Abstract
We tested the hypothesis that de novo copy number variation (CNV) is associated with autism spectrum disorders (ASDs). We performed comparative genomic hybridization (CGH) on the genomic DNA of patients and unaffected subjects to detect copy number variants not present in their respective parents. Candidate genomic regions were validated by higher-resolution CGH, paternity testing, cytogenetics, fluorescence in situ hybridization, and microsatellite genotyping. Confirmed de novo CNVs were significantly associated with autism (P = 0.0005). Such CNVs were identified in 12 out of 118 (10%) of patients with sporadic autism, in 2 out of 77 (3%) of patients with an affected first-degree relative, and in 2 out of 196 (1%) of controls. Most de novo CNVs were smaller than microscopic resolution. Affected genomic regions were highly heterogeneous and included mutations of single genes. These findings establish de novo germline mutation as a more significant risk factor for ASD than previously recognized.
Autism spectrum disorders (ASDs) [Mendelian Inheritance in Man (MIM) 209850] are characterized by language impairments, social deficits, and repetitive behaviors. The onset of symptoms occurs by the age of 3 and usually requires extensive support for the lifetime of the afflicted. The prevalence of ASD is estimated to be 1 in 166 (1), making it a major burden to society.
Genetics plays a major role in the etiology of autism. The concordance rates in monozygotic twins are 70% for autism and 90% for ASD, whereas the concordance rates in dizygotic twins are 5% and 10%, respectively. Previous studies suggest autism displays a high degree of genetic heterogeneity. Efforts to map disease genes using linkage analysis have found evidence for autism loci on 20 different chromosomes. Regions implicated by multiple studies include 1p, 5q, 7q, 15q, 16p, 17q, 19p, and Xq (2). Moreover, microscopy studies have identified cytogenetic abnormalities in >5% of affected children, involving many different loci on all chromosomes (3). In some rare syndromic forms of autism, such as Rett syndrome (4) and tuberous sclerosis (5), mutations in a single gene have been identified. Otherwise, neither linkage nor cytogenetics has unambiguously identified specific genes involved.
Genetic heterogeneity poses a considerable challenge to traditional approaches for gene mapping (6). Some of these limitations are overcome by methods that rely on the direct detection of functional variants, which in most cases are de novo events. New array-based technologies can detect differences in DNA copy number at much higher resolution than cytogenetic methods (7) and, hence, might reveal spontaneous mutations that were previously unidentified. These techniques have shown an abundance of copy number variants (CNVs) in humans (8, 9), and the same methods have been used to find de novo chromosome aberrations below the resolution of microscopy in children with mental retardation and dysmorphic features (10–14), including patients with syndromic forms of autism (15). Yet, the association of spontaneous CNVs in idiopathic autism has not been systematically investigated. Thus, a large-scale study of genome copy number variation in ASD was needed. We have performed high-resolution genomic microarray analysis on a sample of 264 families to determine the rate of de novo copy number mutation in unaffected and affected children.
Our study focused on a sample of 264 families, including 118 “simplex” families containing a single child with autism, 47 “multiplex” families with multiple affected siblings, and 99 control families with no diagnoses of autism. The majority of patients came from the Autism Genetic Resource Exchange (AGRE) and from the National Institute of Mental Health (NIMH) Center for Collaborative Genetic Studies of Mental Disorders. Additional families were obtained through the authors (T.C.G., J.S.S., J.B., and D.S). Efforts were made at all of the collecting sites to exclude cases of syndromic autism (i.e., those with severe mental retardation or other congenital anomalies) and to exclude known cytogenetic abnormalities. Identities of all subjects and their parents were coded so that analysis could be done blind to affected status while maintaining knowledge of the parent-child relations. We performed whole-genome scans on all parents, patients, and unaffected children. Affected or unaffected siblings of many patients were included in the study as independent cases or controls, respectively; thus, the entire sample yielded a complete parent-child “trio” for each of 195 patients and 196 healthy individuals. (See supplementary methods and table S1 for more extensive details on the patient sample.)
