Abstract
Few intraspecific genetic linkage maps have been reported for cultivated tomato, mainly because genetic diversity within Solanum lycopersicum is much less than that between tomato species. Single nucleotide polymorphisms (SNPs), the most abundant source of genomic variation, are the most promising source of polymorphisms for the construction of linkage maps for closely related intraspecific lines. In this study, we developed SNP markers based on expressed sequence tags for the construction of intraspecific linkage maps in tomato. Out of the 5607 SNP positions detected through in silico analysis, 1536 were selected for high-throughput genotyping of two mapping populations derived from crosses between ‘Micro-Tom’ and either ‘Ailsa Craig’ or ‘M82’. A total of 1137 markers, including 793 out of the 1338 successfully genotyped SNPs, along with 344 simple sequence repeat and intronic polymorphism markers, were mapped onto two linkage maps, which covered 1467.8 and 1422.7 cM, respectively. The SNP markers developed were then screened against cultivated tomato lines in order to estimate the transferability of these SNPs to other breeding materials. The molecular markers and linkage maps represent a milestone in the genomics and genetics, and are the first step toward molecular breeding of cultivated tomato. Information on the DNA markers, linkage maps, and SNP genotypes for these tomato lines is available at http://www.kazusa.or.jp/tomato/.
Keywords: DNA marker, linkage map, single nucleotide polymorphism, Solanum lycopersicum, tomato
1. Introduction
Genetics in tomato (Solanum lycopersicum) and its wild relatives, including S. chilense, S. habrochaites, S. pimpinellifolium, and S. pennellii, have been greatly advanced since molecular markers have become available.1 During the past two decades, several genetic maps in tomato have been reported, with a total of more than 2000 loci detected by restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), cleaved amplified polymorphic sequence (CAPS), and simple sequence repeat (SSR) markers based on the mapping of populations derived from crosses between tomato and related wild species.2–6 Recently, 1282 novel SSR markers and 151 intronic polymorphic markers were mapped onto an interspecific map, ‘Tomato-EXPEN 2000’ derived from a cross between S. lycopersicum and S. pennellii.7 Such efforts have resulted in the identification of a number of quantitative trait loci (QTLs) and genes for fruit morphology,8–11 disease resistance,12–15 and other agronomical traits.16 The identified genes, e.g. Cf-4, Tm-2, and Sw-5, have already been used for tomato breeding through advanced-backcross and introgression-line strategies using molecular markers.1 Though significant advances in molecular genetics and breeding have been reported in tomato, most of them were based on interspecific crosses because genetic diversity in the cultivated tomato is lower than in its wild relatives.17 Meanwhile, intraspecific maps are required to identify QTLs for agronomically important traits, which are the targets of practical breeding programs. However, only one intraspecific map, based on AFLP, RFLP, and random amplified polymorphic DNA (RAPD) markers, has been reported for S. lycopersicum.18
Single nucleotide polymorphisms (SNPs) are the most abundant source of variation in the genome for both intragenic and intergenic regions. They therefore represent a valuable basis for the development of molecular markers for identification of polymorphisms among closely related lines. Previous studies have suggested that DNA markers developed from intergenic regions tend to cluster in heterochromatic portions of chromosomes, while those derived from genic regions disperse along entire chromosomes.7,19–22 Therefore, SNPs, especially those located in intragenic regions, are expected to distribute randomly along the whole genome. In addition, novel techniques based on the DNA microarray method allow high-throughput SNP genotyping.23 For these reasons, SNP markers derived from intragenic regions are the most informative markers for genome-wide genetic analysis in intraspecific tomato populations. By comparing expressed sequence tags (ESTs) in tomato and related wild species, approximately 40 000 candidate SNPs have been identified.24–27 Since then, the number of ESTs derived from several tomato cultivars has increased to approximately 300 000, all of which are available in the public DNA databases, e.g. DNA Data Bank of Japan (DDBJ: http://www.ddbj.nig.ac.jp/), Sol Genomics Network (SGN: http://solgenomics.net/), and MiBASE (http://www.pgb.kazusa.or.jp/mibase/).
