Abstract
Kainate receptors (KARs) modulate synaptic transmission and plasticity, and their dysfunction has been linked to several disease states such as epilepsy and chronic pain. KARs are tetramers formed from five different subunits. GluK1–3 are low affinity kainate binding subunits, whereas GluK4/5 bind kainate with high affinity. A number of these subunits can be present in any given cell type, and different combinations of subunits confer different properties to KARs. Here we report the characterization of a new GluK1 subunit-selective radiolabeled antagonist (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxythiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione ([3H]UBP310) using human recombinant KARs. [3H]UBP310 binds to GluK1 with low nanomolar affinity (KD = 21 ± 7 nM) but shows no specific binding to GluK2. However, [3H]UBP310 also binds to GluK3 (KD = 0.65 ± 0.19 μM) but with ∼30-fold lower affinity than that observed for GluK1. Competition [3H]UBP310 binding experiments on GluK1 revealed the same rank order of affinity of known GluK1-selective ligands as reported previously in functional assays. Nonconserved residues in GluK1–3 adjudged in modeling studies to be important in determining the GluK1 selectivity of UBP310 were point-mutated to switch residues between subunits. None of the mutations altered the expression or trafficking of KAR subunits. Whereas GluK1-T503A mutation diminished [3H]UBP310 binding, GluK2-A487T mutation rescued it. Likewise, whereas GluK1-N705S/S706N mutation decreased, GluK3-N691S mutation increased [3H]UBP310 binding activity. These data show that Ala487 in GluK2 and Asn691 in GluK3 are important determinants in reducing the affinity of UBP310 for these subunits. Insights from these modeling and point mutation studies will aid the development of new subunit-selective KAR antagonists.
Introduction
Kainate receptors (KARs) are members of the ionotropic glutamate receptor family, which also include the (S)-2-amino-3-hydroxy-5-methyl-4-isoxazolepropanoic acid (AMPA) and N-methyl-d-aspartate receptors (Lerma, 2006; Jane et al., 2009). KARs modulate neurotransmitter release presynaptically, and they contribute to the slow component of the EPSC and inhibit K+ channels (metabotropic action) (Lerma, 2006) postsynaptically. There are five KAR subunits, named GluK1–5 (formerly GluR5–7 and KA-1, KA-2) (Bettler et al., 1990, 1992; Egebjerg et al., 1991; Jane et al., 2009). The development and functional characterization of novel potent and selective compounds is essential for understanding the contribution of KAR subtypes in neuronal processes and for the identification of new drug targets. For instance, GluK1-selective KAR antagonists have been used to investigate the involvement of GluK1 in a number of neurological disorders such as epilepsy, chronic pain, migraine, anxiety, and neurodegeneration (Jane et al., 2009). We have reported previously that (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxythiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione (UBP310) is a potent antagonist of GluK1-containing KARs at low nanomolar concentrations with high selectivity for GluK1 versus GluK2 and GluK3 (Dolman et al., 2007; Dargan et al., 2009). However, a recent study raised the possibility that UBP310 also potently antagonizes recombinant homomeric GluK3 receptors (Perrais et al., 2009), although the affinity of this ligand for GluK1 was not characterized in this study, and so the selectivity for GluK1 over GluK3 could not be estimated.
The ligand binding domain (LBD) of KARs is formed by two segments (S1 and S2) that flank the pore domain (Mayer, 2005; Jane et al., 2009). The LBD can be recombinantly expressed and purified as a soluble protein, which has enabled the determination of X-ray crystal structures for KAR ligand binding sites (Mayer, 2005). The crystal structure of the antagonist UBP310 bound to the GluK1 LBD has been determined (Mayer et al., 2006). Whereas molecular modeling of ligand-KAR interactions has been used to design more potent and subunit-selective antagonists (Dolman et al., 2007; Dargan et al., 2009; Jane et al., 2009), the involvement of individual amino acid residues in the subunit-selective binding of antagonists to KARs has not been established experimentally. In addition, a complication in the drug design process is that the degree of opening of the LBD of GluK1, the conformation of flexible side chains of residues in the LBD, and the position of water molecules in the binding site can vary with the structure of the interacting ligand (Mayer et al., 2006; Alushin et al., 2010), and so information regarding the involvement of nonconserved residues in ligand binding from site-directed mutagenesis studies would facilitate the design of KAR subunit-selective antagonists. Site-directed mutagenesis of key amino acid residues likely to be involved in ligand binding is often used to confirm predictions made by molecular modeling. However, it has been reported that mutations in the LBDs of KARs that disrupt interaction with glutamate or interfere with desensitization lead to endoplasmic reticulum (ER) retention of subunit proteins (Mah et al., 2005; Valluru et al., 2005; Fleck, 2006; Gill et al., 2009). Therefore, potential changes in the biosynthesis and trafficking of binding site mutant subunits need to be investigated carefully when site-directed mutagenesis is used for the mapping of ligand binding sites in KAR subunits.
In this study, we describe the binding characteristics of a novel radiolabeled KAR antagonist [3H]UBP310 using human recombinant KAR subtypes. To date, no high-resolution structures of the LBDs of GluK2 or GluK3 in complex with antagonists have been described. Herein we describe homology models of the LBDs of GluK2 and GluK3 and their use to investigate the involvement of certain nonconserved residues in the LBDs of KARs in subunit-selective interactions with antagonists. We have used site-directed mutagenesis and ligand binding assays with the newly developed radioligand [3H]UBP310 to investigate hypotheses generated as a result of our modeling studies.
Materials and Methods
Synthesis of [3H]UBP310.
The precursor for radiolabeling, (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxy-4,5-dibromothiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione (UBP315), was synthesized by the method published previously (Dolman et al., 2007). Before attempting tritiation of the ligand, we established that it was possible to replace the bromo groups on the thiophene ring with hydrogen atoms by stirring an aqueous solution of the disodium salt of UBP315 and a 10% palladium on carbon catalyst under an atmosphere of hydrogen for 24 h. A 1H NMR spectrum of the product showed complete replacement of the bromo groups by hydrogen atoms. The same conditions were used by GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK) to conduct the tritiation of UBP315. [3H]UBP310 was purified by high-performance liquid chromatography using a Prodigy ODS(2) column (Phenomenex, Torrance, CA) and an eluent of 0.1% trifluoroacetic acid in water with a gradient of 10 to 90% of 0.1% trifluoroacetic acid in acetonitrile. Detection was by UV at a wavelength of 270 nm.
Analysis of Ligand Binding Sites.
The X-ray crystal structure of UBP310 in complex with the LBD of rat GluK1 (Protein Data Bank identification number 2F34) was used to build homology models of the LBDs of GluK2 and GluK3 using SWISS-MODEL (Peitsch, 1995; Arnold et al., 2006; Kiefer et al., 2009). The structure of UBP310 was built with Maestro, a module of the Schrödinger molecular modeling suite (Schrödinger LLC, New York, NY), and energy was minimized using the MMF94s force field in Macromodel. Docking of UBP310 into the homology models of the LBDs of GluK2 and GluK3 was carried out using the Induced Fit workflow (Schrödinger Suite 2009 Induced Fit Docking protocol; Glide version 5.5, Prime version 2.1; both from Schrödinger LLC) within Maestro, which carries out an initial docking using Glide, followed by optimization with Prime and then a more precise Glide docking in XP mode. The grid for Glide docking into GluK2 was calculated using the following residues: Tyr457, Ala487, Arg492, Val654, Gly657, Ala658, Asn690, Thr709, and Thr710 (residue numbers exclude the signal peptide). The corresponding residues in the LBD of GluK3 were used to set up the grid for Glide when docking UBP310 into this subunit. The following constraints were used for GluK2: NHs of Arg492 and Ala487 and C=O of Pro485 (residues corresponding to these were selected for hydrogen bonding constraints when docking into the LBD of GluK3). The best poses from ligand docking studies and the X-ray crystal structure of the complex of the GluK1 LBD with UBP310 were examined to select potential nonconserved residues that may determine selectivity of UBP310 for GluK1. Ligand poses shown in Fig. 6 were prepared using Accelrys DS Visualizer 2.0 (Accelrys, Inc., San Diego, CA).
