Abstract
Thiopurine drugs such as 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG) are used to treat acute lymphoblastic leukemia of childhood. To test the hypothesis that variation in the expression of genes within the “thiopurine pathway” might influence 6-MP and 6-TG sensitivity, we generated basal gene expression profiles and IC50 values for both of these thiopurine drugs using a model system consisting of 194 Human Variation Panel lymphoblastoid cell lines. Association analysis showed that thiopurine S-methyltransferase, ecto-5′-nucleotidase (NT5E), and multidrug resistance protein 4 (ABCC4) expression were correlated with thiopurine cytotoxicity. Those observations suggested the possible existence of a “thiopurine cellular circulation” involving nucleotide efflux by ABCC4, hydrolysis of thiopurine nucleotide monophosphates outside of the cell by NT5E, and subsequent transport of thiopurine nucleosides back into the cell by nucleoside transporters. The existence of this cellular circulation was confirmed by a series of functional experiments performed with cultured cells stably or transiently transfected with ABCC4 and/or NT5E. Because of the central role of NT5E in this cellular circulation, the NT5E gene was resequenced using 287 DNA samples from three ethnic groups, with the identification of 68 single nucleotide polymorphisms (SNPs), 46 of which were novel. Several SNPs in the 5′-flanking region of NT5E were highly correlated with expression, rs9450278 having the lowest p value (p = 2.4 × 10−10, R = −0.376). The thiopurine cellular circulation and genetic polymorphisms for genes encoding the proteins involved should be incorporated into future studies of thiopurine drug therapy and effect.
Introduction
Thiopurine drugs such as 6-mercaptopurine (6-MP) and 6-thioguanine (6-TG) are used to treat acute lymphoblastic leukemia (ALL) of childhood (Lennard, 1992). 6-MP and 6-TG are prodrugs that undergo metabolic activation to form 6-thioguanine nucleotides (6-TGNs), followed by their incorporation into DNA (Lennard, 1992). Red blood cell (RBC) 6-TGN concentrations have been used as an index for the therapeutic and toxic effects of these drugs (Lennard et al., 1983, 1987; Lennard, 1987). Higher concentrations of 6-TGNs can be associated with profound life-threatening myelosuppression (Lennard et al., 1983, 1987). Several genes encoding proteins within the known “thiopurine pathway” (http://www.pharmgkb.org) can contribute to individual variation in thiopurine response. For example, inherited variation in the activity of the thiopurine-metabolizing enzyme thiopurine S-methyltransferase (TPMT) is a major factor responsible for individual variation in RBC 6-TGN concentrations and thiopurine response (Wang and Weinshilboum, 2006). Patients homozygous for TPMT alleles associated with low activity have elevated RBC 6-TGN concentrations when treated with standard doses of thiopurines, and they are also at increased risk for life-threatening myelosuppression (Lennard et al., 1987; Wang and Weinshilboum, 2006). However, TPMT activity does not account for all of the variation in response to thiopurine therapy (Gearry et al., 2003). For example, a recent study showed that human leukemic lymphoblasts expressing high levels of the multidrug resistance protein 4 (ABCC4/MRP4) had decreased intracellular 6-TGN concentrations (Krishnamurthy et al., 2008), consistent with observations that ABCC4/MRP4 can mediate the transport of thiopurine nucleotide monophosphates out of cells (Wielinga et al., 2002; Peng et al., 2008).
In the present study, in an attempt to determine whether thiopurine pathway genes beyond TPMT might contribute to thiopurine-induced cytotoxicity, we have used 194 ethnically defined “Human Variation Panel” lymphoblastoid cell lines as a model system to explore the relationship between variation in gene expression and thiopurine cytotoxicity (Li et al., 2008). Although lymphoblastoid cells are not tumor-derived, they have been used successfully to determine the association of genome-wide expression data with antineoplastic drug response, and their use has provided novel insights into mechanisms of drug action (Huang et al., 2007, 2008; Li et al., 2008; Pei et al., 2009). In our study, we correlated basal gene expression profiles with thiopurine-dependent cytotoxicity (IC50 values) in these 194 cell lines to identify genes within the known thiopurine pathway that might contribute to thiopurine cytotoxicity, followed by functional validation of the candidates identified. The results highlighted ecto-5′-nucleotidase (NT5E), an extracellular membrane purine monophosphate-hydrolyzing ecto-enzyme, in variation in thiopurine cytotoxicity and suggested the existence of a “thiopurine cellular circulation” involving the ABCC4/MRP4 transporter. Because of its central role in this process, we resequenced the NT5E gene and observed a large number of novel single nucleotide polymorphisms (SNPs), including a series of 5′-flanking region (5′-FR) SNPs that were highly associated with NT5E expression. This thiopurine cellular circulation and genes encoding the proteins involved, in particular NT5E, should be taken into account in future studies of thiopurine drug therapy and effect.
Materials and Methods
Chemicals.
6-MP, 6-TG, and 6-thioguanine ribonucleoside (TGrib) were purchased from Sigma-Aldrich (St. Louis, MO). 6-Thioguanine monophosphate (TGMP) was obtained from Jena Bioscience (Jena, Germany).
Cell Lines.
Lymphoblastoid cell lines from 58 European-American (EA), 53 African-American (AA), 60 Han Chinese-American (HCA), and 23 Centre d'Étude du Polymorphisme Humain EA unrelated subjects were purchased from the Coriell Institute (Camden, NJ). HEK293 parental cells and HEK293/4.3 cells stably transfected with ABCC4 were provided by Dr. Piet Borst, Netherlands Cancer Institute, Amsterdam, The Netherlands (Wielinga et al., 2002).
DNA Samples, NT5E Resequencing, Genotyping, and SNP Imputation.
DNA samples from 95 AA, 96 EA, and 96 HCA unrelated individuals were obtained from the Coriell Cell Repository (Camden, NJ). This DNA was used to PCR-amplify all NT5E exons, splice junctions, and ∼2000 base pairs of 5′-FR. PCR primer sequences and reaction conditions for those amplifications are listed in Supplemental Table 1. These amplicons were sequenced on both strands in the Mayo Clinic Genomics Core facility using BigDye terminator sequencing chemistry (Perkin-Elmer Life and Analytical Science, Waltham, MA) and an ABI 3700 DNA sequencer (Applied Biosystems, Foster City, CA). Any polymorphisms seen only once, as well as those with ambiguous chromatograms, were reamplified and resequenced. The sequence chromatograms were analyzed using Mutation Surveyor (version 2.2; Soft Genetics, State College, PA), with GenBank sequence NT007299.12 as the reference. All NT5E DNA sequence data were deposited in the National Institutes of Health PharmGKB database (accession number PA31804).
Additional polymorphism information for NT5E 5′- and 3′-FRs and for introns in the 287 DNA samples used to resequence the gene was obtained using Illumina 550K and 510S SNP arrays (Illumina Inc., San Diego, CA) as well as publicly available Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA) data. SNPs within 100 kb on either side of NT5E as well as those within NT5E introns were imputed using currently available genotype imputation methods (see subsequent description of imputation under Statistical Analysis). Finally, one imputed SNP, rs7772233, was genotyped in all of the DNA samples using a TaqMan 5′-allelic discrimination assay (Assays-by-Design; Applied Biosystems, Foster City, CA).
TPMT Genotyping.
Genotyping of the major functional TPMT nonsynonymous SNPs was performed using DNA from the 194 lymphoblastoid cell lines that were used to perform cytotoxicity studies with TaqMan 5′-allelic discrimination assays. In particular, the TPMT SNPs rs1800462 (*2), rs1800460 (*3B), and rs1142345 (*3C) were genotyped.
Expression Array and Quantitative RT-PCR.
Total RNA was extracted from the 194 lymphoblastoid cell lines that were used to perform cytotoxicity studies and was hybridized to Affymetrix U133 Plus 2.0 GeneChips, as described previously (Li et al., 2008). Expression data were normalized by GC Robust Multi-array Average (Wu et al., 2004). qRT-PCR was also performed for ABCC4 and NT5E using RNA from 16 randomly selected cell lines by using the QuantiFast SYBR Green RT-PCR kit (QIAGEN, Valencia, CA).