We analyzed DNA samples, prepared from either whole blood or Epstein-Barr virus (EBV) immortalized B cells or both, collected from subjects and their biological parents. Genome scans were performed by ROMA, a form of comparative genomic hybridization described previously (8, 16). We performed two-color assays by cohybridizing each sample to an oligonucleotide array, and we used a standard reference genome, SKN1, for comparison. Assays were performed in duplicate with dye-swap. The array consisted of 85,000 probes, providing a mean resolution of one probe every 35 kb. Log intensity ratios from duplicate scans were averaged, and normalized ratio data were segmented by a Hidden Markov Model to define CNVs relative to reference (8) (with minor modifications).
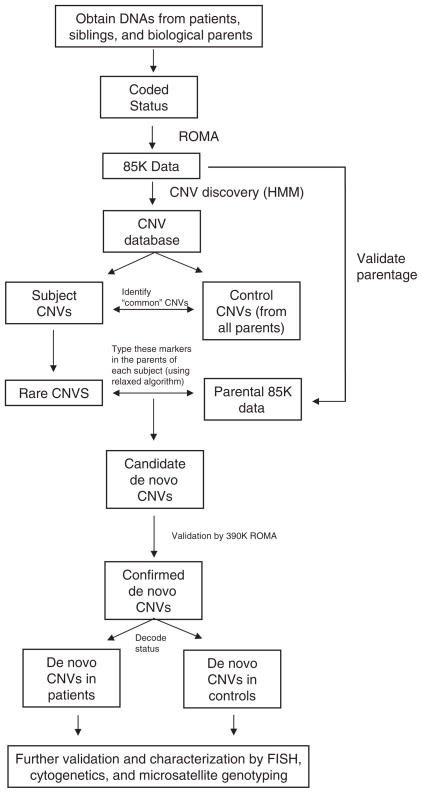
Detecting copy number variation from array data is an error-prone process, and so procedures were followed to ensure that events we detected in subjects were in fact de novo: not false-positive in the subject, and not false-negative in either biological parent. A flow chart of our procedure for finding and testing de novo mutations is depicted in Fig. 1. CNV regions detected in subjects were considered only if they involved at least three consecutive probes and had an overall likelihood measure >0.9 (8). Then, CNVs were disregarded if they were 60% similar in probe content to a variant detected in the set of all parents, where similarity between two CNVs is defined as (the number of common probes)/(the total number of probes in either CNV). This step was done in order to simultaneously filter out any CNVs present in the biological parents and to eliminate common polymorphic loci that would incorrectly appear to be de novo. The latter can occur, for example, when the parents and the reference are all heterozygous for a deletion and 0 or 2 copies are transmitted to the child. These two procedures greatly reduce the total number of candidates requiring validation.
Fig. 1.
Procedure for the detection of de novo CNVs. The flow diagram describes the step-by-step procedure for identifying regions of altered copy number that are present in a child and not in the biological parents.
We then further examined each candidate variant by a more careful assessment of the parents for the presence of the CNV, using a relaxed set of criteria for its presence (see legend to Fig. 2), to rule out false-negatives. If at that point the variant in the subject still appeared to be de novo, that is, present in the child but not in either parent, we tested parentage using multiple informative genetic markers. We then conducted additional validation of the suspect de novo lesion in parents and subjects, including Dpn II–ROMA using 390K arrays, CGH using Agilent 244K arrays, cytogenetics, and micro-satellite genotyping. When de novo mutations were detected in DNA derived from an EBV-immortalized cell line, we sought to repeat analysis on DNA derived from an independent blood sample and found confirmation in 11 out of 12 available cases.