The tomato is regarded as a model plant not only for the Solanaceae but also for other fruiting plants.28 A miniature dwarf cultivar, ‘Micro-Tom’, originally bred for home gardening purposes,29 has drawn attention as a model tomato line because of its small plant size, short life cycle, easy transformation, and availability of transposon-tagging systems for use in reverse genetics.30 Various genomic and genetic resources have been developed for ‘Micro-Tom’. These include mutagenized lines,31,32 effective transformation systems,33,34 metabolite annotations,35 full-length cDNAs,36 and BAC-end sequences (Asamizu et al., released in the public DNA database with accession numbers: FT227487–FT321168). ‘Micro-Tom’ seeds are available through two seed stock centers: the Tomato Genetics Resource Center at the University of California, Davis (USA, accession no. LA3911) and the National Bio-Resource Project at the University of Tsukuba (Japan, accession no. TOMJPF00001).
In this study, we developed SNP markers using publicly available ESTs from several tomato cultivars and designed an SNP-genotyping platform using the GoldenGate® assay (Illumina, San Diego, CA, USA) in order to accelerate genetic studies and molecular breeding in tomato. SNP markers, along with SSR markers and intronic polymorphic markers, which were developed and mapped onto the interspecific map Tomato-EXPEN 2000 by Shirasawa et al.,7 were applied to create linkage maps using two mapping populations derived from crosses between ‘Micro-Tom’ and ‘Ailsa Craig’, a greenhouse-type tomato, and between ‘Micro-Tom’ and ‘M82’, a processing tomato. In addition, the polymorphism of the SNP markers was investigated in cultivated tomato lines in order to estimate the transferability of the SNPs to breeding materials.
2. Materials and methods
2.1. Plant materials
Two F2 mapping populations, AMF2 and MMF2, each derived by crossing two S. lycopersicum lines, were used for the construction of the linkage maps. AMF2 (n = 120) was derived from a cross between the ‘Ailsa Craig’ and ‘Micro-Tom’ lines, while MMF2 (n = 135) was derived from a cross between the ‘M82’ and ‘Micro-Tom’ lines. AMF2 and MMF2 were generated in the National Institute of Vegetable and Tea Science, Japan, and in the Institut National de la Recherche Agronomique, France, respectively (Table 1). To address potential residual heterozygosity in the parental ‘Micro-Tom’ lines used to create AMF2 and MMF2, they are distinguished in this study by the designations ‘Micro-Tom_AM’ and ‘Micro-Tom_MM’, respectively. Along with the four parental lines of the mapping populations, 22 lines, including 16 inbred and 6 hybrid tomato lines, and an S. pennellii line (‘LA716’) were used for polymorphic analysis of SNPs (Table 1). Total DNA for each line was extracted using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany).
Table 1.