Cell Proliferation Assays.
Cell proliferation assays were performed with MTS reagent as described previously (Li et al., 2008). 6-MP and 6-TG were dissolved in dimethyl sulfoxide and were diluted with culture media to perform these experiments. To perform these cytotoxicity studies, lymphoblastoid and HEK293 cells were plated in triplicate at densities of 5 × 104 and 3 × 103 cells/well, respectively.
6-TG Metabolites.
Extracellular 6-TG metabolites were assayed using a reverse-phase ion-pairing HPLC assay modified from the procedure described by Wielinga et al. (2002). In particular, HEK293 and HEK293/4.3 cells stably transfected with ABCC4 cDNA were seeded in six-well plates at a density of 1 × 106 per well. Twenty-four hours later, the cells were transfected with empty vector or with an NT5E mammalian expression construct (Origene, Rockville, MD). After 48 h, the cells were treated with 5 μM 6-TG and were incubated at 37°C for 4 h. The incubation medium was collected and used for HPLC assay. Intracellular metabolites were assayed by HPLC as described previously (Dervieux and Boulieu, 1998a). Media used to study the possible involvement of SLC28 and SLC29 family members in TGrib transport was assayed as described by Jakobs et al. (1990).
Fluorescence Microscopy.
HEK293 and HEK293/4.3 cells were transfected with an NT5E/CD73 expression construct and grown on coverslips for 48 h. The cells were then fixed with 4% paraformaldehyde and stained for 30 min with an anti-NT5E monoclonal antibody (BD Biosciences, San Jose, CA), followed by incubation for 30 min with fluorescein isothiocyanate-conjugated anti-mouse secondary antibody (Invitrogen, Carlsbad, CA). The cells were visualized with a Zeiss LSM 510 confocal laser scanning microscope.
Statistical Analysis.
Cytotoxicity experiments were performed in triplicate, and averages of the three values were calculated. To determine IC50 values, a logistic function was fitted to the cytotoxicity data using the R package drc (http://cran.r-project.org/web/packages/drc/index.html). From the fitted logistic dose-response curve, the IC50 value for each cell line was computed. Normalized expression array data were then regressed on gender, race, and time since the Coriell Institute had acquired the cell line (dichotomized at 10 years). Residuals from this regression were then standardized by subtracting mean expression levels for individual probe sets and dividing by the standard deviation to derive a “standardized adjusted expression value.” IC50 values were rank-transformed using the Van der Waerden transformation and were adjusted in a fashion similar to that for the expression data. Pearson correlation coefficients were calculated for IC50 and expression values, followed by a Wald test of association. To adjust for multiple testing, q-values for all probes (Storey, 2003) and a permutation-based adjustment of the top p value, using 10,000 permutations, were computed. Percent variation in IC50 values explained by variation in mRNA expression was calculated on the basis of the coefficient of determination (R2) using a multiple regression model between IC50 values and individual probesets for each gene. Associations among metabolite concentrations, as well as associations of qRT-PCR with microarray data, were summarized using Spearman's rank correlation coefficient and were tested with a Wald test of ρ = 0. Population-specific linkage disequilibrium was analyzed, and haplotypes were determined using Haploview v4.2 (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview). Analysis of associations between SNPs and NT5E mRNA expression were performed with PLINK (http://pngu.mgh.harvard.edu/∼purcell/plink/), using the least-squares regression method. The Alibaba 2.1 (http://www.gene-regulation.com/pub/programs.html) and TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) programs were used to predict binding motifs that might be created or disrupted by NT5E SNPs. Last, imputation for untyped markers available from HapMap Phase II was completed using MACH v1.0 software (Li and Abecasis, 2006). For the imputation, reference population haplotypes were used from the HapMap populations, with CEU haplotypes for imputation of markers within the EA cell lines, CEU and YRI haplotypes for imputation of markers within the AA cell lines, and JPT and CHB haplotypes for imputation of markers within the HCA cell lines. Imputation was implemented using a two-step process in which the model parameters used for imputation were estimated in the first step (Settings: r = 150, −greedy) followed by the imputation of the expected genotype (or “dosage”) in the second step.
Results
6-MP and 6-TG Cytotoxicity.
IC50 values for the 194 human lymphoblastoid cell lines studied were determined using the MTS assay. Average IC50 values for these 194 cell lines were 1.77 ± 1.10 (mean ± S.D.) and 0.53 ± 0.33 μM for 6-MP and 6-TG, respectively. IC50 values for the three ethnic groups studied showed significant differences in mean IC50 values, with cells from HCA subjects being more sensitive to both drugs than were those obtained from EA and AA subjects (Supplemental Fig. 1). For example, compared with all of the other samples, IC50 values in the HCA samples differed from the others with p = 0.002 for 6-MP and p = 7.9 × 10−11 for 6-TG.
TPMT Genotyping.
Because the TPMT genotype is known to influence thiopurine drug response, we genotyped all cell lines used in the cytotoxicity studies for the most common TPMT variant nonsynonymous SNPs, *2 (A80P), *3B (A145T), and *3C (Y240C) to determine whether TPMT genotypes for these known functional variants might affect IC50 values for the two thiopurine drugs studied. Five of 194 human lymphoblastoid cell lines were heterozygous for TPMT*3A (a combination of the SNPs present in *3B and *3C), and six cell lines were heterozygous for TPMT*3C. As anticipated, the frequencies of variant alleles differed among ethnic groups, with TPMT*3A observed only in samples from EA subjects, whereas TPMT*3C was present in five samples from AAs and one from a HCA subject. However, these TPMT variant genotypes were not significantly associated with 6-MP or 6-TG IC50 values (6-MP, p = 0.95; 6-TG, p = 0.63). Therefore, subsequent analyses were not adjusted for TPMT genotype.
Thiopurine Drug Pathway-Based Expression Association Studies.
The thiopurine pathway includes genes known to be involved in thiopurine drug transport, metabolism, or metabolic activation (http://www.pharmgkb.org). We focused our analysis on those genes and performed a “pathway-based” association study. Eighteen genes targeted by 32 expression probesets on the Affymetrix U133 2.0 Plus GeneChip are known to participate in the thiopurine pathway. Therefore, as a first step in our analysis, we determined correlations between the expression of these 18 thiopurine pathway genes and IC50 values for 6-TG and 6-MP (Table 1). The expression of NT5E/CD73 showed the most significant association with 6-MP IC50 (p = 0.009) and the second most significant association with 6-TG IC50 (p = 0.010), whereas the expression of TPMT showed the most significant association with 6-TG IC50 (p = 0.003). Because IC50 values were significantly lower for cell lines from HCA subjects, we asked whether those subjects might have higher NT5E/CD73 expression and, thus, lower IC50 values, at least in part as a result of NT5E expression. The NT5E/CD73 1553995_a_at probeset did not display significantly elevated expression in HCA samples (p = 0.173), but probeset 203939_at did show significantly higher expression (p = 0.041). After adjustment for multiple testing, none of the p values for the top genes shown in Table 1 were statistically significant at the 0.05 level, with only TPMT remaining marginally significant for 6-TG (permutation p = 0.08).
TABLE 1.