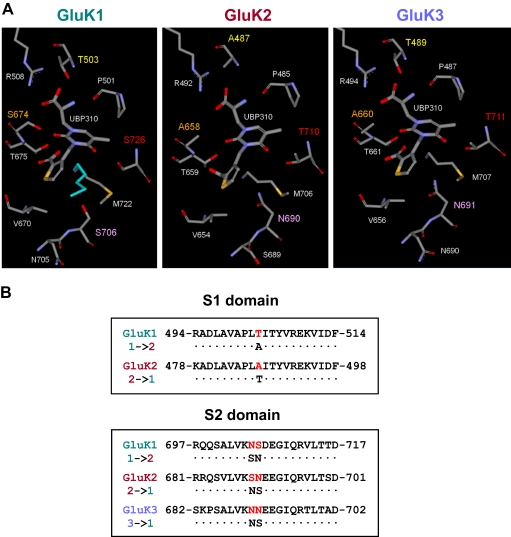
Fig. 6.
Conversion of subunit-selective characteristics of KAR ligand binding sites by mutations of key amino acid residues. A, comparison of the binding pockets of GluK1–3, with the four nonconserved residues highlighted (Thr503, Ser674, Ser706, and Ser726 in GluK1). Note the two alternative conformations for Met722 (the conformation that forms van der Waals interactions with the thiophene ring of UBP310 is colored cyan) that were modeled in the X-ray crystal structure of the GluK1 LBD-UBP310 complex. Middle and right, UBP310 docked into homology models of GluK2 and GluK3, respectively. B, differing residues in sites S1-T/A and S2-S/N were changed using site-directed mutagenesis to analyze their significance in subunit selectivity of UBP310. Mutants created were GluK1-T503A, GluK1-N705S/S706N, GluK2-A487T, GluK2-S689N/N690S, and GluK3-N691S. Residue numbers exclude the signal peptides.
Mutagenesis.
Mutagenesis was carried out on human GluK1–2a(Q), GluK2a(Q), and GluK3a encoding cDNAs using the QuikChange XL Site-Directed Mutagenesis kit from Stratagene (La Jolla, CA) according to the manufacturer's protocol. All residue numbering excludes the signal peptide. All mutants were verified by full-length sequencing (Geneservice, Oxford, UK).
Cell Culture and Transfection.
Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco's modified Eagle's medium (Sigma, Gillingham, UK) supplemented with 10% (v/v) fetal calf serum (Invitrogen Ltd., Paisley, UK), 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin (all from Invitrogen) at 37°C in a humidified atmosphere of 5% CO2. Cells were transiently transfected using linear polyethylenimine (PEI; molecular weight ∼25,000; Polysciences, Warrington, PA) (Durocher et al., 2002). In brief, DNA and PEI were mixed in 150 mM NaCl at an N/P ratio of 5:8, incubated for 20 min, and added dropwise to cells. For endoglycosidase assays, 5 μg of DNA were mixed with 10 μl of PEI (1 mg/ml) in 500 μl of NaCl (150 mM), added to cells in a 25-cm2 flask, and incubated for 24 h. For membrane binding assays, 10 μg of DNA was mixed with 40 μl of PEI (1 mg/ml) in 500 μl of NaCl (150 mM), added to cells in a 175-cm2 flask, and incubated for 48 h. HEK 293 cells stably transfected with either GluK1, GluK2, or GluK3 were grown in the presence of 0.2 mg/ml hygromycin B (Invitrogen) as described previously (Hoo et al., 1994; Nutt et al., 1994; Korczak et al., 1995).
Preparation of Membrane Fractions from HEK 293 Cells and Rat Brains.
Membrane fractions were prepared from both stably and transiently transfected cells harvested in hypotonic solution (10 mM NaHCO3 and complete protease inhibitor cocktail; Roche Diagnostics GmbH, Mannheim, Germany) and disrupted using sonication (2 × 10-s bursts at 10 W, with 20-s incubation on ice in between). Cell homogenates were then spun at 1000g for 20 min at 4°C to remove cellular debris. Membranes were pelleted at 40,000g for 20 min at 4°C and washed three times by resuspension in binding buffer (50 mM Tris, buffered with citric acid, pH 7.4) and centrifugation (40,000g, 20 min, 4°C). After the measurement of protein concentrations using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hemel Hempstead, UK), membrane fractions were separated into aliquots, snap-frozen in liquid nitrogen, and stored at −80°C.
Unless indicated otherwise, native brain membrane fractions were prepared from postnatal day 1 (P1) Wistar rat pups (B&K Universal Ltd., Hull, UK). Whole brains were homogenized in 10× volume of 0.32 M sucrose, 2 mM EDTA, and 10 mM HEPES, pH 7.4 (all from Sigma), with protease inhibitor cocktail (Roche Diagnostics), using a motor-driven glass-Teflon homogenizer. Tissue homogenate was spun at 1000g for 10 min at 4°C to remove nuclear fraction, and resulting supernatant was spun at 40,000g for 20 min at 4°C to yield crude membrane pellet. The pellet was resuspended in homogenization buffer, centrifuged at 40,000g for 20 min at 4°C again, and washed three times using binding buffer, with one freeze-thaw cycle between washes to disrupt glutamate-containing vesicles. The resulting pellet was resuspended in binding buffer and stored at −80°C.
Radioligand Binding Assays.
For the filtration assay, cell membranes were diluted in binding buffer (50 mM Tris, buffered with citric acid, pH 7.4) and 100 to 200 μg of protein was incubated with radioligand on ice, filtered using a Brandel cell harvester (model M-30; Brandel, Gaithersburg, MD), and washed three times using 2 to 4 ml of binding buffer. Radioligands used were [3H]UBP310, described below, and [3H]kainate (37 MBq/ml; PerkinElmer Life and Analytical Sciences, Waltham, MA). Radioligand binding was measured using a scintillation counter (LS6500; Beckman, High Wycombe, UK). For the centrifugation assay, saturation binding to GluK3 was determined using a polyethylene glycol 6000 precipitation-based centrifugation binding assay as described previously (Molnár et al., 1993). (R,S)-1-(2-Amino-2-carboxyethyl)-3-(2-carboxybenzyl)pyrimidine-2,4-dione (UBP296), (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxybenzyl)pyrimidine-2,4-dione (UBP302), and ACET were obtained from Tocris Bioscience (Avonmouth, UK). (S)-1-(2-Amino-2-carboxyethyl)-3-(2-carboxythiophene-3-ylmethyl)pyrimidine-2,4-dione) (UBP304), UBP310, kainate, and NBQX disodium salt were obtained from Ascent Scientific (Avonmouth, UK). [3H]Kainate was purchased from PerkinElmer Life and Analytical Sciences.