Description of plant materials
| Line name | Note | Sourcea | Accession number | SNP validationb |
|---|---|---|---|---|
| Parental lines of mapping populations | ||||
| Micro-Tom_AM | Inbred line | NIVTS | Tested | |
| Ailsa Craig | Inbred line | NBRP | TOMJPF00004 | Tested |
| Micro-Tom_MM | Inbred line | INRA | ||
| M82 | Inbred line | INRA | Tested | |
| Tomato lines for SNP typing | ||||
| Aichi First | Inbred line | NBRP | TOMJPF00003 | |
| Best of All | Inbred line | NIVTS | LS3908 | |
| Earliana | Inbred line | TGRC | LA3238 | Tested |
| Fruit | Inbred line | NIVTS | LS1100 | |
| Furikoma | Inbred line | NIVTS | LS3903 | |
| Heinz 1706-BG | Inbred line | NIVTS | LS461 | Tested |
| LA925 | Inbred line | Cornell University | Tested | |
| Marglobe | Inbred line | TGRC | LA0502 | Tested |
| Money Maker | Inbred line | TGRC | LA2706 | Tested |
| Ponderosa | Inbred line | NIVTS | LS1728 | |
| Rio Grande | Inbred line | TGRC | LA3343 | Tested |
| Rutgers | Inbred line | TGRC | LA1090 | Tested |
| San Marzano | Inbred line | NIVTS | LS4956 | Tested |
| Tomato Chuukanbohon Nou 9 | Inbred line | NIVTS | ||
| Tomato Chuukanbohon Nou 11 | Inbred line | NIVTS | ||
| Geronimo | F1 hybrid | De Ruiter Seeds Co. | Tested | |
| Labell | F1 hybrid | De Ruiter Seeds Co. | Tested | |
| Matrix | F1 hybrid | De Ruiter Seeds Co. | Tested | |
| Momotaro 8 | F1 hybrid | Takii Seeds Co. | Tested | |
| Reika | F1 hybrid | Sakata Seeds Co. | Tested | |
| Regina | Inbred line, cherry type | Sakata Seeds Co. | ||
| Sweet100 | F1 hybrid, cherry type | Vilmorin Seeds Co. | Tested | |
| LA716 | Inbred line, S. pennellii | Cornell University | ||
aNBRP: University of Tsukuba in National Bio-Resource Project of MEXT, Japan; INRA: National Institute for Agricultural Research, France; NIVTS: National Institute of Vegetable and Tea Science, Japan; TGRC: Tomato Genetics Resource Center, University of California, Davis, USA.
bLines that used for validation of 82 eSNPs prior to design a SNP genotyping platform using Illumina GoldenGate® assay.
2.2. Development of SNP markers and polymorphic analysis
A total of 229 086 EST sequences from S. lycopersicum, retrieved from two public databases, SGN (http://solgenomics.net/) and MiBASE (http://www.pgb.kazusa.or.jp/mibase/), were used for identification of eSNPs, i.e. SNPs discovered in silico. The ESTs registered in MiBASE were derived only from ‘Micro-Tom’, while those registered in SGN were developed from 19 tomato lines including ‘Micro-Tom’. The retrieved EST sequences were assembled using the MIRA program.37 The eSNPs were then selected according to the following three criteria: (i) only nucleotides with Phred scores of 15 or more were considered candidates for eSNPs, (ii) a nucleotide at an eSNP site should be identical among multiple sequences within a given line, and (iii) no other SNP candidates should be detected on the flanking sequences 10 bp upstream and downstream of a given candidate.
In order to validate the credibility of the identified eSNP, nucleotide sequences of PCR products containing the eSNP regions were determined by direct sequencing using a DNA sequencer (ABI-3730xl, Applied Biosystems, Foster City, CA, USA). A total of 82 primer pairs were designed in flanking regions of the randomly selected target eSNPs using the Primer3 program.38 PCR was performed for 17 tomato lines listed in Table 1 in a 5-µl reaction mixture containing 0.5 ng genomic DNA, 1× PCR buffer (Bioline, London, UK), 3 mM MgCl2, 0.04 U BIOTAQ™ DNA polymerase (Bioline), 0.2 mM dNTPs, and 0.8 µM of each of the primers. The modified ‘touchdown PCR’ protocol was used as described previously.39
After validation of the 82 eSNPs, a total of 1536 eSNPs were subjected to polymorphic analysis for the two mapping populations and the 23 tomato lines described above using the GoldenGate® assay system (Illumina). Allele- and locus-specific oligonucleotides were designed from the flanking sequences of the 1536 SNP sites using the iCom website (https://icom.illumina.com/). Polymorphic analysis of the SNPs was performed according to the standard protocol of the GoldenGate® assay, and the data analysis was performed using GenomeStudio Data Analysis software (Illumina).
SNPs in DWARF (D) and SELF-PRUNING (SP) were analyzed using the dCAPS and CAPS methods, respectively. PCR was performed under the same conditions as described above. The primer sequences are shown in Supplementary Table S1. The PCR products from the D and SP genes were digested with PstI and MvaI, respectively, and were subjected to electrophoresis on native 10% polyacrylamide gels in 1× TBE buffer. The resulting DNA bands were then stained with ethidium bromide.