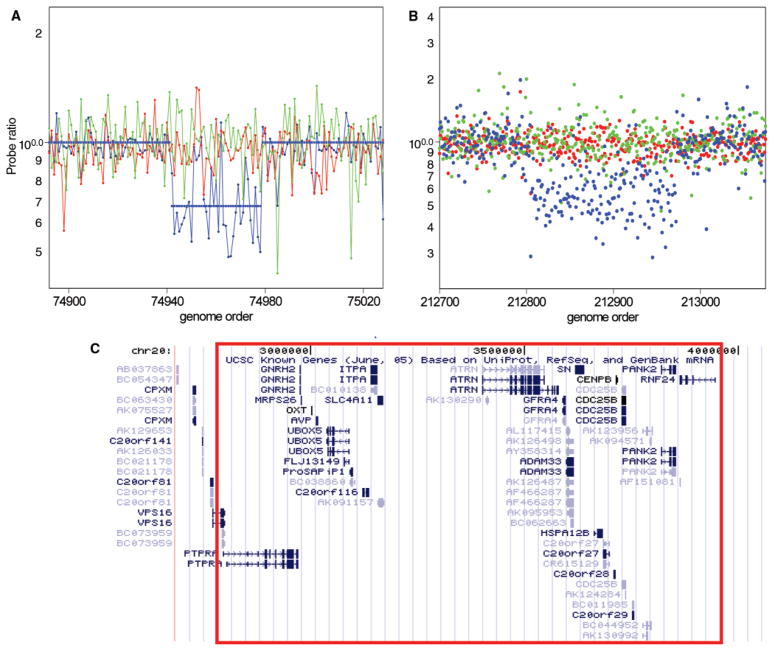
Fig. 2.
Detection and validation of a spontaneous deletion in a patient with Asperger syndrome. CNVs were detected in patient scans using the standard HMM. Parents were ascertained and determined to have no change in copy number using an algorithm with relaxed criteria. These second detection criteria included (1) detection by an HMM with reduced stringency (false-positive expectation set at 1%), and median probe ratio ≤0.91 or ≥1.1. Probe ratio data are shown for the patient SK-135 (blue) and the mother (red) and father (green) for 85K ROMA (A) and 244K Agilent CGH (B) platforms. The map of annotated known genes was obtained from the UCSC Genome Browser, May 2004 assembly (30). The genomic region estimated to be deleted is shown in (C) outlined in red.
One example of the detection and confirmation of a de novo CNV is illustrated in Fig. 2. We detected a 1.1-Mb deletion of 20p13 in a child with the diagnosis of Asperger syndrome. This deletion involves ~27 genes, including the oxytocin gene OXT, a particularly noteworthy candidate in light of studies in humans and rodents that find evidence for the role of oxytocin in regulating social cognition (17, 18). All validated de novo subject variants are listed in Table 1 with a description of each type of mutation, its methods of validation, genomic location, gene content, and information on the subject’s affected status and family type (simplex, multiplex, or control). Additional details regarding these and other variants detected in this study are provided in table S2. Initially, we detected 19 de novo CNVs in 17 individuals. In one family, subsequent analysis of the parental chromosomes by fluorescence in situ hybridization (FISH) determined that the two de novo events detected (a duplication and deletion) were the result of an unbalanced translocation inherited from an unaffected father who carries the balanced reciprocal translocation. In conclusion, 17 CNVs were confirmed to be de novo in 16 individuals (Table 1), consisting of 14 patients and 2 controls. The majority of these mutations are novel, and only the largest of them (all CNVs >4 Mb in size) have been reported previously in the literature (19–21).
Table 1.
Spontaneous CNVs detected by ROMA. A description of 17 de novo CNVs in 16 subjects is provided, along with the methods used for its validation. The number of unique RefSeq genes within each CNV region is indicated, and when the locus apparently encompasses only a single gene, the gene symbol is listed. Types of validation included (A) higher-resolution microarray scans by 390K ROMA or Agilent 244K CGH, (B) G-banded karyotype, (C) FISH, and (D) microsatellite genotyping. References are listed for four cases where similar de novo CNVs were previously reported in the literature.