Thiopurine cytotoxicity and “pathway” gene expression: Correlations between thiopurine pathway gene expression and IC50 values for 6-TG and 6-MP in 194 lymphoblastoid cell lines
Data for genes listed more than once represent results for different expression array probesets. Many genes had multiple probesets on the array. The data have been ordered by p values from lowest to highest.
| Gene | Chr | Probe Sets | 6-TG |
6-MP |
||
|---|---|---|---|---|---|---|
| p Value | Correlation | p Value | Spearman Correlation | |||
| TPMT | 6 | 203672_x_at | 0.003 | 0.211 | 0.248 | 0.083 |
| NT5E | 6 | 1553995_a_at | 0.010 | −0.186 | 0.009 | −0.188 |
| TPMT | 6 | 203671_at | 0.018 | 0.170 | 0.536 | 0.045 |
| NT5E | 6 | 203939_at | 0.025 | −0.161 | 0.016 | −0.174 |
| GART | 21 | 210005_at | 0.062 | −0.134 | 0.130 | −0.109 |
| AOX1 | 2 | 205083_at | 0.065 | −0.133 | 0.268 | −0.08 |
| GART | 21 | 217445_s_at | 0.075 | −0.128 | 0.957 | −0.004 |
| PPAT | 4 | 209434_s_at | 0.090 | −0.122 | 0.869 | −0.012 |
| ABCC4 | 13 | 203196_at | 0.081 | 0.126 | 0.364 | 0.066 |
| IMPDH1 | 7 | 204169_at | 0.115 | −0.114 | 0.091 | 0.122 |
| PRPS1 | X | 209440_at | 0.144 | −0.105 | 0.490 | 0.05 |
| HPRT1 | X | 202854_at | 0.127 | 0.110 | 0.328 | 0.071 |
| GART | 21 | 212379_at | 0.203 | −0.092 | 0.260 | 0.081 |
| ADK | 10 | 204119_s_at | 0.207 | −0.091 | 0.177 | −0.097 |
| ABCC5 | 3 | 226363_at | 0.308 | −0.074 | 0.930 | −0.006 |
| ADA | 20 | 204639_at | 0.383 | −0.063 | 0.483 | −0.051 |
| ADA | 20 | 216705_s_at | 0.384 | −0.063 | 0.494 | −0.049 |
| XDH | 2 | 210301_at | 0.433 | −0.057 | 0.920 | 0.007 |
| PPAT | 4 | 209433_s_at | 0.523 | −0.046 | 0.386 | −0.063 |
| GMPS | 3 | 214431_at | 0.499 | 0.0490 | 0.058 | 0.136 |
| IMPDH2 | 3 | 201892_s_at | 0.620 | −0.036 | 0.145 | 0.105 |
| AOX1 | 2 | 205082_s_at | 0.611 | −0.037 | 0.926 | 0.007 |
| ABCC4 | 13 | 1555039_a_at | 0.731 | 0.0250 | 0.864 | −0.012 |
| SLC29A1 | 6 | 201802_at | 0.604 | −0.037 | 0.581 | 0.039 |
| SLC29A1 | 6 | 201801_s_at | 0.697 | 0.028 | 0.454 | 0.054 |
| GART | 21 | 216990_at | 0.727 | 0.0250 | 0.165 | 0.1 |
| GART | 21 | 212378_at | 0.808 | −0.018 | 0.175 | 0.098 |
| ITPA | 20 | 209171_at | 0.761 | −0.022 | 0.662 | 0.032 |
| ADK | 10 | 204120_s_at | 0.829 | −0.016 | 0.284 | −0.077 |
| AHCY | 20 | 200903_s_at | 0.803 | −0.018 | 0.086 | 0.124 |
| ABCC4 | 13 | 1554918_a_at | 0.839 | 0.015 | 0.279 | −0.078 |
| ABCC5 | 3 | 209380_s_at | 0.937 | 0.006 | 0.096 | 0.12 |
Because enzymes and transporters within the thiopurine pathway are involved, in part, in regulating intracellular levels of active drug metabolites, 6-TGNs, we randomly selected 16 cell lines from among the 194 studied and used HPLC to measure intracellular 6-TGN levels after treatment with 6-MP or 6-TG. Concentrations of 6-TGNs in the majority of the cell lines treated with 6-MP were less than levels that could be measured accurately. However, levels of 6-TGNs could be determined after 6-TG treatment, and those concentrations were significantly correlated with expression array data for several of the pathway genes (Table 2). The expression of TPMT, NT5E/CD73, ABCC4/MRP4, and SLC29A1 showed the most significant associations with intracellular concentrations of 6-TGNs in these 16 cell lines (p = 0.0002, 0.0064, 0.0078, and 0.0100, respectively). Finally, we also performed quantitative RT-PCR to validate gene expression levels for NT5E/CD73 and ABCC4/MRP4 as determined by microarray analysis. mRNA levels determined by microarray analysis and qRT-PCR were significantly related for both genes (p < 0.05) (data not shown).
TABLE 2.
Correlations between thiopurine pathway gene expression and levels of 6-TGNs in 16 randomly selected cell lines after 6-TG treatment
Data for genes listed more than once represent results for different expression array probesets. Many genes had multiple probesets on the array. The data have been ordered by p values from lowest to highest.
| Gene | Chromosome | Probe Set | p Value | Spearman Correlation |
|---|---|---|---|---|
| TPMT | 6 | 203672_x_at | 0.0002 | −0.80 |
| NT5E | 6 | 203939_at | 0.0064 | 0.65 |
| ABCC4 | 13 | 203196_at | 0.0078 | −0.64 |
| SLC29A1 | 6 | 201802_at | 0.0100 | −0.61 |
| NT5E | 6 | 1553995_a_at | 0.0108 | 0.62 |
| ABCC5 | 3 | 209380_s_at | 0.0273 | −0.55 |
| GART | 21 | 212378_at | 0.0283 | −0.55 |
| PPAT | 4 | 209433_s_at | 0.0350 | −0.53 |
| ADK | 10 | 204119_s_at | 0.0441 | −0.51 |
| GART | 21 | 212379_at | 0.0602 | −0.48 |
| ABCC5 | 3 | 226363_at | 0.1130 | 0.41 |
| PPAT | 4 | 209434_s_at | 0.1290 | −0.40 |
| HPRT1 | X | 202854_at | 0.1577 | −0.37 |
| IMPDH1 | 7 | 204169_at | 0.2168 | 0.33 |
| TPMT | 6 | 203671_at | 0.2172 | −0.33 |
| GART | 21 | 217445_s_at | 0.2541 | −0.30 |
| ADK | 10 | 204120_s_at | 0.2589 | −0.30 |
| ABCC4 | 13 | 1554918_a_at | 0.2999 | −0.28 |
| ABCC4 | 13 | 1555039_a_at | 0.3622 | −0.24 |
| GART | 21 | 210005_at | 0.3743 | −0.24 |
| GART | 21 | 216990_at | 0.4035 | −0.22 |
| ADA | 20 | 216705_s_at | 0.5204 | 0.17 |
| XDH | 2 | 210301_at | 0.5276 | 0.17 |
| AOX1 | 2 | 205082_s_at | 0.5554 | 0.16 |
| SLC29A1 | 6 | 201801_s_at | 0.6900 | 0.11 |
| IMPDH2 | 3 | 201892_s_at | 0.7125 | 0.10 |
| AHCY | 20 | 200903_s_at | 0.7207 | −0.10 |
| AOX1 | 2 | 205083_at | 0.7969 | 0.07 |
| PRPS1 | X | 209440_at | 0.8541 | −0.05 |
| ITPA | 20 | 209171_at | 0.8968 | −0.04 |
| ADA | 20 | 204639_at | 0.9139 | 0.03 |
| GMPS | 3 | 214431_at | 0.9139 | 0.03 |
Thiopurine Cellular Circulation.
The ATP-binding cassette transporter ABCC4/MRP4 is localized to the plasma membrane and mediates the outward transport from cells of TGMP (Wielinga et al., 2002). Our study of 16 randomly selected cell lines had shown a negative correlation between ABCC4/MRP4 expression and intracellular 6-TGN concentrations (ρ = −0.64, p = 0.0078) (Table 2), indicating that elevated expression of ABCC4/MRP4 was associated with decreased intracellular 6-TGN concentrations. This result was consistent with previous reports that overexpression of ABCC4/MRP4 enhances transport of nucleotide monophosphates out of cells (Schuetz et al., 1999; Wielinga et al., 2002). In contrast, there was a positive correlation between the expression of NT5E/CD73 and intracellular 6-TGN concentrations. NT5E/CD73, ecto-5′-nucleotidase (5′-NT, EC 3.1.3.5), is an ecto-enzyme that is anchored to the outer plasma membrane by glycosyl-phosphatidylinositol (Zimmermann, 1992). This enzyme catalyzes the hydrolysis of extracellular ribonucleotide 5′-monophosphates to form their corresponding nucleosides (Zimmermann, 1992). Our results for the randomly selected cell lines showed a positive correlation between NT5E/CD73 expression and intracellular 6-TGN levels (ρ = 0.65, p = 0.0064), indicating that elevated expression of NT5E/CD73, an enzyme localized to the extracellular cell membrane, was associated with elevated intracellular 6-TGN levels, opposite in direction to the association of ABCC4/MRP4 with intracellular 6-TGN concentrations.