2.3. Mapping of SSR and intronic polymorphic markers on AMF2
A total of 3510 tomato genomic SSR (TGS), 2047 tomato EST-SSR (TES), and 166 tomato EST-derived intronic polymorphic (TEI) markers, developed by Shirasawa et al.,7 were used for segregation analysis of the AMF2 population (Supplementary Table S1). The polymorphic analyses of the markers were performed as described previously.7 Primer information for the tested markers is available at http://www.kazusa.or.jp/tomato/.
2.4. Linkage analysis
Linkage analysis was performed using the JoinMap® program, version 4.40 The segregated data were classified into 12 linkage groups, which corresponded to the Tomato-EXPEN 2000 map,7 using the grouping module of JoinMap® with LOD scores of 4.0–10.0. The marker order and relative genetic distances were calculated by the regression-mapping algorithm with the following parameters: Haldane's mapping function, recombination frequency ≤0.35, and LOD score ≥2.0.
3. Results
3.1. In silico SNP mining and validation
A total of 170 586 and 58 500 EST sequences available in SGN and MiBASE, respectively, along with data on their quality, were used for in silico SNP mining. The name of the original tomato line for each EST was obtained from the DDBJ database (http://www.ddbj.nig.ac.jp/). In total, 229 086 ESTs derived from 20 tomato cultivars, the average length of which was 497 bp, were used for assembly (Table 2).
Table 2.
Number of ESTs and their original sources used for assembling
| Line namea | No. of ESTs |
|---|---|
| TA496 | 106 142 |
| Micro-Tom | 101 157 |
| Rio Grande PtoR | 8803 |
| R11-13 | 5031 |
| R11-12 | 4925 |
| TA492 | 2120 |
| West Virginia 106 | 861 |
| Money Maker | 11 |
| Ailsa Craig | 7 |
| VF36 | 7 |
| Momotaro | 4 |
| Zhongshu 4 | 4 |
| Betterboy | 3 |
| Vendor | 3 |
| House Odoriko | 2 |
| Rutgers | 2 |
| M82 | 1 |
| Pera | 1 |
| Rio Grande | 1 |
| UC82B | 1 |
| Total | 229 086 |
aNames of tomato lines used for EST generation.
Assembly was performed using nucleotides with Phred scores ≥15. As a result, a total of 20 274 contigs, the average length of which was 775 bp, and 29 698 singletons were generated. From initial alignment data from all 20 274 contigs, a total of 5607 eSNP sites were identified in 2634 of these contigs (Supplementary Tables S2 and S3). We gave an SNP code to each eSNP according to the following rule: contig name and position of the eSNP on the contig, linked with an underscore, e.g. the 112th position on contig 2758 was given the following SNP code: 2758_112.
Before designing the SNP genotyping platform (using the Illumina GoldenGate® assay), 82 randomly selected eSNPs were tested in 17 tomato lines (Table 1) by direct sequencing of fragments amplified by PCR. As a result, 55 (67%) out of the 82 examined eSNP candidates were experimentally confirmed as SNPs at the predicted positions, indicating that approximately 67% of the 5607 eSNPs detected in silico represent true SNPs in the tomato lines used in the present study. In addition, 40 (49%) and 50 (61%) of the 82 eSNPs segregated between the two mapping parents for AMF2 and MMF2, respectively.
For SNP genotyping, a total of 1536 SNPs were selected from the 5607 eSNPs, as follows: (i) one eSNP was selected from each contig and the Selected-BAC-Mixture contig released from the Kazusa Tomato SBM & Marker Database (http://www.kazusa.or.jp/tomato/); (ii) an SNP score of more than 0.6, as determined by the iCom website of Illumina (https://icom.illumina.com/), was required for each of these eSNPs. As reported by the GoldenGate® assay, 1338 (87%) out of the 1536 SNPs could be properly genotyped in the 279 plants. These included the two mapping populations (AMF2 and MMF2) and 23 other tomato lines. The remaining 198 (13%) eSNPs failed to be genotyped because fluorescent signals for these eSNPs did not form clusters pursuant to the criteria required by the GenomeStudio Data Analysis software (Illumina).