Endoglycosidase Digestion.
HEK 293 cells expressing the appropriate KAR subunits were maintained for 24 h before solubilization in lysis buffer (1% Triton X-100 and 0.1% SDS in phosphate-buffered saline, pH 7.4). Cell lysates were cleared of cellular debris by centrifugation at 28,000g for 15 min at 4°C. Equal aliquots of samples were incubated with either endoglycosidase H (EndoH; 5 mU/100 μl of lysate) or endoglycasidase F (EndoF; 1 U/100 μl of lysate) (Roche Diagnostics) overnight at 37°C as described previously (Molnár et al., 1995; Ball et al., 2010) and analyzed by immunoblotting.
Immunoblot Analysis.
Protein samples were separated using 7% (w/v) SDS-polyacrylamide gel electrophoresis gels and transferred electrophoretically onto polyvinylidene difluoride membranes as described previously (Gallyas et al., 2003). Rabbit polyclonal antibodies to GluK1 (20 μg/ml) and GluK2/3 (1:2000 dilution; Millipore, Watford, UK) with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5000 dilution; Sigma) were used to identify KAR subunit proteins as described previously (Gallyas et al., 2003; Ball et al., 2010). A series of antigen concentrations, primary and secondary antibody dilutions, and enhanced chemiluminescence (Roche Diagnostics) exposure times were used to optimize our experimental conditions for the linear sensitivity range of the autoradiography film (HyperfilmTM MP; GE Healthcare). Immunoreactive bands were subjected to densitometry using ImageJ (http://rsbweb.nih.gov/ij/) to determine pixel volume (pixel intensity × area). For figures, we used more exposed images in which all immunopositive bands are clearly visible.
Statistics.
Statistical analysis was performed using an unpaired Student's t test. For nonequal n values, a two-sample equal variance t test was performed. For comparison of multiple sets of data, an analysis of variance (SPSS; SPSS Inc., Chicago, IL) was used. A p value less than 0.05 was considered significant (*), and a p value less than 0.01 was considered highly significant (**). Results are presented as mean ± S.E.M., and n indicates the number of independent experiments.
Results
Synthesis of [3H]UBP310.
Previous functional studies using Ca2+ fluorescence assays have shown that UBP310 and its derivative ACET are high-affinity antagonists of GluK1 homomeric receptors whereas having no effect on GluK2 homomeric receptors or GluK2/GluK5 heteromers at concentrations of up to 1 mM (Dolman et al., 2007; Dargan et al., 2009). In addition, an electrophysiological assay on human homomeric GluK3 receptors showed that ACET had no effect at a concentration of 1 μM (Dargan et al., 2009). Although a recent study raised the possibility that UBP310 effectively blocks recombinant homomeric GluK3 receptors (Perrais et al., 2009), the affinity of this ligand for GluK3 and the molecular mechanisms of subunit-selective interactions of UBP310 with KARs have not been investigated in detail. Tritiation of UBP315 proceeded successfully and [3H]UBP310 was obtained with a specific activity of 39.0 Ci/mmol and a radiochemical purity of 99.3% by high-performance liquid chromatography. The mass spectrum of the purified product was consistent with the proposed structure (Fig. 1). The availability of [3H]UBP310 allowed us to characterize the binding properties of UBP310 directly, without interference from confounding factors associated with the use of radiolabeled agonists such as desensitization.
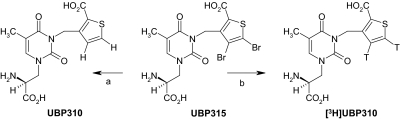
Fig. 1.
Tritiation procedure for production of [3H]UBP310. An analog of UBP310 was synthesized in which two bromo groups have been added to the thiophene ring (UBP315). a, catalytic hydrogenation of the dibromo analog UBP315 lead to replacement of the bromo groups by hydrogen atoms to give UBP310 with a 100% conversion, as adjudged by 1H NMR. Reagents for a: 2 equivalents of NaOH (aq), H2, 10% Pd/C. b, under the same conditions, UBP315 can also be converted to a radiolabeled form of UBP310 simply by switching tritium for hydrogen in the reaction. Reagents for b: 2 equivalents of NaOH (aq), T2, 10% Pd/C.
Differential Binding of [3H]UBP310 to GluK1, GluK3, and GluK2 Kainate Receptor Subunits.
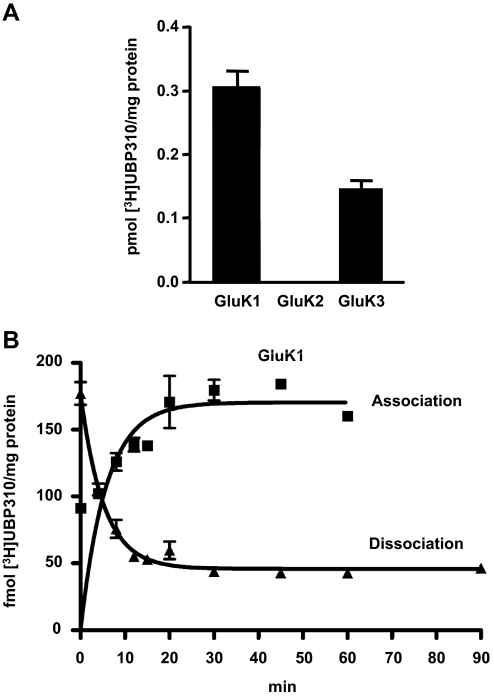
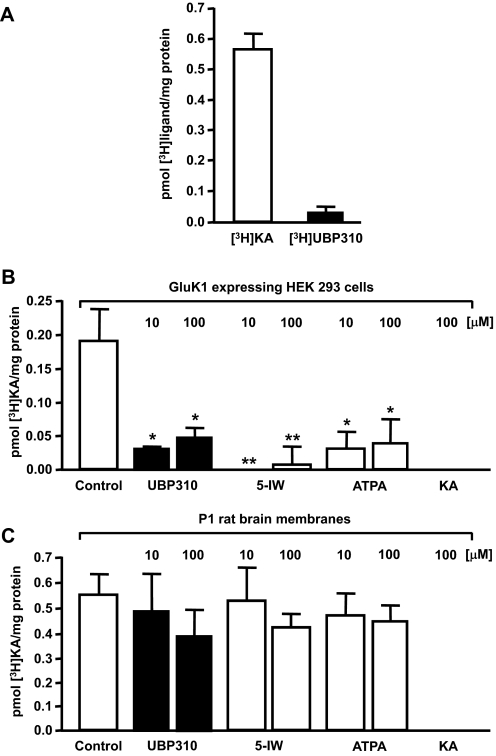
Binding of [3H]UBP310 to the low-affinity KAR subunits GluK1, GluK2. and GluK3 was analyzed using cell membranes from stably transfected HEK 293 cell lines (Fig. 2A) (Hoo et al., 1994; Nutt et al., 1994; Korczak et al., 1995). Although GluK1 shows high level of specific binding (0.30 ± 0.03 pmol [3H]UBP310/mg protein), no specific binding is detected with GluK2 (Fig. 2A). GluK3 shows clearly detectable specific [3H]UBP310 binding at 100 nM concentration (0.14 ± 0.02 pmol [3H]UBP310/mg protein). These results indicate that in addition to GluK1, UBP310 also interacts with GluK3 but not with GluK2.
Fig. 2.