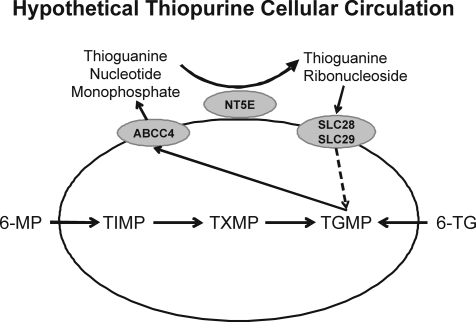
The functions and cellular localizations of ABCC4/MRP4 and NT5E/CD73, as well as the association of their expression with intracellular 6-TGN levels, suggested the possible existence of a “cellular thiopurine circulation” (Fig. 1). In this hypothetical model, ABCC4/MRP4 extrudes TGMP (Wielinga et al., 2002), a compound impermeable to cells that, as a result, would otherwise remain in the extracellular environment. NT5E/CD73 then catalyzes the dephosphorylation of TGMP to form TGrib. The final component(s) of this hypothetical cellular thiopurine cellular circulation would be members of the concentrative (SLC28) and equilibrative (SLC29) transporter families, membrane-bound proteins that transport nucleosides and nucleoside analogs into cells (Baldwin et al., 2004; Gray et al., 2004). Therefore, TGrib formed as a result of the dephosphorylation of TGMP could potentially reenter cells as a result of the action these transporters (Fig. 1).
Fig. 1.
Cellular thiopurine circulation. The figure shows schematically the “transport” of TGMP out of the cell by ABCC4, its hydrolysis by NT5E on the exterior of the cell to form thioguanine nucleosides, and the transport of those nucleosides back into the cell by SLC28/SLC29 nucleoside transporters. TXMP, thioxanthosine monophosphate.
HEK293 Cells and Thiopurine Cellular Circulation.
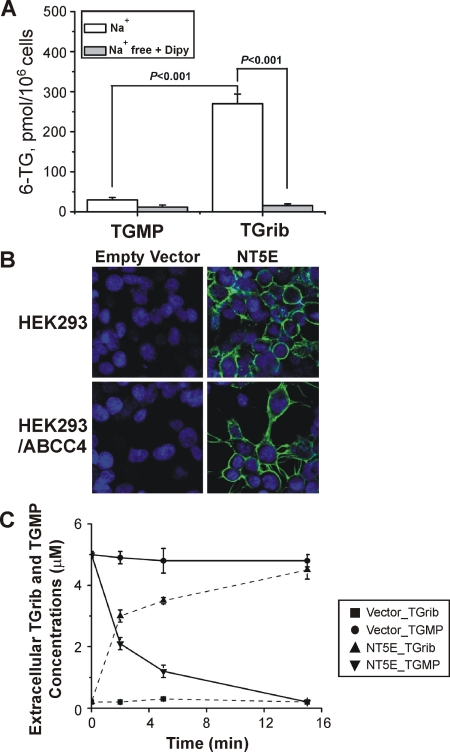
We next took advantage of the availability of HEK293 cells that have been stably transfected with ABCC4 (Wielinga et al., 2002) to test the hypothesis that a thiopurine cellular circulation might exist. We used HEK293 cells to perform these experiments because of the difficulty of transfecting the lymphoblastoid cell lines and because “parental” HEK293 cells do not express ABCC4/MRP4, but HEK293 cells stably transfected with this transporter are available and have been well characterized (Wielinga et al., 2002). As a first step, we measured levels of intracellular 6-TGNs after treating HEK293 cells with TGrib or TGMP. The assay involved determination of the concentration of free 6-TG after the hydrolysis of 6-TGNs with increased temperature and acidic pH, a commonly used approach (Lennard, 1987; Erdmann et al., 1990, 1991; Lennard and Singleton, 1992; Dervieux and Boulieu, 1998b; Mawatari et al., 1998). After exposure of the cells to 5 μM TGrib, the intracellular 6-TGN concentration was 250 pmol/106 cells (Fig. 2A). However, levels of intracellular 6-TGNs in cells treated with 5 μM TGMP were 10-fold lower than those for cells treated with TGrib, confirming that TGMP is unable to enter the cell (Fig. 2A). The trace amount of 6-TGNs in cells treated with TGMP might be due to TGrib contamination of the TGMP used to perform these experiments (approximately 2–3% based on HPLC assay).
Fig. 2.
A, permeability of HEK293 cells to TGMP and TGrib. HEK293 cells were treated with 5 μM TGMP or TGrib for 1 h in Na+-containing medium or in NMG+ medium that contained dipyridamole (Dipy, 10 μM), followed by HPLC assay of intracellular 6-TGN. B, HEK293 cells and HEK293 cells stably transfected with ABCC4/MRP4 stained for NT5E/CD73 before and after transfection with an NT5E/CD73 expression construct. C, extracellular TGMP and TGrib levels at different time points after incubation of the cells shown in B with 5 μM TGMP.
Members of the SLC28 and SLC29 transporter families mediate the transport of nucleosides and nucleoside analogs into cells (Baldwin et al., 2004; Gray et al., 2004). Down-regulation of expression or inhibition of the activity of these transporters reduces the uptake of thiopurines and decreases the accumulation of 6-TGNs in cells (Zaza et al., 2005; Fotoohi et al., 2006). Transport mediated by the SLC29 family members, SLC29A1/ENT1 and SLC29A2/ENT2, is inhibited by 10 μM dipyridamole (Ward et al., 2000). SLC28 family members, SLC28A1/CNT1, SLC28A2/CNT2, and SLC28A3/CNT3, transport nucleosides and nucleoside analogs by coupling with the transmembrane sodium gradient. Therefore, the activity of SLC28 family members is dramatically reduced in Na+-free media. To determine whether SLC28 and/or SLC29 family members might mediate TGrib uptake, we measured intracellular concentrations of 6-TGNs after incubating HEK293 cells with TGrib or TGMP in Na+-free medium that contained dipyridamole. Levels of 6-TGNs were 90% lower in cells treated with TGrib and dipyridamole in the absence of Na+ than those in cells treated with TGrib alone (Fig. 2A). The Affymetrix U133 2.0 Plus microarray assay of mRNA isolated from HEK293 cells confirmed that SLC29A1 is expressed in those cells (data not shown).
Therefore, although the HEK293 cells appeared to be impermeable to TGMP, uptake of TGrib did occur, apparently as a result of transport mediated by SLC29A1 (Fig. 2A). We next determined whether NT5E/CD73 was able to catalyze the dephosphorylation of extracellular TGMP to form TGrib. HEK293 cells and HEK293 cells stably transfected with ABCC4 were transfected with either an NT5E/CD73 expression construct or empty vector. Immunofluorescence showed the anticipated surface location of NT5E/CD73 in these cells (Fig. 2B). The time course of extracellular TGrib formation by these cells during incubation with TGMP is shown in Fig. 2C. In particular, after incubation of HEK293 cells that expressed NT5E/CD73 with 5 μM TGMP, TGMP levels decreased rapidly with a proportional rise in TGrib (Fig. 2C). In contrast to the rapid hydrolysis of TGMP outside of HEK293 cells that expressed NT5E/CD73, TGMP levels outside of HEK293 cells transfected with empty vector were unchanged over the same time interval (Fig. 2C). These results demonstrated that NT5E/CD73 on the extracellular membrane is able to catalyze the dephosphorylation of extracellular TGMP to form TGrib.