3.2. Mapping of SNP, SSR, and intronic markers
In the AMF2 population, 648 of the 1338 available SNPs (48.4%) generated segregation data, a similar ratio to that determined in the validation of the 82 eSNPs. Two SNP markers designed in the D and SP genes, for which ‘Micro-Tom’ has mutant alleles,41 showed polymorphism between ‘Ailsa Craig’ and ‘Micro-Tom’. Along with the SNP markers, a total of 5723 previously reported markers, including 2047 EST-SSR (TES), 3510 genomic-SSR (TGS), and 166 intronic (TEI) markers, were used for the polymorphic analysis. As a result, 96 TES (4.7%), 223 TGS (6.3%), and 28 TEI (16.8%) markers exhibited polymorphism between the parental lines. In total, 997 markers were used to construct the AMF2 linkage map.
In the MMF2 population, 640 of the 1338 available SNPs (47.9%) segregated. This ratio was over 10% less than that determined in the validation of the 82 eSNPs, suggesting that the result of the eSNP validation was overestimated. The SNP on the D gene showed polymorphism in the MMF2 mapping population, while two parental lines detected the mutated sp allele for the SP gene. In total, 641 segregated markers were used to construct the MMF2 map.
3.3. Construction of linkage maps
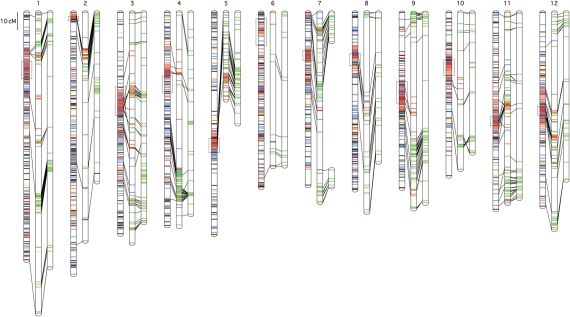
For AMF2, a total of 989 of the 997 segregated loci (99.2%) formed 12 linkage groups (LGs), while 637 of the 641 segregated loci (99.4%) formed 13 linkage groups for MMF2. The total sizes of the LGs of the AMF2 and MMF2 maps were 1467.8 and 1422.7 cM, respectively (Table 3, Fig. 1, Supplementary Table S1). Combining the two maps yielded a total of 1137 markers, including 793 SNP, 221 TGS, 93 TES, 28 TEI, and 2 gene markers, located on the intraspecific map. Among these, 488 SNP markers were commonly located on both linkage maps, while 157 and 148 marker loci were specific to the AMF2 map and the MMF2 map, respectively. Chromosome 7 (Chr07) of MMF2 divided into two linkage groups, Chr07p and Chr07q, which were located at the upper and the lower portions, respectively, of Chr07 of Tomato-EXPEN 2000. The average lengths of the intervals between two loci on the AMF2 and the MMF2 maps were calculated to be 1.5 and 2.2 cM, respectively.
Table 3.