| Individual | Locus | Start position | Length | CN change | Family type | Diagnosis | Gender | Validation | # Genes | Single-gene targets | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 63-144-2575 and 2667 | 2q24.2 | 162,212,720 | 99,252 | Loss | Simplex | Autism | Female | A | 1 | SLC4A10 | |
| 61-2710-3 | 2q37.2-q37.3 | 236,414,455 | 6,286,648 | Loss | Simplex | Autism | Male | A, B, D | 50 | (19) | |
| Van69-258900 | 2q37.3 | 238,217,066 | 4,484,037 | Loss | Simplex | Autism | Male | A, D | 43 | (19) | |
| 89-3507-1 | 3p14.2 | 60,746,033 | 101,507 | Loss | Simplex | Autism | Male | A | 1 | FHIT | |
| 63-562-6612 | 3p14.2 | 61,072,100 | 293,096 | Gain | Simplex | Autism | Male | A | 1 | FHIT | |
| AU010604 | 6p23 | 13,997,280 | 1,264,651 | Loss | Multiplex | Autism | Male | A, D | 2 | ||
| 13q14.12-q14.13 | 44,199,441 | 1,943,737 | Loss | A, D | 13 | ||||||
| AU072203 | 7p21.1 | 15,160,118 | 151,880 | Loss | Simplex | Autism | Male | A | 1 | FLJ16237 | |
| AU032903 | 10q11.23-q21.2 | 50,562,149 | 10,916,362 | Gain | Multiplex | Autism | Male | A, B | 23 | ||
| 60-3061-4 | 15q11-q13.33 | 18,526,971 | 12,229,800 | Gain | Simplex | Autism | Male | A, B | 30 | (21) | |
| AU077504 | 16p13.3 | 5,992,836 | 207,980 | Loss | Simplex | Autism | Female | A, B, C, D | 1 | A2BP1 | |
| CG2061 | 16p11.2 | 29,578,715 | 502,574 | Loss | Simplex | Asperger’s | Female | A, C, D | 27 | ||
| 71-259100 | 20p13 | 75,912 | 291,959 | Loss | Simplex | Autism | Female | A, C, D | 7 | ||
| SK-135-C | 20p13 | 2,785,194 | 1,169,205 | Loss | Simplex | Asperger’s | Male | A, D | 23 | ||
| 89-3524-100 | 22q13.31-q13.33 | 45,144,027 | 4,321,856 | Loss | Simplex | Autism | Female | A, B, C, D | 30 | (20) | |
| NA10857 | 2p16.1 | 58,394,177 | 2,786,284 | Gain | Control | Unaffected | Male | A | 7 | ||
| AU070807 | 20p13-p12.3 | 111,824 | 5,316,286 | Gain | Simplex | Unaffected | Female | A | 69 |
These data show that spontaneous copy number changes are more frequent in patients with ASD (14 out of 195) than in unaffected individuals (2 out of 196), with an association that is statistically significant (P = 0.0005). The frequency of spontaneous mutation was 10% (12 out of 118) in our sample of sporadic cases and 3% (2 out of 77) in our sample of cases from multiplex families (Table 2). The frequency of spontaneous mutation in unaffected individuals was 1% (2 out of 196). Most mutations in persons with autism were deletions (12 out of 15); however, the two mutations detected in controls were both duplications.
Table 2.
Increased frequency of de novo CNVs in autism. The numbers of de novo events are listed for our autism sample and for each category of family separately (simplex, multiplex, and nonautism control). The difference between cases and controls was examined, and the statistical significance was determined using Pearson’s chi-square test with simulated P value from 2000 replicates. The P value for the difference in frequency between cases from simplex and multiplex families was also determined.
| Sample group | n | CNVs de novo | Ratio |
P value |
|
|---|---|---|---|---|---|
| χ2 | Multiplex/simplex | ||||
| Simplex autism | 118 | 12 | 0.102 | 0.0005 | 0.043 |
| Multiplex autism | 77 | 2 | 0.026 | 0.59 | |
| Simplex + multiplex | 195 | 14 | 0.072 | 0.0035 | |
| Controls | 196 | 2 | 0.010 | ||
The strong association of de novo CNVs with ASD is consistent with such mutations being a primary cause in most cases rather than merely contributory. A further line of evidence to support this claim is the higher proportion of females among cases with de novo mutations, where the genders of patients consisted of 9 males and 5 females (1.8:1) compared with 163 males and 32 females (5:1) in our overall sample. This reduced gender ratio suggests that de novo CNVs that are detectable by our method have increased penetrance and, thus, contribute to disease more equally in females and males.
