and midazolam (C) by cryopreserved primary human hepatocyte cultures was monitored by a substrate depletion method. Aliquots of the incubation media were taken at 0, 5, 10, 15, 20 30, 45, and 60 min, and the quenched samples were analyzed by LC-MS/MS. Ketoconazole (1 μM) was used as a competitive inhibitor of CYP3A4.
Discussion
Of the four proteasome inhibitors evaluated in this study, only bortezomib was effective in inhibition of proteasome activity in PB-treated human hepatocyte cultures. The preponderance of evidence suggests that the lack of efficacy of MG132 and epoxomicin under these conditions is due to their rapid CYP3A-dependent metabolism. Even in inducer-naive hepatocytes, MG132 and epoxomicin were only partially effective for the same reason.
The metabolic stabilities of MG132, epoxomicin, and lactacystin have not been described previously. MG132 is the proteasome inhibitor most commonly used as a mechanistic tool to define proteasome function. Here we provide strong evidence that MG132 is rapidly metabolized by cultured human hepatocytes, limiting its efficacy, and that this clearance is catalyzed primarily by CYP3A enzymes. The metabolism of MG132 by human hepatocytes and by human liver microsomes is blocked by the CYP3A-specific inhibitor ketoconazole. Furthermore, MG132 inhibits the metabolism of a known CYP3A4 substrate by human liver microsomes, CYP3A4 Supersomes, and cultured hepatocytes. Thus, the fast turnover of MG132 by CYP3A affects its action in cells, as demonstrated by the facts that inhibition of CYP3A with ketoconazole potentiates and induction of CYP3A4 with PB or RIF reduces its efficacy.
We reported previously that 24 h of ILmix treatment caused a 50% down-regulation of CYP3A4 protein in primary human hepatocyte cultures from two different sources (Aitken et al., 2008). However, Fig. 7 shows that 90% of the MG132 is metabolized by uninduced hepatocytes within 60 min, with a half-life of approximately 15 min. Thus, it is likely that the great majority of the drug would be cleared long before CYP3A4 would be significantly down-regulated.
Epoxomicin is a more specific inhibitor of the proteasome than is MG132 (Meng et al., 1999), but it was ineffective at 10 μM in PB-induced hepatocytes, a concentration that is effective in other cell types (Meng et al., 1999; Kikuchi et al., 2003). Epoxomicin was also rapidly cleared by human liver microsomes in a ketoconazole-sensitive process and also inhibited the activity of CYP3A4 in microsomes, CYP3A4 Supersomes, and cultured hepatocytes. Thus, our findings suggest that the efficacy of this compound, like that of MG132, is limited by CYP3A-dependent metabolism.
In our study, bortezomib was metabolized relatively slowly by human liver microsomes and not detectably by hepatocytes after 30 min of incubation. That it inhibits CYP3A4 activity with potency similar to that of MG132 and epoxomicin suggests that the catalytic efficiency of CYP3As for bortezomib is relatively low. We observed that bortezomib clearance in human liver microsomes was approximately half of the value described previously (Uttamsingh et al., 2005), which could easily be explained by different microsomal preparations. Uttamsingh et al. (2005) identified the contributions of the microsomal P450 isoforms to bortezomib metabolism as CYP3A4 (38.4%) > CYP2C19 (30.1%) > CYP1A2 (10.5%) > CYP2D6 (7.1%) > CYP2C9 (1.2%) and concluded that the sum of the contributions of those five P450s was approximately 87% (Uttamsingh et al., 2005). They further suggested that the “clearance of bortezomib in humans at the recommended dose of 1.0 mg/m2 is approximately 0.4 l/h/kg, which is one-third the liver blood flow rate; thus, CYP3A4-based drug-drug interactions may not be observed in patients.” Thus, even though CYP3A4-dependent metabolism clearly is important in vivo, it is not fast enough to affect the efficacy of bortezomib in cultured hepatocytes.
Lactacystin also failed to inhibit NOSII induction in the PB-treated hepatocytes, but unlike epoxomicin and MG132 it was not significantly metabolized by human liver microsomes. It also failed to inhibit CYP3A4 in microsomes, Supersomes, or cultured hepatocytes. More work is needed to determine the reason for the inefficacy of lactacystin in the hepatocytes. Lactacystin could be inactivated by non-P450 enzymes in the cytoplasm or mitochondria, or it might be extruded from the cell by an efflux transporter.
We note that MG132 has been used successfully by us and others to inhibit proteasome activity in rat hepatocytes (Wang et al., 1999; Lee et al., 2008, 2009). Indeed, MG132 effectively blocked IL-1β-evoked NO production in PB-treated rat hepatocytes (Lee et al., 2008, 2009). In studies not shown, we determined that MG132 was metabolized by rat and human liver microsomes at approximately equal rates. It is possible that the difference in efficacy of MG132 between rat and human hepatocytes could be due to a lower expression in cultured hepatocytes of the rat enzymes responsible for its metabolism, relative to CYP3A expression in human hepatocytes.
In conclusion, our finding that MG132 and epoxomicin have limited efficacies in human hepatocytes because of their metabolism by CYP3A enzymes has important implications for the interpretation of studies using these proteasome inhibitors in human hepatocytes or other cells that express CYP3A enzymes (e.g., intestinal cells). A negative result could mean that the drug is being metabolized by the cells, rather than a lack of involvement of the proteasome in the process being investigated. The results suggest that bortezomib is a better investigational tool for cells with high drug-metabolizing activities.