Length, number of mapped loci, and segregation distortion of the AMF2 and MMF2 maps
| Chromosome | AMF2 |
MMF2 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (cM) | Number of loci |
Segregation distortion ratio (%) | Length (cM) | Number of loci |
Segregation distortion ratio (%) | ||||||||
| TGS | TES | TEI | SNP | Gene | Total | SNP | Gene | Total | |||||
| Chr01 | 187.0 | 18 | 8 | 5 | 62 | 0 | 93 | 0.0 | 158.4 | 57 | 0 | 57 | 3.5 |
| Chr02 | 141.9 | 24 | 6 | 2 | 39 | 1 | 72 | 2.8 | 105.1 | 60 | 1 | 61 | 9.8 |
| Chr03 | 143.2 | 29 | 6 | 3 | 70 | 0 | 108 | 11.1 | 130.4 | 100 | 0 | 100 | 4.0 |
| Chr04 | 132.0 | 15 | 15 | 4 | 114 | 0 | 148 | 3.4 | 125.3 | 97 | 0 | 97 | 4.1 |
| Chr05 | 54.2 | 45 | 8 | 1 | 51 | 0 | 105 | 2.9 | 69.2 | 70 | 0 | 70 | 0.0 |
| Chr06 | 95.5 | 3 | 0 | 1 | 9 | 1 | 14 | 7.1 | 95.1 | 18 | 0 | 18 | 16.7 |
| Chr07 | 117.9 | 8 | 17 | 1 | 91 | 0 | 117 | 3.4 | 29.0a | 54a | 0 | 54 | 1.9 |
| Chr08 | 124.0 | 5 | 5 | 0 | 22 | 0 | 32 | 0.0 | 92.8 | 23 | 0 | 23 | 0.0 |
| Chr09 | 121.7 | 13 | 7 | 7 | 53 | 0 | 80 | 15.0 | 118.2 | 53 | 0 | 53 | 17.0 |
| Chr10 | 97.3 | 6 | 3 | 0 | 22 | 0 | 31 | 6.5 | 87.2 | 26 | 0 | 26 | 11.5 |
| Chr11 | 118.4 | 25 | 11 | 3 | 62 | 0 | 101 | 55.4 | 115.3 | 43 | 0 | 43 | 2.3 |
| Chr12 | 134.7 | 30 | 7 | 1 | 50 | 0 | 88 | 0.0 | 104.0 | 35 | 0 | 35 | 2.9 |
| Total | 1467.8 | 221 | 93 | 28 | 645 | 2 | 989 | 9.8 | 1422.7 | 636 | 1 | 637 | 5.3 |
aChr07 of the MMF2 were divided into two linkage groups, Chr07p and Chr07q. The numbers indicate total value of two linkage groups.
Figure 1.
Intraspecific linkage maps for tomato. The middle and right bars of each linkage group represent maps of the AMF2 and MMF2 lines, respectively. The left bar represents a linkage map of Tomato-EXPEN 2000, which is an interspecific map constructed in our previous study.7 The lines on each linkage group show SNP (green), TGS (red), TES (blue), TEI (yellow), and the other available (black) markers. Common markers between two maps are connected with black lines. Boxes on the Tomato-EXPEN 2000 map indicate heterochromatic regions. Detailed information on the markers, including marker positions, is shown in Supplementary Table S1, and is also available at http://www.kazusa.or.jp/tomato/.
Segregation distortions were observed in the two maps. In the AMF2 map, 9.8% of the marker loci showed segregation distortions, ranging from 0.0% for Chr01, Chr08, and Chr12, to 55.4% for Chr11 (Table 3). In the MMF2 map, 5.3% of the marker loci were distorted, ranging from 0.0% for Chr05 and Chr08, to 17.0% for Chr09 (Table 3). The linkage groups harboring severe segregation distortions were different between the two mapping populations, especially between Chr11 of AMF2 (55.4%) and that of MMF2 (2.3%), suggesting Chr11 of ‘Ailsa Craig’ might have transmission ratio distorters.