[3H]UBP310 binds to GluK1 and GluK3 but not to GluK2. A, membranes from HEK 293 cells stably transfected with either GluK1, GluK2, or GluK3 were incubated with 100 nM [3H]UBP310 for 1 h, with nonspecific binding determined in the presence of 100 μM kainate. Both GluK1 and GluK3 show specific binding, whereas GluK2 shows none at 100 nM [3H]UBP310 concentration. B, binding of [3H]UBP310 to GluK1 shows relatively fast on and slow off rates. Membranes from HEK 293 cells stably transfected with GluK1 were incubated with 25 nM [3H]UBP310 and harvested at the indicated time points. Nonspecific binding was determined in the presence of 100 μM kainate and was subtracted from the association curve. Dissociation was initiated by the addition of 100 μM kainate after 1-h incubation with 25 nM [3H]UBP310. Graphs show a representative example of three independent determinations. Kob was 0.32 ± 0.07 min−1, Koff was 0.26 ± 0.05 min−1, and the derived Kon was 2.5 × 106 min−1 · M−1.
Analysis of [3H]UBP310 Binding to Recombinant GluK1 and GluK3.
The time course of [3H]UBP310 binding to GluK1 is shown in Fig. 2B. The observed association rate Kob was 0.32 ± 0.07 min−1, whereas the dissociation rate, Koff, was 0.26 ± 0.05 min−1. The derived association rate constant Kon was calculated to be 2.5 × 106 min−1 · M−1 assuming a simple bimolecular interaction. The calculated equilibrium constant derived from the Kon and Koff values was 104 nM. The KD for [3H]UBP310 binding on GluK1 was determined to be 24 ± 6 nM from a saturation binding assay (Fig. 3). The 4-fold discrepancy between the calculated KD value and the one obtained from the saturation binding assay may be explained by experimental error or it may indicate that the mechanism of binding is more complex than a simple bimolecular interaction.
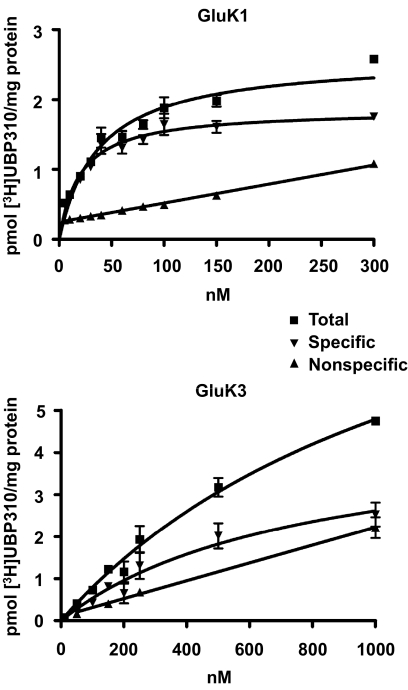
Fig. 3.
Saturation binding analysis indicates that GluK1 has approximately 30-fold higher affinity for [3H]UBP310 than GluK3. Membranes prepared from HEK 293 cells stably transfected with either GluK1 or GluK3 were incubated with increasing concentration (0–1 μM) of [3H]UBP310 for 1 h. Nonspecific binding was determined in the presence of 100 μM kainate. GluK1-containing samples were filtered in a Brandel cell harvester, whereas GluK3-containing samples were analyzed using a centrifugation assay. The graphs show a representative example of three independent determinations. The KD of [3H]UBP310 on GluK1 was 24 ± 6 nM, whereas on GluK3, it was 653 ± 186 nM.
The low affinity of [3H]UBP310 binding to GluK3 ruled out an investigation of its binding kinetics and made the determination of KD difficult using filtration assays; therefore, we used a polyethylene glycol 6000 precipitation-based centrifugation assay (Molnár et al., 1993). The KD obtained for [3H]UBP310 binding to GluK3 with the centrifugation binding assay was 0.65 ± 0.19 μM. GluK1 produced comparable results using the centrifugation assay (KD = 21 ± 7 nM) as with the filtration assay (KD = 24 ± 6 nM).
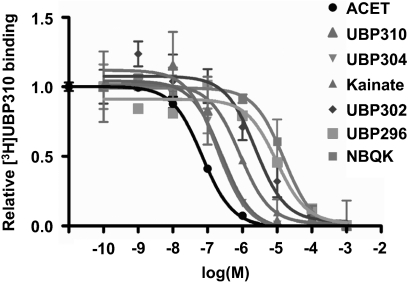
Competition binding analysis on GluK1 with [3H]UBP310 and a range of known KAR ligands (Fig. 4) produced the same rank order of affinity (see Fig. 4 legend for Ki values) as reported previously using a functional Ca2+ fluorescence assay: UBP310≈ACET>UBP304>kainate>UBP302>UBP296> NBQX (Dolman et al., 2007; Dargan et al., 2009; Jane et al., 2009).
Fig. 4.
Competition binding curves for KAR ligands against [3H]UBP310 on GluK1. Cell membranes from stably transfected GluK1 expressing HEK 293 cells were incubated with 25 nM [3H]UBP310 and increasing concentrations of known KAR ligands. Each value is expressed as relative to specific [3H]UBP310 binding as determined in the absence of a competing ligand. Nonspecific binding was determined using 1 mM kainate. Curves were drawn using GraphPad's one-site competition binding function (GraphPad Software Inc., San Diego, CA), and Ki values were determined using the previously determined KD value of 24 nM for [3H]UBP310 (Fig. 3). Ki values were as follows: ACET, 48.5 ± 8.24 nM; UBP310, 46.7 ± 14.8 nM; UBP304, 0.23 ± 0.07 μM; kainate, 0.53 ± 0.10 μM; UBP302, 2.02 ± 0.58 μM; UBP296, 4.10 ± 1.83 μM; NBQX, 29.80 ± 14.56 μM. All data are averages ± S.E.M. of three independent determinations.
Binding of [3H]UBP310 to Native KARs.
Previous studies identified that the expression levels of GluK1 (Bettler et al., 1990) and other KAR subunits (Lilliu et al., 2002) are relatively high in P1 rat brains compared with other postnatal developmental time points. Therefore, we prepared whole-brain membrane fractions from P1 brains for ligand binding assays. Although specific [3H]kainate binding to native receptors is clearly detectable (0.56 ± 0.07 pmol [3H]kainate/mg protein), [3H]UBP310 shows only very modest specific binding (0.023 ± 0.022 pmol [3H]UBP310/mg protein) to the same P1 brain membrane samples in parallel experiments (Fig. 5A). Furthermore, although UBP310- and GluK1-preferring agonists [such as (S)-5-iodowillardiine or (R,S)-2-amino-3-(3-hydroxy-5-tert-butylisoxazol-4-yl)propanoic acid] (Jane et al., 2009) displaced most of the [3H]kainate binding to homomeric GluK1 KARs (Fig. 5B), only a small reduction of [3H]kainate binding to P1 rat brain membranes was observed in parallel experiments (Fig. 5C). [3H]Kainate binds exclusively to KARs in rat brain membranes, because (S)-2-amino-3-hydroxy-5-methyl-4-isoxazole propanoic acid does not inhibit binding (Bunch et al., 2001). In our experiments 100 μM (S)-5-iodowillardiine did not inhibit [3H]kainate binding. If AMPA receptors contributed to [3H]kainate binding, then a high concentration of (S)-5-iodowillardiine should have inhibited [3H]kainate binding, given the moderately high affinity of this agonist for recombinant AMPA receptors (Jane et al., 1997). In addition to P1 rat brain membranes (Fig. 5A), we have also analyzed [3H]UBP310 binding to embryonic day 18 whole brain and dissected adult cerebellar and hippocampal membrane fractions (data not shown). Specific [3H]UBP310 binding was either undetectable or it was similar to values obtained with P1 membranes (Fig. 5A). These results indicate that GluK1 and GluK3 levels with active binding sites are of low abundance, and therefore other KAR subunits are responsible for the relatively high kainate binding activity in native brain samples.