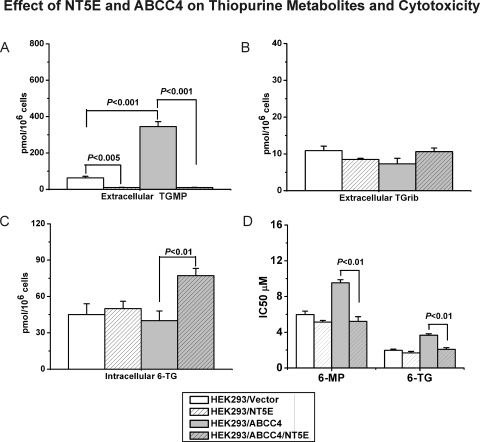
Previous studies showed that ABCC4/MRP4 was localized to the cell membrane and that TGMP excretion was increased in HEK293 cells stably transfected with ABCC4/MRP4 compared with the parental cell line (Wielinga et al., 2002). As described in the preceding paragraph, expression of NT5E/CD73 resulted in the surface localization of NT5E/CD73 in cells stably transfected with ABCC4/MRP4 (Fig. 2B). After exposure to 5 μM 6-TG, the excretion of TGMP was increased in cells stably transfected with ABCC4/MRP4, consistent with previous reports (Wielinga et al., 2002), whereas the expression of NT5E/CD73 dramatically reduced the extracellular concentration of TGMP (Fig. 3A). Under the same conditions, extracellular levels of TGrib did not change significantly among these cell lines (Fig. 3B). However, levels of intracellular 6-TGNs were significantly higher in cells expressing both ABCC4/MRP4 and NT5E/CD73 than in cells expressing only ABCC4/MRP4 or only NT5E (Fig. 3C), suggesting that TGMP exported by ABCC4/MRP4 might undergo dephosphorylation catalyzed by NT5E/CD73 to form TGrib that could then be “circulated” back into cells, ultimately resulting in elevated 6-TGN concentrations. Finally, cells expressing both ABCC4/MRP4 and NT5E/CD73 were also more sensitive to thiopurine drug-induced cytotoxicity, i.e., they had lower IC50 values than did cells expressing ABCC4/MRP4 alone (Fig. 3D). All of these observations were compatible with the hypothetical thiopurine cellular circulation model shown graphically in Fig. 1.
Fig. 3.
A–C, HEK293 cells stably transfected with ABCC4 cDNA and parental HEK293 cells were transfected with an NT5E/CD73 construct or with empty vector. Levels of extracellular TGMP and TGrib and intracellular 6-TGNs were determined after treatment with 5 μM 6-TG. D, cytotoxicity after incubation of the same cells with 5 μM 6-MP or 5 μM 6-TG.
The development of this “model” began with our study of the expression of thiopurine pathway genes and their association with thiopurine-induced cytotoxicity. Although the role of nucleotide and nucleoside transporters in thiopurine drug effect has been explored previously (Wielinga et al., 2002; Peng et al., 2008), the present experiments led to the conclusion that NT5E also plays a pivotal role. For example, variation in NT5E expression (probeset 1553995_a_at) explained 3.44 and 3.52% of the variation in IC50 values for 6-TG and 6-MP in the cell lines, respectively. Therefore, we next turned our attention to NT5E and resequenced this gene using DNA from 287 lymphoblastoid cell lines to make it possible to determine the possible relationship between sequence variation in NT5E and its expression, the phenotype that we had originally found to be correlated with thiopurine-induced cytotoxicity (Tables 1 and 2).
NT5E Resequencing and Functional Genomic Studies.
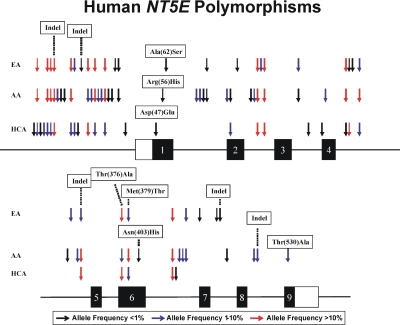
The areas of NT5E resequenced included exons, splice junctions, and a portion of the 5′-FR using 287 DNA samples from three ethnic groups. Sixty-eight polymorphisms, 63 SNPs, and 5 insertion/deletion events were identified (Table 3; Fig. 4). Forty-six of these polymorphisms were “novel,” i.e., not available in public database. Seven nonsynonymous (141G>C, N47E; 167G>A, R56H; 184G>T, A62S; 1126G>A, T376A; 1136T>C, M379T; 1207A>C, N403H; and 1587A>G, T530A) and 6 synonymous (1221C>G; 897C>T; 840C>T; 555A>G; 528C>T; and 342 A>G) SNPs were identified within the open reading frame (Table 3). Two of the nonsynonymous SNPs had minor allele frequencies (MAFs) greater than 1.0% in at least one ethnic group, whereas the remainder were observed in only a single DNA sample. All of the NT5E resequencing data listed in Table 3 were deposited in the National Institutes of Health PharmGKB database with accession number PA31004.
TABLE 3.
NT5E polymorphisms
NT5E polymorphism locations, alterations in nucleotide and amino acid sequences, and frequencies of polymorphisms observed are listed for each of the three ethnic groups studied. If a polymorphism had already been deposited in a public database (dbSNP or HapMap), an rs number is indicated. All other polymorphisms listed are unique to this study. Polymorphisms within exons are in italics. The numbering scheme for nucleotide positions is based on assignment of the (+1) position to the A in the translation initiation codon, with nucleotides 5′ to that position assigned negative numbers and those 3′ within the cDNA assigned positive numbers. IVS is intervening sequence (intron). Nucleotide positions in introns are numbered from the nearest splice site, with distances from 3′ splice junctions assigned positive numbers and distances from 5′ splice junctions assigned negative numbers.
| Location | Nucleotide | Sequence Change | Amino Acid | Frequency of Variant
Allele |
RefSNP Identification | ||
|---|---|---|---|---|---|---|---|
| AA | EA | HCA | |||||
| 5′-FR | −1855 | A→G | 0.000 | 0.000 | 0.005 | ||
| 5′-FR | −1698 | T→G | 0.279 | 0.137 | 0.068 | rs4463232 | |
| 5′-FR | −1570 | T→G | 0.000 | 0.000 | 0.005 | ||
| 5′-FR | −1551 | A→T | 0.000 | 0.000 | 0.010 | ||
| 5′-FR | −1516 | G→A | 0.374 | 0.137 | 0.068 | rs9450278 | |
| 5′-FR | −1440 | C→T | 0.371 | 0.137 | 0.063 | rs4599602 | |
| 5′-FR | −1431 | deletion of GTTAA | 0.231 | 0.574 | 0.255 | rs10565593 | |
| 5′-FR | −1407 | A→G | 0.032 | 0.000 | 0.000 | ||
| 5′-FR | −1368 | T→C | 0.005 | 0.000 | 0.000 | ||
| 5′-FR | −1364 | G→A | 0.005 | 0.000 | 0.000 | ||
| 5′-FR | −1343 | T→C | 0.371 | 0.137 | 0.064 | rs4458647 | |
| 5′-FR | −1271 | C→T | 0.000 | 0.011 | 0.011 | ||
| 5′-FR | −930 | A→C | 0.000 | 0.000 | 0.005 | ||
| 5′-FR | −824 | insertion of TC | 0.000 | 0.005 | 0.000 | ||
| 5′-FR | −715 | G→A | 0.068 | 0.223 | 0.421 | rs2065114 | |
| 5′-FR | −637 | C→T | 0.026 | 0.000 | 0.000 | ||
| 5′-FR | −634 | C→T | 0.374 | 0.137 | 0.064 | rs9450279 | |
| 5′-FR | −482 | G→A | 0.047 | 0.000 | 0.000 | rs6919558 | |
| 5′-FR | −417 | G→A | 0.021 | 0.000 | 0.000 | ||
| 5′-FR | −396 | G→C | 0.347 | 0.128 | 0.064 | rs2295890† | |
| 5′-FR | −220 | C→A | 0.005 | 0.000 | 0.000 | ||
| 5′-FR | −93 | C→T | 0.005 | 0.000 | 0.000 | ||
| 5′-FR | −85 | C→A | 0.000 | 0.005 | 0.000 | ||
| 5′-FR | −79 | C→T | 0.005 | 0.005 | 0.000 | ||