3.4. Polymorphic analysis of the SNP markers in tomato cultivars and S. pennellii
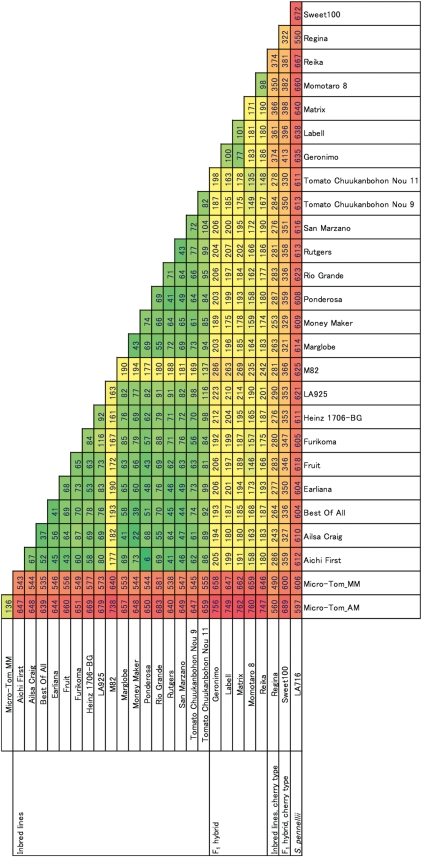
A total of 916 (68.5%) out of the 1338 SNP markers showed polymorphisms in at least one line among the 27 tomato lines listed in Table 1 (Supplementary Table S4). The polymorphic ratio was similar to the ratio determined during the PCR-based validation of the 82 eSNPs. In ‘LA719’ (S. pennellii) and ‘Sweet 100’, no data were obtained for 229 (17.1%) and one SNP markers, respectively. The polymorphic ratios differed according to the combination of tomato lines (Fig. 2), and the number of segregated SNPs between any two lines among the 27 lines was 255.0 (19.1%) on average. A total of 608.2 SNPs (45.5%) were identified between ‘Micro-Tom’ and the other inbred lines, on average, while only 80.8 SNPs (6.0%) were identified among the 17 inbred tomato lines. Within the 17 inbred tomato lines, ‘M82’ showed the highest number of polymorphisms: 176.3 SNPs (13.2%) on average, which was twice as high as that of the other lines. SNPs between the F1 hybrid cultivars and the inbred lines were found at 190.6 loci (14.2%) on average. The two cherry-type tomato cultivars showed higher polymorphisms than the inbred tomato lines, with 310.1 (23.2%) SNPs on average. When 26 S. lycopersicum lines were compared with S. pennellii, on average, 618.5 out of the 1338 loci (46.2%) were polymorphic. Heterozygosity was observed at multiple SNP sites in all six F1 hybrid cultivars, ranging from 69 (5.2%) in ‘Matrix’ to 229 (17.1%) in ‘Sweet 100’. In the inbred line ‘Rio Grande’, 25 (1.9%) heterozygous SNPs were identified.
Figure 2.
Number of SNP markers identified by the GoldenGate® assay among 27 tomato and S. pennellii lines. The numbers show the loci of the 1338 SNPs genotyped in this study. The colors in each cell represent a continuum of polymorphic ratios: lower ratios are represented by green color, higher ratios by red color, and middle ratios by yellow color.
It is noteworthy that 136 SNPs (10.2%) were identified between ‘Micro-Tom_AM’ and ‘Micro-Tom_MM’, the parental lines of AMF2 and MMF2, respectively (Fig. 2). Out of these 136 SNP loci, 134 mapped onto the AMF2 and/or the MMF2 maps, mainly on Chr04 (44 loci), Chr07 (38 loci), and Chr12 (36 loci) (Supplementary Table S1). ‘Micro-Tom_AM’ had a higher number of polymorphisms, in comparison with the other 25 examined lines, than ‘Micro-Tom_MM’ (Fig. 2). For example, 738 loci (55.2%) showed polymorphisms between ‘Micro-Tom_AM’ and ‘M82’, while only 640 SNPs (47.8%) were found between ‘Micro-Tom_MM’ and ‘M82’. It is likely that this difference resulted in an overestimation of the number of segregated loci between ‘Micro-Tom’ and ‘M82’ in the 82-eSNP PCR-based validation.
4. Discussion
To our knowledge, the two genetic linkage maps presented here are the first intraspecific maps for S. lycopersicum with SNPs and other PCR-based co-dominant markers. The AMF2 and MMF2 genetic linkage maps comprise a total of 989 and 637 DNA marker loci, respectively, including SNP, SSR, and intron polymorphic and gene markers. Because the SNP markers developed in this study showed a higher degree of polymorphism among the tomato cultivars than SSR markers, SNP information is greatly important to be utilized for genetic analyses in cultivated tomato, including gene mapping, QTL analysis, population genetics, and marker-assisted breeding. In addition, the genomic tools developed in this study will be valuable for exploiting the extensive artificially induced genetic variability created by ethyl methane sulfonate (EMS) mutagenesis in ‘Micro-Tom’ mutant collections. For example, they could allow, by forward genetic approaches, the identification of the causal mutations for remarkable fruit and plant phenotypes.