Fig. 5.
Binding of [3H]UBP310 to native KARs. A, P1 rat whole-brain membranes were incubated with 100 nM concentration of either [3H]kainate ([3H]KA) or [3H]UBP310 for 1 h, with nonspecific binding determined in the presence of 1 mM kainate. Robust specific binding was observed with [3H]KA, but specific [3H]UBP310 binding was very low using the same samples under identical conditions. B, the majority of [3H]KA binding at 100 nM was displaced by UBP310 (10–100 μM), (S)-5-Iodowillardiine (5-IW; 10–100 μM), and (R,S)-2-amino-3-(3-hydroxy-5-tert-butylisoxazol-4-yl)propanoic acid (ATPA; 10–100 μM) in GluK1-expressing HEK 293 cell membranes. C, in contrast, these GluK1-preferring ligands (Jane et al., 2009) produced only modest reduction in [3H]KA binding to P1 rat brain membranes. Data presented are averages ± S.E.M. from three independent experiments. *, p < 0.05; **, p < 0.01.
Docking of UBP310 into the LBDs of GluK2 and GluK3.
To date, no X-ray crystal structures of the LBDs of GluK2 or GluK3 in complex with antagonists have been reported. We therefore produced homology models of the open form of the LBDs of GluK2 and GluK3 based on the X-ray crystal structure of the GluK1 LBD-UBP310 complex (Mayer et al., 2006). Docking of UBP310 into the homology models of the LBDs of GluK2 and GluK3 using the Induced Fit workflow in Maestro placed UBP310 in a position similar to that observed in the X-ray crystal structure of the GluK1 LBD-UBP310 complex (Fig. 6A). However, there are differences in key nonconserved residues that bind to UBP310 in GluK2 and GluK3 compared with GluK1 that may explain the observed lower affinity of UBP310 for these two subunits. A major difference is the switch from Thr503 in GluK1 to Ala487 in GluK2, which results in a loss of binding interactions for the α-amino and α-carboxyl groups of UBP310.
In the X-ray crystal structure of the GluK1 LBD-UBP310 complex, Met722 was modeled in two conformations with occupancies of 0.65 and 0.35, and only the latter conformation (colored cyan in Fig. 6A) allows a favorable van der Waals contact of Met722 with the thiophene ring of UBP310 (Mayer et al., 2006). However, inspection of the homology models suggests that this favorable interaction is not possible in GluK2 and GluK3, because the bulky residues Asn690 in GluK2 and Asn691 in GluK3 force the methionine residue to adopt the alternative conformation (Fig. 6A), which cannot form an interaction with the thiophene ring of UBP310. In addition, these asparagine residues in the GluK2 and GluK3 homology models are much closer to the thiophene ring of UBP310 than the corresponding residue Ser706 in GluK1 and are likely to hinder the adoption of the optimal conformation of UBP310 for interaction of the carboxylate group attached to the thiophene ring with Thr659 (GluK2) or Thr661 (GluK3). Although we have identified two reasons why these asparagine residues may play a detrimental role in antagonist binding to GluK2 and GluK3, their influence on binding affinity will depend on the degree of opening of the LBDs, and therefore, confirmation of their role in determining UBP310 binding affinity requires point mutation studies.
The switch from Ser674 in GluK1 to Ala658 in GluK2 and Ala660 in GluK3 is likely to play a minor role in explaining the lower affinity of UBP310 for GluK2 and GluK3. Inspection of the X-ray crystal structure suggests that the OH group of Ser674 in GluK1 only forms an indirect hydrogen bond via a water molecule with the 2-oxo group on the uracil ring of UBP310 and is therefore not significantly contributing to the binding affinity (Mayer et al., 2006). The switch from Ser726 in GluK1 to Thr710 in GluK2 and Thr711 in GluK3 (Fig. 6A) would not be expected to affect the binding of UBP310, because this residue is a considerable distance away from the ligand.
Mutations of Key Amino Acids Involved in UBP310 Binding at the Ligand Binding Sites of KAR Subunits.
A comparison of the binding modes of UBP310 to GluK1, GluK2, and GluK3 suggested that Thr503 in the S1 domain of GluK1 is critical for UBP310 binding, because it forms hydrogen bonds with the α-amino and α-carboxyl groups of UBP310 (Fig. 6A). The corresponding site in GluK2 is occupied by Ala487, whereas the GluK3 subunit like GluK1, contains a threonine (Thr489). This site was mutated in both GluK1 and GluK2 to produce GluK1-T503A and GluK2-A487T, respectively, thereby converting their subunit specificity at this site (Fig. 6).
The second residue we identified that may play a role in ligand binding was Ser706 in the S2 domain of GluK1 (Fig. 6A). This site is occupied by an asparagine in both GluK2 (Asn690) and GluK3 (N691), and inspection of the binding modes of UBP310 in the homology models of GluK2 and GluK3 suggested that these asparagine residues may reduce binding affinity through steric hindrance (Fig. 6A). The residue immediately preceding this site is an asparagine (Asn705) in GluK1 and GluK3 (Asn690) and a serine (Ser689) in GluK2. Although this residue is not believed to participate directly in binding, it was also targeted to fully convert the subunit selectivity at this site (Fig. 6). Thus, the double mutants GluK1-N705S/S706N and GluK2-S689N/N690S and the single mutant GluK3-N691S were created.
Comparison of the Expression Levels and Intracellular Trafficking of Wild-Type and Ligand Binding Site Mutant KAR Subunits.