A lower rate of de novo mutation in multiplex families is also consistent with a causal role for the mutations reported in this study. An alternative hypothesis is that de novo CNVs are associated with autism indirectly, the consequence of a “fragile-genome disorder” in which many lesions in addition to the ones we detected occur due to an unknown environmental or heritable factor. We regard this alternative as unlikely; first, because we would expect evidence for such a disorder to be present equally in multiplex or simplex families. Another observation that is inconsistent with this alternative hypothesis is that we do not see patients with copy number mutations littered throughout the genome. Instead, de novo CNVs typically involve a single mutational event.
Two of the patients mentioned in Table 1 have a formal diagnosis of Asperger syndrome, which suggests that spontaneous chromosomal imbalances are common across the whole spectrum of the disorder. We examined whether there were many cases of mental retardation [defined as having a nonverbal intelligence quotient (IQ) less than 70] among patients in whom we detected de novo mutations. Clinical data on 60 patients were obtained, and these data included five of the patients in Table 1. The average nonverbal IQ of five cases was 85 and the minimum was 70. Although this average was lower than the average for all patients (100), these data indicate that the de novo mutations identified in our study were not found generally in patients with mental retardation.
Because the difference in rate between autism and control is so marked, we can make a fair presumption that many of the lesions we observed contribute to the disorder. However, the observation of a de novo mutation in a single family is not sufficient evidence to prove that a mutation is causal, nor does it provide unequivocal evidence for the involvement of a specific gene in autism. When an individual gene candidate can be identified, because the mutation affects a single gene or a small number of functional candidates, a straightforward path to validation can be planned, involving sequencing and higher-resolution CNV analysis in additional samples. The principle is illustrated in the recent study by Durand et al. where an intensive survey of variation in a candidate gene, SHANK3, revealed multiple additional variants, including de novo and inherited mutations (22). SHANK3 is one of the genes within the 4.3-MB deletion on chromosome 22q13 that we reported in Table 1, and this region is also a site of recurrent deletions in autism (20). Thus, an aggregate of deletions and coding variants that occurs in patients and not in controls can provide further evidence of a gene’s role in disease once that candidate gene is identified by copy number mutation.
Some of the genes contained within the de novo CNVs we identified are good candidates for further study. A list of all RefSeq genes that overlap with the de novo mutations identified in this study is shown in table S3. Five of the de novo events we detected involved only a single gene and are worthy of mention. A spontaneous deletion was identified involving exons 2–8 of the putative sterol desaturase FLJ16237. Little is known about the function of FLJ16237, but its expression has been detected by in situ hybridization in the superior temporal gyrus of fetal brain (D.H.G.). In a pair of monozygotic twins concordant for autism, we detected a spontaneous deletion of exon 1 of the putative sodium bicarbonate cotransporter SLC4A10. Mutations of the related gene, SLC4A4, are associated with renal tubular acidosis and mental retardation (23). Other single-gene mutations were detected affecting ataxin 2–binding protein 1 (A2BP1/FOX-1) and the fragile histidine triad gene (FHIT) (Table 1). A2BP1 is known to interact with the SCA2, the gene for spinocerebellar ataxia type 2, and A2BP1 mutations have been identified in mental retardation and epilepsy (24). We observed two independent spontaneous mutations of FHIT, a locus that is one of the most fragile sites in the human genome (25), but one of these was not detected in an extract of the original blood from which the cell line was derived.