| 5′-FR | −77 | G→T | 0.000 | 0.000 | 0.005 | ||
| Exon 1 | 141 | C→G | D(47)E | 0.000 | 0.000 | 0.005 | |
| Exon 1 | 167 | G→A | R(56)H | 0.005 | 0.000 | 0.000 | |
| Exon 1 | 184 | G→T | A(62)S | 0.000 | 0.005 | 0.000 | |
| IVS 1 | 13 | C→T | 0.021 | 0.000 | 0.000 | ||
| IVS 1 | 103 | G→A | 0.032 | 0.000 | 0.000 | ||
| IVS 1 | −26 | C→G | 0.005 | 0.000 | 0.000 | ||
| IVS 1 | −14 | T→C | 0.005 | 0.005 | 0.000 | ||
| Exon 2 | 342 | A→G | 0.063 | 0.000 | 0.000 | rs35694460 | |
| Exon 2 | 528 | C→T | 0.005 | 0.000 | 0.063 | ||
| Exon 2 | 555 | A→G | 0.000 | 0.005 | 0.000 | ||
| IVS 2 | 37 | T→C | 0.005 | 0.000 | 0.000 | ||
| IVS 2 | 53 | C→T | 0.036 | 0.000 | 0.000 | ||
| IVS 2 | −223 | A→C | 0.271 | 0.609 | 0.245 | rs10944728 | |
| IVS 2 | −194 | T→C | 0.000 | 0.016 | 0.000 | ||
| IVS 2 | −29 | A→G | 0.271 | 0.609 | 0.247 | rs10944129 | |
| IVS 3 | −225 | G→C | 0.005 | 0.036 | 0.000 | ||
| IVS 3 | −109 | G→A | 0.000 | 0.000 | 0.005 | ||
| Exon 4 | 840 | C→T | 0.000 | 0.000 | 0.005 | ||
| Exon 4 | 897 | C→T | 0.073 | 0.000 | 0.000 | rs35478984 | |
| IVS 4 | 70 | T→A | 0.000 | 0.000 | 0.005 | ||
| IVS 4 | 148 | G→A | 0.401 | 0.261 | 0.693 | rs9450282 | |
| IVS 4 | −350 | C→T | 0.000 | 0.005 | 0.000 | ||
| IVS 4 | −209 | T→C | 0.000 | 0.005 | 0.000 | ||
| IVS 4 | −63 | C→G | 0.375 | 0.094 | 0.073 | rs3812139† | |
| IVS 5 | 87 | G→A | 0.005 | 0.000 | 0.000 | ||
| IVS 5 | 160 | C→G | 0.000 | 0.016 | 0.000 | ||
| IVS 5 | −117 | A→G | 0.011 | 0.000 | 0.000 | ||
| IVS 5 | I5m46 | deletion of GAAG | 0.158 | 0.063 | 0.255 | rs60666442 | |
| Exon 6 | 1126 | G→A | T(376)A | 0.063 | 0.226 | 0.427 | rs2229523 |
| Exon 6 | 1136 | T→C | M(379)T | 0.395 | 0.053 | 0.073 | rs2229524 |
| Exon 6 | 1207 | A→C | N(403)H | 0.005 | 0.000 | 0.000 | |
| IVS 6 | 12 | G→C | 0.379 | 0.684 | 0.500 | rs9450284 | |
| IVS 6 | 33 | A→T | 0.000 | 0.000 | 0.005 | ||
| IVS 6 | −371 | C→T | 0.081 | 0.000 | 0.000 | ||
| IVS 6 | −79 | A→T | 0.048 | 0.000 | 0.000 | ||
| IVS 6 | −77 | C→T | 0.011 | 0.026 | 0.000 | rs41271619 | |
| Exon 7 | 1221 | C→G | 0.000 | 0.005 | 0.000 | ||
| IVS 7 | 11 | C→T | 0.000 | 0.005 | 0.000 | ||
| IVS 7 | 12 | deletion of ATCCTGTAGG | 0.000 | 0.005 | 0.000 | ||
| IVS 7 | −81 | A→G | 0.005 | 0.000 | 0.000 | ||
| IVS 8 | −218 | C→G | 0.097 | 0.000 | 0.000 | rs58285927 | |
| IVS 8 | −101 | deletion of T | 0.026 | 0.000 | 0.000 | ||
| Exon 9 | 1587 | A→G | T(530)A | 0.010 | 0.000 | 0.000 | rs34160251 |
Fig. 4.
Human NT5E polymorphisms. Arrows indicate the locations of polymorphisms, with different colors indicating minor allele frequencies. Black rectangles represent portions of exons encoding the open reading frame. Open rectangles represent portions of exons encoding 5′- and 3′-untranslated regions. Indel, insertion/deletion event.
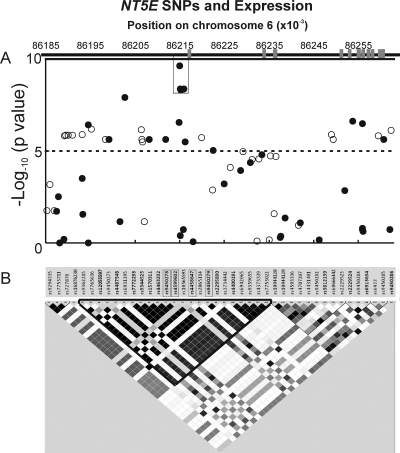
MAFs for NT5E SNPs, both those observed during the resequencing experiments and those present on GWAS platforms spanning a region ∼270 kb in length that included the gene (chr6:86,100,000–86,370,000), are listed in Supplemental Table 2, A and B. Finally, additional SNPs within this area were imputed (as described under Statistical Analysis) based on Hapmap Phase II data. All SNPs in this region with MAFs >0.05 that were observed during NT5E resequencing either were genotyped using the Illumina and Affymetrix genome-wide SNP platforms or were imputed and used to determine population-specific linkage disequilibrium (Fig. 5). Those data were also used to determine whether any of the SNPs might be associated with basal NT5E expression, the original phenotype that we had correlated with thiopurine-induced cytotoxicity (Table 1).
Fig. 5.
Correlation of SNPs with NT5E expression in a portion of chromosome 6 that includes NT5E is shown, aligned with the gene structure at the top (A) and an LD map of SNPs genotyped or imputed in the same region for EA subjects (B).
Association of SNPs with NT5E Expression.
Data for the association of SNPs in and surrounding NT5E with basal expression of the gene in the lymphoblastoid cells used to perform the cytotoxicity studies are depicted graphically in Fig. 5A and are listed in Supplemental Table 2. Many SNPs across a 100-kb region that included NT5E showed highly significant correlations (p ≤ 10−6) with NT5E expression (Supplemental Table 3), with the most significant SNPs mapping to the 5′-FR (Fig. 5A). The rs9450278 SNP displayed the lowest p value (p = 2.4 × 10−10, R = −0.376). This SNP was in linkage disequilibrium with rs9450279, rs4599602, rs4458647, and rs2295890 and an imputed SNP, rs7772233 (r2 > 0.97) in AA and EA subjects. We will refer to this group of SNPs as “LD block A.” This LD block was also present, but not as clearly defined, in DNA from HCA subjects (r2 = 0.82–0.84). Because the rs7772233 SNP was imputed, we also genotyped this polymorphism to obtain the “true” genotype, not merely an estimate of the genotype and MAF. The negative correlations of these six SNPs with NT5E expression (r = −0.283 to −0.376) indicated that as the number of minor alleles increased we observed a decrease in NT5E expression.
Discussion
Thiopurines are a major component of the drug therapy of childhood ALL (Lennard, 1992). Thiopurines are also immunosuppressants that are used to treat patients undergoing solid organ transplantation and patients with autoimmune disorders (Lennard, 1992). There are large individual variations in response to these drugs, both in terms of efficacy and the occurrence of adverse drug reactions. The discovery three decades ago of the TPMT genetic polymorphism, initially at the biochemical genetic level (Weinshilboum and Sladek, 1980), and subsequently, after the cloning of TPMT cDNA (Honchel et al., 1993) and gene (Szumlanski et al., 1996), at the level of individual SNPs (Szumlanski et al., 1996; Tai et al., 1996), provided one of the first examples of the potential clinical value of pharmacogenetics. However, even though TPMT is a major factor responsible for clinical variation in response to thiopurines, mainly through the effects of nonsynonymous SNPs that result in protein misfolding and accelerated degradation of TPMT protein (Lennard et al., 1983, 1987; Wang et al., 2003, 2005; Wang and Weinshilboum, 2006; Li et al., 2008), TPMT does not explain all of the variation in thiopurine response.