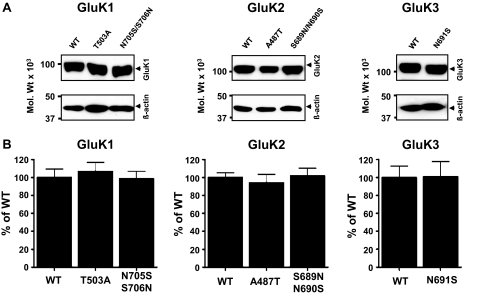
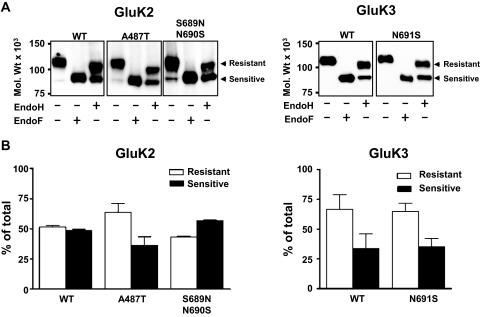
Recent studies reported that mutations in the ligand binding sites of KARs that disrupt interaction with glutamate or prevent conformational changes can block the export of subunit proteins from the ER to the cell surface (Fleck, 2006; Gill et al., 2009). Although based on our molecular modeling studies of the KAR ligand binding sites, it is unlikely that any of the introduced mutations (Fig. 6) disrupt glutamate binding and/or conformational changes in the receptor subunits; however, it was important to investigate potential changes in KAR biogenesis. Therefore, we compared the expression levels (Fig. 7) and trafficking (Fig. 8) of wild-type (WT) and mutant GluK1, GluK2, and GluK3 subunits. To identify potential differences in GluK2 or GluK3 trafficking, we analyzed their glycosylation status by EndoH. EndoH is a highly specific endoglycosidase that cleaves asparagine-linked mannose rich oligosaccharides, typically associated with unprocessed glycoproteins in the ER. EndoH is unable to cleave highly processed complex oligosaccharides from more mature glycoproteins after they are processed in the Golgi apparatus. Therefore, EndoH sensitivity is routinely used to monitor the maturation state of glycoproteins during their biosynthesis and trafficking through the secretory pathway. Our previous study established that EndoH resistance directly correlates to the cell surface expression of KAR subunit proteins (Ball et al., 2010). After EndoH treatment, both GluK2 and GluK3 yielded double bands in immunoblots (Fig. 8A). The lower EndoH-sensitive band displayed a molecular mass of ∼95 kDa. EndoH insensitive upper band displayed only a minor molecular mass shift in response to EndoH digestion, which is probably due to the presence of a residual high mannose glycan in the mature subunit. Quantitative comparison of the EndoH-insensitive bands revealed no detectable differences in the proportions of WT and corresponding mutant GluK2 or GluK3 proteins that were EndoH-insensitive (Fig. 8B). Full deglycosylation of either GluK2 or GluK3 with EndoF resulted in a single band with molecular masses (∼95 kDa) similar to the EndoH-sensitive component (Fig. 8A). The GluK1–2a splice variant used here does not traffic well on its own when expressed in heterologous systems and was therefore not analyzed in detail. These results indicate that the expression levels, trafficking, and degradation rates of mutant subunit proteins are not significantly different from WT, and they are able to fold and assemble to KARs, which is required for their ER export.
Fig. 7.
Binding site mutations do not alter KAR subunit expression levels. A, representative immunoblots of the WT and mutated GluK1, GluK2, and GluK3 subunits show similar expression levels. Top, the KAR subunits indicated; bottom, β-actin in the same samples as loading controls. B, quantitative analysis of the WT and mutated subunits shows no significant differences in expression levels (p > 0.05). Graphs are averages ± S.E.M. of four to six independent experiments in which each sample was assessed in triplicate.
Fig. 8.
Binding site mutations do not alter KAR trafficking. A, lysates from HEK 293 cells transiently transfected with either WT or mutated GluK2 and GluK3 subunits were treated with either EndoH or EndoF and analyzed using immunoblotting. Arrows show EndoH-resistant and -sensitive bands, respectively. Mutated subunits do not show an altered glycosylation pattern from WT. B, graphs show relative densities of bands from EndoH-treated WT and mutated GluK2 and GluK3 subunits. Numbers are averages from three independent experiments and are presented as mean ± S.E.M. There were no significant differences between WT and mutated subunits (p > 0.05).
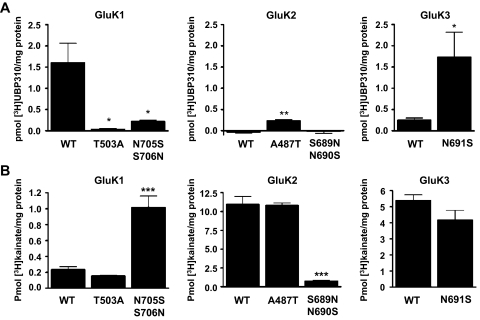
Mutation-Induced Changes in [3H]UBP310 and [3H]Kainate Binding to KAR Subunits.
The binding properties of mutant subunits were analyzed using 100 nM [3H]UBP310, which is a saturating concentration for GluK1 subunits (Fig. 9A) (Jane et al., 2009). The GluK1-T503A and GluK1-N705S/S706N mutants show significantly reduced binding to [3H]UBP310. Although the WT GluK2 does not interact with [3H]UBP310, the GluK2-A487T mutant shows significantly increased binding activity (Fig. 9A). The GluK3-N691S mutant shows considerably increased binding to [3H]UBP310 compared with the WT subunit. These results indicate that switching amino acid residues between subunits at the S1-T/A and S2-S/N sites dramatically transforms their subunit-specific antagonist binding properties. It is noteworthy that although all mutated subunits were able to bind [3H]kainate, the affinities of GluK1-N705S/S706N and GluK2-S689N/N690S were shifted from the parent subunit toward that of the subunit the mutation was designed to imitate (Fig. 9B). Therefore, residues that interact with UBP310 also participate in kainate binding.
Fig. 9.
The S1-T/A and S2-S/N sites determine the subunit-selective binding of [3H]UBP310 to KARs. A, membranes from HEK 293 cells transiently transfected with either WT or mutated subunits were incubated with 100 nM [3H]UBP310 for 1 h, with nonspecific binding determined in the presence of 100 μM kainate. The GluK1-T503A and GluK1-N705S/S706N mutants show significantly reduced [3H]UBP310 binding compared with WT. On the other hand, GluK2-A487T displayed significantly increased [3H]UBP310 binding, whereas GluK2 WT and GluK2-S689N/N690S show no specific binding. The N691S mutation substantially increased the binding of [3H]UBP310 to GluK3 compared with WT. Data are presented as mean ± S.E.M., n = 3–4; *, p < 0.05, **, p < 0.01, Student's t test. Immunoblots of membrane fractions used for ligand binding assays confirmed that the expression levels of all mutants are similar to WT (Fig. 7). B, membranes identical with the ones in A were incubated in the same way with 100 nM [3H]kainate ([3H]KA). GluK1-T503A shows no difference in binding to WT, whereas GluK1-N705S/S706N shows a significant increase in [3H]KA binding. Likewise, the GluK2-A487T mutant shows no difference in [3H]KA binding, whereas GluK2-S689N/N690S shows a significant decrease. The GluK3-N691S shows only a moderate change from WT.
Discussion
UBP310 has been shown to be a highly potent antagonist of GluK1-containing KARs but not of GluK2 or GluK3 (Dolman et al., 2007; Dargan et al., 2009). However, a recent study showed that UBP310 and a related antagonist, ACET, are potent antagonists of homomeric GluK3 with IC50 values of 23 and 92 nM, respectively, for blocking currents mediated by rapid application of glutamate (Perrais et al., 2009). In the latter study, UBP310 and ACET were found to have no effect on GluK2/GluK3 heteromeric receptors, but neither compound was tested on homomeric GluK1 in the same assay. GluK3 receptors are rapidly desensitized upon the application of glutamate. In our earlier studies, we used either competition binding assays using [3H]kainate (Dolman et al., 2007) or electrophysiological assays with much slower application times for glutamate (Dargan et al., 2009) than the study by Perrais et al., (2009), suggesting that in our earlier studies, we were primarily assaying antagonists on the desensitized state of GluK3. The present study provides a method of assaying KAR ligands using the radiolabeled antagonist [3H]UBP310 without the confounding factor of receptor desensitization. Kinetic and saturation binding analyses demonstrated that [3H]UBP310 has a high affinity for GluK1 and approximately 30-fold lower affinity for GluK3. The fast-on and slow-off rate for the kinetics of [3H]UBP310 binding to GluK1 are consistent with the high affinity of UBP310 for this subunit. Competition binding assays on GluK1 using [3H]UBP310 confirmed the rank order of potencies of known KAR ligands (Jane et al., 2009). [3H]UBP310 therefore represents a useful new tool for the direct study of ligand binding to GluK1 and GluK3 subunit-containing KARs.