All five single-gene mutations we detect involve unusually large genes, the smallest of which (SLC4A10) spans 359 kb of the genome. All four target genes rank among the top 3% of human genes by length. This is consistent with previous observations that large genes are frequently located within unstable regions of the genome (26). Our simulations, made by randomly permuting the location of our CNV regions, indicate that this result may simply reflect that large genes, by virtue of their size alone, are more likely to be affected by random rearrangements. Whatever the explanation, large genes do play prominent roles in spontaneous genetic disorders in humans, such as Duchenne muscular dystrophy (27), retinoblastoma (28), and neurofibromatosis (29); and the same could be true for autism.
These studies do not address the mechanisms by which structural mutations of genes contribute to autism. Changes in dosage or structure of genes within a lesion could have quantitative effects on gene function, including haploinsufficiency or altered transcription patterns. Alternatively, hemizygous deletion could result in total loss of function if it is compounded by recessive mutation or monoallelic exclusion of the remaining allele. A genomic rearrangement may also disrupt regulatory elements that influence the expression of neighboring genes; thus, in some cases, a gene related to autism may lie adjacent to the deletion or duplication.
Our findings have implications for an understanding of the genetic basis for ASDs. An important feature of the de novo CNVs we report is that each is individually rare in the population of patients. None of the genomic variants we detected were observed more than twice in our sample, and most were seen but once. Although our sample size is small, these results suggest that lesions at many different loci can contribute to autism, a result consistent with the findings from cytogenetics, as well as consistent with the failure to find common heritable variants with a major effect on disease risk. Lack of recurrence may in fact reflect an underlying reality that autistic behavior can result from many different genetic defects. This would be consistent with the hypothesis that the common features of autism such as failure to develop social skills and repetitive and obsessive behavior may in fact be the consequence of a reaction to many different cognitive impairments, drawing their “commonality” from a normal but maladaptive programmed response of humans early in development to those diverse impairments.
We do not know the full contribution of spontaneous mutation to autism. Population studies divide autism into sporadic and familial or “multiplex.” Our work provides clear evidence that these two classes are indeed genetically distinct. The rate of de novo mutation in multiplex families was significantly lower than for sporadic cases (Table 2, P = 0.04), as would be expected if there were two different genetic mechanisms contributing to risk: spontaneous mutation and inheritance, with the latter being more frequent in families that have multiple affected children.
The rate of spontaneous mutation that we detect in autism is an underestimate. Adding the known rate of cytogenetically visible abnormalities, the total frequency of de novo variation detectable in sporadic cases is ~15% at our current resolution. Because of the limited resolution of genome microarray scans, we expect that we fail to detect the vast majority of CNVs. Much smaller deletions or even point mutations can produce the same consequences as the larger, more easily detectable events. As technology for discovering spontaneous germline mutation in children improves, the proportion of autism cases with detectable events is bound to rise.
We can incorporate a high rate of spontaneous mutation in a genetic model that accounts for both sporadic and familial forms of the disease, based on new mutations that cause autism by haploinsufficiency but have incomplete penetrance, especially in females. Such individuals who escape the phenotypic consequences can then pass on the mutation in an apparently dominant fashion to their children. This model makes very clear predictions that can be tested in the short term.
Our findings highlight how methods for directly detecting CNVs genomewide provide a powerful alternative to traditional gene-mapping approaches for discovering genetic risk factors in autism and in other disorders of complex etiology. Improved technologies for mutation detection, such as high-throughput DNA sequencing and tiling-resolution oligonucleotide arrays, promise to improve our power to identify new mutations associated with disease.
Supplementary Material
Acknowledgments
We wish to thank patients and families for their valuable contributions and the NIMH Center for Collaborative Genetic Studies of Mental Disorders, the Autism Genetic Resource Exchange (AGRE), the Centre d’Etude du Polymorphisme Humain (CEPH), and D. Levy for providing materials or data for the study. We are grateful to G. Fischbach, C. Lord, N. Heinz, E. Cook, L. Brzustowicz, J. and M. Simons, and L. Iakoucheva for helpful discussions throughout the study, and we thank C. Reed, P. Roccanova, J. Lloyd, V. Grubor, L. Rodgers, D. Esposito, L. Hufnagel, X. Zhao, E. Thorolfsdottir, K. A. Olafsdottir, and the staff of Lingen EHF for technical assistance. This work was supported by a grant from the Simons Foundation and by NIMH grant MH076431 to J.S., which reflects cofunding from Autism Speaks, Cure Autism Now, and the Southwestern Autism Research and Resource Center. Additional support was provided by NIMH grant MH61009 to J.S.S., grants to D.S. from Nancy Lurie Marks Family Foundation and the National Alliance for Autism Research (NAAR), and a grant to K.P. and T.L. from the University of Tampere Hospital Medical Fund. AGRE is a program of Cure Autism Now and is supported in part by NIMH grant MH64547 to D.H.G.