In the present study, we set out to use a data-rich Human Variation Panel lymphoblastoid cell line model system in an attempt to move beyond TPMT to other genes that might contribute to individual variation in thiopurine drug response. The Human Variation Panel cell lines, although not tumor-derived, have demonstrated their value for the identification of novel candidate genes and molecular mechanisms underlying variation in drug effect. A particularly striking example is provided by their successful application to study variation in response to another antineoplastic drug, the cytidine analog gemcitabine (Li et al., 2008; Pei et al., 2009). We focused our experiments on variation in the expression of genes encoding proteins in the known thiopurine pathway and found that TPMT expression was highly correlated with 6-TG but not 6-MP cytotoxicity in this in vitro cell line-based model system. However, that analysis also served to highlight the potential importance of variation in NT5E expression for variation in thiopurine-induced cytotoxicity, a possibility that was first raised, purely on “theoretical” grounds, more than 20 years ago (Pieters et al., 1987). The observation that ABCC4 expression also contributed to cytotoxicity suggested that a thiopurine cellular circulation might exist, a possibility that we verified during a series of experiments performed with HEK293 cells.
We then focused our attention on NT5E, resequencing the gene encoding this protein and discovering SNPs that were highly associated with variation in NT5E expression (Fig. 5). The conversion of 6-MP and 6-TG to 6-TGNs is required for the primary antineoplastic effect of thiopurines. These drugs are converted, respectively, into thioinosine monophosphate (TIMP) and TGMP by hypoxanthine-guanine phosphoribosyl transferase. Both TIMP and TGMP can then be methylated by TPMT to form methyl-TIMP and methyl-TGMP, respectively. Methyl-TIMP is an efficient inhibitor of de novo purine biosynthesis (Tay et al., 1969; Hill and Bennett, 1969; Dervieux et al., 2001). Those observations suggest that two distinct mechanisms, incorporation of 6-TGNs into DNA and RNA and inhibition of de novo purine biosynthesis, contribute to 6-MP-dependent cytotoxicity. Therefore, any mechanisms that might modulate intracellular concentration of 6-TGNs could potentially influence thiopurine drug response.
In the present study, we found that variation in the expression of TPMT was significantly associated with 6-TG but not with 6-MP IC50 values (Table 1). However, significant correlations were also observed between expression levels of other thiopurine pathway genes, particularly NT5E and intracellular levels of 6-TGNs after 6-TG exposure (Table 2). It is obvious that variation in mRNA expression does not always correlate directly with variation in protein levels or enzyme activity. In addition, our observation of a positive association in cytotoxicity with NT5E expression might initially seem puzzling because NT5E is an ecto-nucleotidase located on the exterior of the cell membrane. However, we also observed that ABCC4/MRP4 expression was correlated with intracellular 6-TGN levels after incubation of cells with 6-TG. We then confirmed that ABCC4/MRP4 mediated the cellular efflux of TGMP, a compound that is impermeable to cells (Figs. 2 and 3). As a result, ABCC4/MRP4 may contribute to the cellular effects of thiopurine drugs, as suggested previously by both clinical and cell system-based data (Fig. 3D) (Wielinga et al., 2002). When this action of ABCC4 is linked to the activity of NT5E, the relationship of NT5E to thiopurine cytotoxicity begins to become clearer and supports the thiopurine cellular circulation depicted graphically in Fig. 1.
It should be emphasized that the in vivo situation may not be identical with the process observed in these cell line model systems. The transport of TGMP by ABCC4 out of the cell, the dephosphorylation of TGMP by NT5E, and the transport of TGrib back into the cell would all be expected to be tissue-specific processes and not all cells will express all components of the “circulation” shown in Fig. 1. However, our thiopurine pathway studies have provided insight into possible mechanisms modulating intracellular 6-TGN concentrations. Central to the proposed thiopurine circulation is NT5E. Therefore, we also resequenced NT5E to determine whether polymorphisms in or around the gene might be related to variation in its expression. A series of SNPs in the 5′-FR of NT5E were significantly correlated with expression (Fig. 5). Those SNPs could explain 12 to 14% of the total variation in NT5E expression, and, as a result, they should be included in future genotyping studies of thiopurine drug effect. Variation in expression may not correlate with either level of enzyme protein or activity. The results reported here for a cell line-based model system complement and extend those reported during previous studies of the association of gene expression profiles with intracellular 6-TGN concentration in ALL cells (Zaza et al., 2005).
Finally, it should be emphasized once again that lymphoblastoid cells are not tumor cell lines. However, although the tumor genome is important for variation in response to antineoplastic drugs, polymorphisms of germline DNA have repeatedly been shown to play a critical role in variation in antineoplastic drug response (Weinshilboum and Wang, 2006). The cell line-based model system used in our studies has already demonstrated its power to identify novel candidate genes and mechanisms that contribute to individual variation in antineoplastic drug response (Huang et al., 2008; Li et al., 2008; Pei et al., 2009). The results reported here indicate that future studies of thiopurine drug effect should consider the thiopurine cellular circulation that we observed and, in particular, the possible role of NT5E in variation in response to this important class of drugs.
Supplementary Material
Acknowledgments.
We thank Professor Piet Borst, Netherlands Cancer Institute, Amsterdam, The Netherlands, for his generous gift of the HEK293 cells stably transfected with human ABCC4 cDNA. We thank Dr. Lynne Lennard, University of Sheffield, Sheffield, UK, for her valuable suggestions and Luanne Wussow for her assistance with this manuscript.
This study was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM28157, U01-GM61388 (The Pharmacogenomics Research Network)]; the National Institutes of Health National Cancer Institute [Grants R01-CA132780, R01-CA138461, K22-CA130828]; an American Society for Pharmacology and Experimental Therapeutics Astellas Award; and a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.035220.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- 6-MP
- 6-mercaptopurine
- 6-TG
- 6-thioguanine
- ALL
- acute lymphoblastic leukemia
- 6-TGN
- 6-thioguanine nucleotide
- RBC
- red blood cell
- TPMT
- thiopurine S-methyltransferase
- ABCC4/MRP4
- multidrug resistance protein 4
- NT5E
- ecto-5′-nucleotidase
- SNP
- single nucleotide polymorphism
- FR
- flanking region
- TGrib
- 6-thioguanine ribonucleoside
- TGMP
- 6-thioguanine monophosphate
- EA
- European-American
- AA
- African-American
- HCA
- Han Chinese-American
- PCR
- polymerase chain reaction
- kb
- kilobase
- RT
- reverse transcriptase
- q
- quantitative
- HPLC
- high-performance liquid chromatography
- HEK
- human embryonic kidney
- SLC
- solute carrier
- MAF
- minor allele frequency
- LD
- linkage disequilibrium
- TIMP
- thioinosine monophosphate.