Under the equilibrium conditions used in the saturation binding assay, UBP310 had a much lower affinity for GluK3 than that estimated in the electrophysiological assay using rapid application of glutamate (Perrais et al., 2009). This suggests that the potency of UBP310 as a GluK3 antagonist has been considerably overestimated in the electrophysiological assay. This is probably due to the conditions used in the rapid agonist application assay in which the agonist is not at equilibrium with the receptor.
Limited information is available about the interaction of GluK1 selective antagonists with endogenous KARs in the brain and the overall contribution of GluK1 to the total kainate binding activity in native receptors. The results of this study indicate that [3H]UBP310 binding to native KARs was relatively low compared with [3H]kainate binding activities measured in the same samples. Furthermore, UBP310- and GluK1-preferring KAR agonists displaced only a small proportion (<20–30%) of [3H]kainate binding to P1 brain samples. These results indicate that GluK1 and GluK3 represent relatively minor components of KARs, and other KAR subunits are probably responsible for the majority of kainate binding activity in native brain samples, as suggested in previous studies (Lerma, 2006; Jane et al., 2009). These results are consistent with the hypothesis that GluK2/GluK5 heteromers are the most abundant KAR subtype in the brain (Lerma, 2006). The low abundance of GluK1-containing KARs could be due to their more specific roles in a subset of synapses (Jane et al., 2009).
High-resolution X-ray crystal structures of the GluK1 LBD in complex with UBP310 and ACET have been reported previously (Mayer et al., 2006; Dargan et al., 2009). Inspection of these structures and comparison with the structures of UBP310 bound to homology models of the LBDs of GluK2 and GluK3 suggest that only two nonconserved amino acids are likely to have a major influence on the binding affinity of UBP310 for GluK2 and GluK3 (Fig. 6). We predicted that the switch from Thr503 in GluK1 to Ala487 at the equivalent position in GluK2 (S1-T/A site) would be detrimental to antagonist binding to GluK2, because Thr503 plays an important role in interacting with the α-amino and α-carboxyl groups of UBP310 (Fig. 6). In addition, we considered that the change from Ser706 in GluK1 to Asn690 in GluK2 (S2-S/N site) may reduce UBP310 binding affinity to GluK2 due to steric hindrance by the asparagine residue in GluK2, resulting in reduced ability of the carboxylate group on the thiophene ring to access its site of interaction on domain S2. The LBD of GluK3 is a hybrid between GluK1 and GluK2, because it has the equivalent threonine residue to that present in GluK1 at the S1-T/A site but an asparagine residue at the S2-S/N site as found in GluK2 (Fig. 6). Inspection of the binding mode of UBP310 in our homology model of GluK3 suggested that both of these residues would play a role in determining the binding affinity of [3H]UBP310 for GluK3. We sought to investigate these hypotheses by point-mutating these nonconserved residues in GluK1–3 and noting their effect on the affinity of binding of [3H]UBP310. We established that the threonine at the S1-T/A site in GluK1 is crucial for [3H]UBP310 binding. Although the mutation of this residue in GluK1 reduces ligand binding, introduction of a threonine residue in the corresponding site of GluK2 increased [3H]UBP310 binding. Thus, point mutation studies back up our conclusion from modeling studies that the replacement of the S1-T residue by alanine in GluK2 is a major contributing factor to the low affinity of UBP310 for GluK2. Mutation of the serine at the S2-S/N site to asparagine reduces [3H]UBP310 binding to GluK1. However, introduction of a serine to GluK2 at this site did not increase [3H]UBP310 binding activity, presumably because of the lack of the threonine at the S1-T/A site that is important for optimal binding of the amino acid moiety of UBP310. GluK3, which has a threonine in the S1-T/A site, shows increased [3H]UBP310 binding after the introduction of serine at the S2-S/N site. The asparagine is probably interfering with [3H]UBP310 binding to GluK3 through steric hindrance. It is noteworthy that because of steric occlusion by these asparagine residues, Met706 in GluK2 and Met707 in GluK3 would not be able to adopt the conformation observed for Met722 in GluK1 that leads to favorable van der Waals interaction with the thiophene ring of UBP310. The GluK2-A487T mutation has the same critical binding site residues as wild-type GluK3, and therefore, a A487T + S689N/N690S switch in GluK2 would probably have the same effect as a GluK3-N691S mutation in restoring UBP310 binding affinity to the level observed in wild-type GluK1.
All mutated subunits bound [3H]kainate, but interestingly, the S2-S/N site seems to confer some of the subunit specific difference in kainate affinity. Inspection of the X-ray crystal structure of kainate bound to the LBD of GluK2 (Mayer, 2005) shows that Asn690 can form hydrogen bond interactions via water molecules with the distal carboxylate of kainate. In addition, Asn690 in the S2 domain forms a direct interdomain hydrogen bond contact with Glu409 in the S1 domain that would help to stabilize the closed agonist-bound conformation. The S1-T/A site does not seem to be as important for kainate binding. This site is less important for agonists because there is a direct ionic interaction between the α-amino group of kainate and the terminal carboxylate of a glutamate residue (Glu707 in GluK2), and therefore, the switch from threonine to alanine in GluK2 is not as detrimental. In the antagonist-bound conformation, the distance between the S1 and S2 domains is too great to permit direct interaction of the α-amino group of UBP310 with Glu707. Thus, the switch from threonine to alanine in GluK2, which results in a loss of hydrogen bonds to the α-amino and α-carboxyl groups, is much more important in reducing the binding affinity of antagonists such as UBP310.
Agonist binding causes KARs to adopt alternative conformation states that drive ER export (Fleck, 2006). Mutations in the functionally significant ligand-receptor interaction sites produced ER retention of receptors without disrupting assembly. These observations indicate that glutamate binding site mutants are fully assembled in the ER, and the trafficking is interrupted at a stage following the assembly process. Recent studies established that ionotropic glutamate receptors exist naturally in their glutamate-bound state and normally traffic from the ER to Golgi en route to the plasma membrane in a glutamate-bound conformation (Fleck, 2006). In this study, we established that biosynthesis, trafficking, and degradation of GluK1–3 are unaffected by mutations at the S1-A/T and S2-S/N sites, and mutant subunits are able to fold and assemble to functional KARs, which is required for their ER export.
In this study, we have developed a novel radioligand [3H]UBP310 and shown that it binds with high affinity to GluK1, although displaying 30-fold lower affinity for GluK3 and negligible binding to GluK2. Thus, [3H]UBP310 is a useful ligand for studying the pharmacology of GluK1 and GluK3 KAR subunits. Furthermore, we have produced models of the interaction of UBP310 with the LBDs of GluK2 and GluK3 and have used point mutation studies to validate conclusions made by inspection of these models regarding the roles of the S1-A/T and S2-S/N sites in the LBDs of KARs in determining ligand selectivity. Targeting of these key residues should facilitate the design and development of new ligands with greater subunit selectivity for individual KAR subunits.
Acknowledgments
We thank Ascent Scientific (Avonmouth, UK) and GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK) for carrying out the analysis of the purity of the precursor UBP315 and the synthesis and purification of [3H]UBP310. We are grateful to Dr. David Bleakman, Eli Lilly and Company (Indianapolis, IN), for supplying the recombinant human GluK1, GluK2, and GluK3 cell lines.
This work was supported by the UK Medical Research Council [Grants G0601509, G0601812]; and the Biotechnology and Biological Sciences Research Council [Grant BB/F012519/1]. C.L.S. is a UK Medical Research Council-funded PhD student.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.067934.