Supplementary Material
This work was supported in part by the National Institutes of Health National Institute for General Medical Science [Grant R01-GM069971]; and the National Institutes of Health National Institute for Diabetes and Digestive and Kidney Diseases [Contract N01-DK70004/HHSN267200700004C].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.035501.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- NF-κB
- nuclear factor κB
- NOSII
- inducible nitric-oxide synthase
- MG132
- N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal
- P450
- cytochrome P450
- PB
- phenobarbital
- RIF
- rifampicin
- TNF-α
- tumor necrosis factor-α
- IFN-γ
- interferon-γ
- IL
- interleukin
- NO
- nitric oxide
- NOx
- nitrate plus nitrite
- DMSO
- dimethyl sulfoxide
- HLM
- human liver microsomes
- LC
- liquid chromatography
- MS/MS
- tandem mass spectroscopy.
References
- Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. (1998) Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg Med Chem Lett 8:333–338 [DOI] [PubMed] [Google Scholar]
- Aitken AE, Lee CM, Morgan ET. (2008) Roles of nitric oxide in inflammatory downregulation of human cytochromes P450. Free Radic Biol Med 44:1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ, Karin M. (1997) Nuclear factor-κB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med 336:1066–1071 [DOI] [PubMed] [Google Scholar]
- Boxenbaum H. (1999) Cytochrome P450 3A4 in vivo ketoconazole competitive inhibition: determination of Ki and dangers associated with high clearance drugs in general. J Pharm Pharm Sci 2:47–52 [PubMed] [Google Scholar]
- Dick LR, Cruikshank AA, Destree AT, Grenier L, McCormack TA, Melandri FD, Nunes SL, Palombella VJ, Parent LA, Plamondon L, et al. (1997) Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem 272:182–188 [DOI] [PubMed] [Google Scholar]
- Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. (1996) Mechanistic studies on the inactivation of the proteasome by lactacystin: a central role for clasto-lactacystin β-lactone. J Biol Chem 271:7273–7276 [DOI] [PubMed] [Google Scholar]
- Ghosal A, Satoh H, Thomas PE, Bush E, Moore D. (1996) Inhibition and kinetics of cytochrome P4503A activity in microsomes from rat, human, and cDNA-expressed human cytochrome P450. Drug Metab Dispos 24:940–947 [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339 [DOI] [PubMed] [Google Scholar]
- Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. (1982) Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem 126:131–138 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2004) Cytochrome P450: what have we learned and what are the future issues? Drug Metab Rev 36:159–197 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Shinpo K, Tsuji S, Takeuchi M, Yamagishi S, Makita Z, Niino M, Yabe I, Tashiro K. (2003) Effect of proteasome inhibitor on cultured mesencephalic dopaminergic neurons. Brain Res 964:228–236 [DOI] [PubMed] [Google Scholar]
- Lee CM, Kim BY, Li L, Morgan ET. (2008) Nitric oxide-dependent proteasomal degradation of cytochrome P450 2B proteins. J Biol Chem 283:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Pohl J, Morgan ET. (2009) Dual mechanisms of CYP3A protein regulation by proinflammatory cytokine stimulation in primary hepatocyte cultures. Drug Metab Dispos 37:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. (1998) Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8:397–403 [DOI] [PubMed] [Google Scholar]
- McKee CM, Lowenstein CJ, Horton MR, Wu J, Bao C, Chin BY, Choi AM, Noble PW. (1997) Hyaluronan fragments induce nitric-oxide synthase in murine macrophages through a nuclear factor κB-dependent mechanism. J Biol Chem 272:8013–8018 [DOI] [PubMed] [Google Scholar]
- Medzhitov R. (2008) Origin and physiological roles of inflammation. Nature 454:428–435 [DOI] [PubMed] [Google Scholar]
- Mellits KH, Hay RT, Goodbourn S. (1993) Proteolytic degradation of MAD3 (IκBα) and enhanced processing of the NF-κB precursor p105 are obligatory steps in the activation of NF-κB. Nucleic Acids Res 21:5059–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA 96:10403–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Chiao PJ, Verma IM. (1994) Enhanced IκBα degradation is responsible for constitutive NF-κB activity in mature murine B-cell lines. Mol Cell Biol 14:3276–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J, Kim KB, Crews CM. (2001) The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev 21:245–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekol T, Daniels JS, Labutti J, Parsons I, Nix D, Baronas E, Hsieh F, Gan LS, Miwa G. (2005) Human metabolism of the proteasome inhibitor bortezomib: identification of circulating metabolites. Drug Metab Dispos 33:771–777 [DOI] [PubMed] [Google Scholar]
- Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. (1996) Use of human hepatocytes to study P450 gene induction. Methods Enzymol 272:388–401 [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Negishi M. (2001) Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol 41:123–143 [DOI] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. (2003) The orphan nuclear receptor HNF4α determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 9:220–224 [DOI] [PubMed] [Google Scholar]
- Uttamsingh V, Lu C, Miwa G, Gan LS. (2005) Relative contributions of the five major human cytochromes P450, 1A2, 2C9, 2C19, 2D6, and 3A4, to the hepatic metabolism of the proteasome inhibitor bortezomib. Drug Metab Dispos 33:1723–1728 [DOI] [PubMed] [Google Scholar]
- Wang HF, Figueiredo Pereira ME, Correia MA. (1999) Cytochrome P450 3A degradation in isolated rat hepatocytes: 26S proteasome inhibitors as probes. Arch Biochem Biophys 365:45–53 [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky JL, McCracken AA. (1996) Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA 93:13797–13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.