References
- Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. (2004) The equilibrative nucleoside transporter family, SLC29. Pflugers Arch 447:735–743 [DOI] [PubMed] [Google Scholar]
- Dervieux T, Blanco JG, Krynetski EY, Vanin EF, Roussel MF, Relling MV. (2001) Differing contribution of thiopurine methyltransferase to mercaptopurine versus thioguanine effects in human leukemic cells. Cancer Res 61:5810–5816 [PubMed] [Google Scholar]
- Dervieux T, Boulieu R. (1998a) Identification of 6-methylmercaptopurine derivative formed during acid hydrolysis of thiopurine nucleotides in erythrocytes, using liquid chromatography-mass spectrometry, infrared spectroscopy, and nuclear magnetic resonance assay. Clin Chem 44:2511–2515 [PubMed] [Google Scholar]
- Dervieux T, Boulieu R. (1998b) Simultaneous determination of 6-thioguanine and methyl 6-mercaptopurine nucleotides of azathioprine in red blood cells by HPLC. Clin Chem 44:551–555 [PubMed] [Google Scholar]
- Erdmann GR, France LA, Bostrom BC, Canafax DM. (1990) A reversed phase high performance liquid chromatography approach in determining total red blood cell concentrations of 6-thioguanine, 6-mercaptopurine, methylthioguanine, and methylmercaptopurine in a patient receiving thiopurine therapy. Biomed Chromatogr 4:47–51 [DOI] [PubMed] [Google Scholar]
- Erdmann GR, Steury JC, Carleton BC, Stafford RJ, Bostrom BC, Canafax DM. (1991) Reversed-phase high-performance liquid chromatographic approach to determine total lymphocyte concentrations of 6-thioguanine, methylmercaptopurine and methylthioguanine in humans. J Chromatogr 571:149–156 [DOI] [PubMed] [Google Scholar]
- Fotoohi AK, Lindqvist M, Peterson C, Albertioni F. (2006) Involvement of the concentrative nucleoside transporter 3 and equilibrative nucleoside transporter 2 in the resistance of T-lymphoblastic cell lines to thiopurines. Biochem Biophys Res Commun 343:208–215 [DOI] [PubMed] [Google Scholar]
- Gearry RB, Barclay ML, Burt MJ, Collett JA, Chapman BA, Roberts RL, Kennedy MA. (2003) Thiopurine S-methyltransferase (TPMT) genotype does not predict adverse drug reactions to thiopurine drugs in patients with inflammatory bowel disease. Aliment Pharmacol Ther 18:395–400 [DOI] [PubMed] [Google Scholar]
- Gray JH, Owen RP, Giacomini KM. (2004) The concentrative nucleoside transporter family, SLC28. Pflugers Arch 447:728–734 [DOI] [PubMed] [Google Scholar]
- Hill DL, Bennett LL., Jr (1969) Purification and properties of 5-phosphoribosyl pyrophosphate amidotransferase from adenocarcinoma 755 cells. Biochemistry 8:122–130 [DOI] [PubMed] [Google Scholar]
- Honchel R, Aksoy IA, Szumlanski C, Wood TC, Otterness DM, Wieben ED, Weinshilboum RM. (1993) Human thiopurine methyltransferase: molecular cloning and expression of T84 colon carcinoma cell cDNA. Mol Pharmacol 43:878–887 [PubMed] [Google Scholar]
- Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. (2007) A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci USA 104:9758–9763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Kistner EO, Bleibel WK, Delaney SM, Fackenthal DL, Das S, Dolan ME. (2008) Genetic variants contributing to daunorubicin-induced cytotoxicity. Cancer Res 68:3161–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs ES, Van Os-Corby DJ, Paterson AR. (1990) Expression of sodium-linked nucleoside transport activity in monolayer cultures of IEC-6 intestinal epithelial cells. J Biol Chem 265:22210–22216 [PubMed] [Google Scholar]
- Krishnamurthy P, Schwab M, Takenaka K, Nachagari D, Morgan J, Leslie M, Du W, Boyd K, Cheok M, Nakauchi H, et al. (2008) Transporter-mediated protection against thiopurine-induced hematopoietic toxicity. Cancer Res 68:4983–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard L. (1987) Assay of 6-thioinosinic acid and 6-thioguanine nucleotides, active metabolites of 6-mercaptopurine, in human red blood cells. J Chromatogr 423:169–178 [DOI] [PubMed] [Google Scholar]
- Lennard L. (1992) The clinical pharmacology of 6-mercaptopurine. Eur J Clin Pharmacol 43:329–339 [DOI] [PubMed] [Google Scholar]
- Lennard L, Rees CA, Lilleyman JS, Maddocks JL. (1983) Childhood leukaemia: a relationship between intracellular 6-mercaptopurine metabolites and neutropenia. Br J Clin Pharmacol 16:359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard L, Singleton HJ. (1992) High-performance liquid chromatographic assay of the methyl and nucleotide metabolites of 6-mercaptopurine: quantitation of red blood cell 6-thioguanine nucleotide, 6-thioinosinic acid and 6-methylmercaptopurine metabolites in a single sample. J Chromatogr 583:83–90 [DOI] [PubMed] [Google Scholar]
- Lennard L, Van Loon JA, Lilleyman JS, Weinshilboum RM. (1987) Thiopurine pharmacogenetics in leukemia: correlation of erythrocyte thiopurine methyltransferase activity and 6-thioguanine nucleotide concentrations. Clin Pharmacol Ther 41:18–25 [DOI] [PubMed] [Google Scholar]
- Li L, Abecasis GR. (2006) Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet S79:2290 [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, Hildebrandt M, Ames M, Schaid D, Wang L. (2008) Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res 68:7050–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawatari H, Kato Y, Nishimura S, Sakura N, Ueda K. (1998) Reversed-phase high-performance liquid chromatographic assay method for quantitating 6-mercaptopurine and its methylated and non-methylated metabolites in a single sample. J Chromatogr B Biomed Sci Appl 716:392–396 [DOI] [PubMed] [Google Scholar]
- Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. (2009) FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng XX, Shi Z, Damaraju VL, Huang XC, Kruh GD, Wu HC, Zhou Y, Tiwari A, Fu L, Cass CE, et al. (2008) Up-regulation of MRP4 and down-regulation of influx transporters in human leukemic cells with acquired resistance to 6-mercaptopurine. Leuk Res 32:799–809 [DOI] [PubMed] [Google Scholar]
- Pieters R, Huismans DR, Veerman AJ. (1987) Are children with lymphoblastic leukaemia resistant to 6-mercaptopurine because of 5′-nucleotidase? Lancet 2:1471. [DOI] [PubMed] [Google Scholar]
- Schuetz JD, Connelly MC, Sun D, Paibir SG, Flynn PM, Srinivas RV, Kumar A, Fridland A. (1999) MRP4: a previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat Med 5:1048–1051 [DOI] [PubMed] [Google Scholar]
- Storey J. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 31:2013–2035 [Google Scholar]
- Szumlanski C, Otterness D, Her C, Lee D, Brandriff B, Kelsell D, Spurr N, Lennard L, Wieben E, Weinshilboum R. (1996) Thiopurine methyltransferase pharmacogenetics: human gene cloning and characterization of a common polymorphism. DNA Cell Biol 15:17–30 [DOI] [PubMed] [Google Scholar]
- Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, Evans WE. (1996) Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet 58:694–702 [PMC free article] [PubMed] [Google Scholar]
- Tay BS, Lilley RM, Murray AW, Atkinson MR. (1969) Inhibition of phosphoribosyl pyrophosphate amidotransferase from Ehrlich ascites-tumour cells by thiopurine nucleotides. Biochem Pharmacol 18:936–938 [DOI] [PubMed] [Google Scholar]
- Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM. (2005) Human thiopurine S-methyltransferase pharmacogenetics: variant allozyme misfolding and aggresome formation. Proc Natl Acad Sci USA 102:9394–9399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sullivan W, Toft D, Weinshilboum R. (2003) Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics 13:555–564 [DOI] [PubMed] [Google Scholar]
- Wang L, Weinshilboum R. (2006) Thiopurine S-methyltransferase pharmacogenetics: insights, challenges and future directions. Oncogene 25:1629–1638 [DOI] [PubMed] [Google Scholar]
- Ward JL, Sherali A, Mo ZP, Tse CM. (2000) Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J Biol Chem 275:8375–8381 [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Sladek SL. (1980) Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet 32:651–662 [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum RM, Wang L. (2006) Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genomics Hum Genet 7:223–245 [DOI] [PubMed] [Google Scholar]
- Wielinga PR, Reid G, Challa EE, van der Heijden I, van Deemter L, de Haas M, Mol C, Kuil AJ, Groeneveld E, Schuetz JD, et al. (2002) Thiopurine metabolism and identification of the thiopurine metabolites transported by MRP4 and MRP5 overexpressed in human embryonic kidney cells. Mol Pharmacol 62:1321–1331 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99:909–917 [Google Scholar]
- Zaza G, Cheok M, Yang W, Panetta JC, Pui CH, Relling MV, Evans WE. (2005) Gene expression and thioguanine nucleotide disposition in acute lymphoblastic leukemia after in vivo mercaptopurine treatment. Blood 106:1778–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H. (1992) 5′-Nucleotidase: molecular structure and functional aspects. Biochem J 285 (Pt 2):345–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.