- KAR
- kainate receptor
- ACET
- (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxy-5-phenylthiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione
- AMPA
- (S)-2-amino-3-hydroxy-5-methyl-4-isoxazolepropanoic acid
- EndoF
- endoglycasidase F
- EndoH
- endoglycasidase H
- HEK
- human embryonic kidney
- KA
- kainate, (2S,3S,4S)-3-carboxymethyl-4-isopropenyl-pyrrolidine-2-carboxylic acid
- LBD
- ligand binding domain
- NMDA
- N-methyl-d-aspartate
- WT
- wild type
- ER
- endoplasmic reticulum
- PEI
- polyethylenimine
- NBQX
- 2,3-dihydroxy-6-nitro-7-sulfamoylbenzo(f)quinoxaline
- UBP310
- (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxythiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione
- UBP315
- (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxy-4,5-dibromothiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione
- UBP302
- (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxybenzyl)pyrimidine-2,4-dione
- UBP296
- (R,S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxybenzyl)pyrimidine-2,4-dione
- UBP304
- (S)-1-(2-amino-2-carboxyethyl)-3-(2-carboxythiophene-3-ylmethyl)pyrimidine-2,4-dione).
References
- Alushin GM, Jane DE, Mayer ML. (2010) Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists. Neuropharmacology doi:10.1016/j.neuropharm.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- Ball SM, Atlason PT, Shittu-Balogun OO, Molnár E. (2010) Assembly and intracellular distribution of kainate receptors is determined by RNA editing and subunit composition. J Neurochem 114:1805–1818 [DOI] [PubMed] [Google Scholar]
- Bettler B, Boulter J, Hermans-Borgmeyer I, O'Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. (1990) Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 5:583–595 [DOI] [PubMed] [Google Scholar]
- Bettler B, Egebjerg J, Sharma G, Pecht G, Hermans-Borgmeyer I, Moll C, Stevens CF, Heinemann S. (1992) Cloning of a putative glutamate receptor: a low affinity kainate-binding subunit. Neuron 8:257–265 [DOI] [PubMed] [Google Scholar]
- Bunch L, Johansen TH, Bräuner-Osborne H, Stensbøl TB, Johansen TN, Krogsgaard-Larsen P, Madsen U. (2001) Synthesis and receptor binding affinity of new selective GluR5 ligands. Bioorg Med Chem 9:875–879 [DOI] [PubMed] [Google Scholar]
- Dargan SL, Clarke VR, Alushin GM, Sherwood JL, Nisticò R, Bortolotto ZA, Ogden AM, Bleakman D, Doherty AJ, Lodge D, et al. (2009) ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function. Neuropharmacology 56:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolman NP, More JC, Alt A, Knauss JL, Pentikäinen OT, Glasser CR, Bleakman D, Mayer ML, Collingridge GL, Jane DE. (2007) Synthesis and pharmacological characterization of N3-substituted willardiine derivatives: role of the substituent at the 5-position of the uracil ring in the development of highly potent and selective GLUK5 kainate receptor antagonists. J Med Chem 50:1558–1570 [DOI] [PubMed] [Google Scholar]
- Durocher Y, Perret S, Kamen A. (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acid Res 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J, Bettler B, Hermans-Borgmeyer I, Heinemann S. (1991) Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not AMPA. Nature 351:745–748 [DOI] [PubMed] [Google Scholar]
- Fleck MW. (2006) Glutamate receptors and endoplasmic reticulum quality control: looking beneath the surface. Neuroscientist 12:232–244 [DOI] [PubMed] [Google Scholar]
- Gallyas F, Jr, Ball SM, Molnar E. (2003) Assembly and cell surface expression of KA-2 subunit-containing kainate receptors. J Neurochem 86:1414–1427 [DOI] [PubMed] [Google Scholar]
- Gill MB, Vivithanaporn P, Swanson GT. (2009) Glutamate binding and conformational flexibility of ligand-binding domains are critical early determinants of efficient kainate receptor biogenesis. J Biol Chem 284:14503–14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoo KH, Nutt SL, Fletcher EJ, Elliott CE, Korczak B, Deverill RM, Rampersad V, Fantaske RP, Kamboj RK. (1994) Functional expression and pharmacological characterization of the human EAA4 (GluR6) glutamate receptor: a kainate selective channel subunit. Receptors Channels 2:327–337 [PubMed] [Google Scholar]
- Jane DE, Hoo K, Kamboj R, Deverill M, Bleakman D, Mandelzys A. (1997) Synthesis of willardiine and 6-azawillardiine analogs: pharmacological characterization on cloned homomeric human AMPA and kainate receptor subtypes. J Med Chem 40:3645–3650 [DOI] [PubMed] [Google Scholar]
- Jane DE, Lodge D, Collingridge GL. (2009) Kainate receptors: pharmacology, function and therapeutic potential. Neuropharmacology 56:90–113 [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korczak B, Nutt SL, Fletcher EJ, Hoo KH, Elliott CE, Rampersad V, McWhinnie EA, Kamboj RK. (1995) cDNA cloning and functional properties of human glutamate receptor EAA3 (GluR5) in homomeric and heteromeric configuration. Receptors Channels 3:41–49 [PubMed] [Google Scholar]
- Lerma J. (2006) Kainate receptor physiology. Curr Opin Pharmacol 6:89–97 [DOI] [PubMed] [Google Scholar]
- Lilliu V, Perrone-Capano C, Pernas-Alonso R, Diaz Trelles R, Luca Colucci d'Amato G, Zuddas A, di Porzio U. (2002) Ontogeny of kainate receptor gene expression in the developing rat midbrain and striatum. Mol Brain Res 104:1–10 [DOI] [PubMed] [Google Scholar]
- Mah SJ, Cornell E, Mitchell NA, Fleck MW. (2005) Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci 25:2215–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML. (2005) Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron 45:539–552 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. (2006) Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci 26:2852–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár E, Baude A, Richmond SA, Patel PB, Somogyi P, McIlhinney RA. (1993) Biochemical and immunocytochemical characterization of antipeptide antibodies to a cloned GluR1 glutamate receptor subunit: cellular and subcellular distribution in the rat forebrain. Neuroscience 53:307–326 [DOI] [PubMed] [Google Scholar]
- Molnár E, Váradi A, McIlhinney RA, Ashcroft SJ. (1995) Identification of functional ionotropic glutamate receptor proteins in pancreatic β-cells and islets of Langerhans. FEBS Lett 371:253–257 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Hoo KH, Rampersad V, Deverill RM, Elliott CE, Fletcher EJ, Adams SL, Korczak B, Foldes RL, Kamboj RK. (1994) Molecular characterization of the human EAA5 (GluR7) receptor: a high-affinity kainate receptor with novel potential RNA editing sites. Receptors Channels 2:315–326 [PubMed] [Google Scholar]
- Peitsch MC. (1995) Protein modelling by e-mail. Nat Biotechnol 13:658–660 [Google Scholar]
- Perrais D, Pinheiro PS, Jane DE, Mulle C. (2009) Antagonism of recombinant and native GluK3-containing kainate receptors. Neuropharmacology 56:131–140 [DOI] [PubMed] [Google Scholar]
- Valluru L, Xu J, Zhu Y, Yan S, Contractor A, Swanson GT. (2005) Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J Biol Chem 280:6085–6093 [DOI] [PubMed] [Google Scholar]