Footnotes
References and Notes
- 1.Fombonne E. J Clin Psychiatry. 2005;66(suppl 10):3. [PubMed] [Google Scholar]
- 2.Klauck SM. Eur J Hum Genet. 2006;14:714. doi: 10.1038/sj.ejhg.5201610. [DOI] [PubMed] [Google Scholar]
- 3.Vorstman JA, et al. Mol Psychiatry. 2006;11:1. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 4.Amir RE, et al. Nat Genet. 1999;23:185. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 5.European Chromosome 16 Tuberous Sclerosis Consortium. Cell. 1993;75:1305. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard JK. Am J Hum Genet. 2001;69:124. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feuk L, Carson AR, Scherer SW. Nat Rev Genet. 2006;7:85. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 8.Sebat J, et al. Science. 2004;305:525. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 9.Iafrate AJ, et al. Nat Genet. 2004;36:949. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 10.Koolen DA, et al. Nat Genet. 2006;38:999. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 11.Sharp AJ, et al. Nat Genet. 2006;38:1038. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 12.Shaw-Smith C, et al. Nat Genet. 2006;38:1032. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 13.Shaw-Smith C, et al. J Med Genet. 2004;41:241. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vissers LE, et al. Am J Hum Genet. 2003;73:1261. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquemont ML, et al. J Med Genet. 2006;43:843. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucito R, et al. Genome Res. 2003;13:2291. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson JN, et al. Nat Genet. 2000;25:284. doi: 10.1038/77040. [DOI] [PubMed] [Google Scholar]
- 18.Kosfeld M, Heinrichs M, Zak PJ, Fischbacher U, Fehr E. Nature. 2005;435:673. doi: 10.1038/nature03701. [DOI] [PubMed] [Google Scholar]
- 19.Ghaziuddin M, Burmeister M. J Autism Dev Disord. 1999;29:259. doi: 10.1023/a:1023088207468. [DOI] [PubMed] [Google Scholar]
- 20.Manning MA, et al. Pediatrics. 2004;114:451. doi: 10.1542/peds.114.2.451. [DOI] [PubMed] [Google Scholar]
- 21.Veenstra-VanderWeele J, Cook EH., Jr Mol Psychiatry. 2004;9:819. doi: 10.1038/sj.mp.4001505. [DOI] [PubMed] [Google Scholar]
- 22.Durand CM, et al. Nat Genet. 2007;39:25. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi T, et al. J Am Soc Nephrol. 2001;12:713. doi: 10.1681/ASN.V124713. [DOI] [PubMed] [Google Scholar]
- 24.Bhalla K, et al. J Hum Genet. 2004;49:308. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 25.Huebner K, Croce CM. Nat Rev Cancer. 2001;1:214. doi: 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- 26.Smith DI, Zhu Y, McAvoy S, Kuhn R. Cancer Lett. 2006;232:48. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 27.Gospe SM, Jr, et al. Neurology. 1989;39:1277. doi: 10.1212/wnl.39.10.1277. [DOI] [PubMed] [Google Scholar]
- 28.Orye E, Delbeke MJ, Vandenabeele B. Lancet. 1971;2:1376. doi: 10.1016/s0140-6736(71)92394-4. [DOI] [PubMed] [Google Scholar]
- 29.Viskochil D, et al. Cell. 1990;62:187. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 30.Kent WJ, et al. Genome Res. 2002;12:996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.