In the SNP genotyping by the GoldenGate® assay, 1338 (87%) of the 1536 SNPs could be successfully genotyped. In other crops, successful ratio of SNP genotyping by the GoldenGate® assay is reported to be raging from 79 to 92%,42–45 which fits to the result of the present study. In order to improve the ratio, we suggest additional three criteria to select SNPs for genotyping. The first is elimination of SNP positioned near junction site of intron and exon, because intron inhibits hybridization of allele- and locus-specific oligonucleotide to the target sequence based on EST. It can be achieved by comparing the sequences of EST with those of genome, if available. Next is avoidance designing SNP markers on multi-copy genes, which disrupts the fluorescent-signal clusters on the GenomeStudio Data Analysis software. Selection of accurate SNP site, e.g. with high-quality value and/or with highly coverage of sequence fragments, is also important. Large scale of genome analysis by massively parallel DNA sequencers would be convenient to overcome these matters.
In interspecific linkage maps of tomato and its relatives, markers derived from ESTs tend to distribute randomly along the genome, while markers derived from random genomic regions, e.g. RAPD, AFLP, and genomic SSRs, tend to form clusters in heterochromatic regions.7,19–22 In this study, however, the marker loci did not disperse along the two linkage maps derived from AMF2 and MMF2, despite the fact that most markers were developed from ESTs. Comparison between the maps of Tomato-EXPEN 2000 and the two intraspecific mapping populations did not indicate any evidence of an obvious relationship between the marker clusters on the maps and chromosome structures, i.e. the heterochromatic and euchromatic regions (Fig. 1). It can be assumed that the marker clusters correspond to probable integration regions originating from ‘Lycopersicon minutum’, an ancestral line of ‘Micro-Tom’,29 which belongs to the S. chmielewskii and S. neorickii complex.46 Such regions are expected to show higher frequencies of polymorphism than the other regions, which originate from cultivated lines.
Out of the 989 SNP markers on the AMF2 map, 489 were located on the two intraspecific maps generated in this study and 155 had already been located on the interspecific map generated from Tomato-EXPEN 2000.7 The common markers for the three maps allow the alignment and connection of these maps as shown in Fig. 1. Significant translocation and inversion of chromosome were not observed between intra- and interspecific maps, meaning the order of genes would be conserved in two species, and the genome sequence of tomato (S. lycopersicum) could be used as a reference genome of S. pennellii. In addition, it is likely that the AMF2 and MMF2 maps cover the whole tomato genome except for the middle part of Chr07 on the MMF2 map, and that the marker order is mostly conserved in the three maps. One possible approach to connecting the two linkage groups of Chr07 on the MMF2 map would be to develop a novel mapping population between ‘M82’ and ‘Micro-Tom_AM’ instead of ‘Micro-Tom_MM’ because SNPs on Chr07 segregated more frequently between ‘M82’ and ‘Micro-Tom_AM’.
Indeed, we found that 136 of the 1338 tested SNP markers (10.2%) showed polymorphisms between ‘Micro-Tom_AM’ and ‘Micro-Tom_MM’ (Fig. 2), indicating possible residual heterosis in ‘Micro-Tom’. It is assumed that these loci had not been fixed at the time of ‘Micro-Tom’ being released, although ‘Micro-Tom’ seeds are propagated and distributed in the F12 generation after a crossing.29 Theoretically, the heterozygosity of genome in the F12 generation is calculated to be 0.05% [=(1/2)12-1] in self-pollinating plants, which means that most of the genomic regions are expected to be homozygous. Most of the polymorphic markers between the two ‘Micro-Tom’ lines were mapped on Chr04, Chr07, and Chr12. This result suggested that ‘Micro-Tom’ might have been bred under natural and/or artificial selection pressure from the regions under the influence of heterosis or crossing incompatibilities between S. lycopersicum and L. minutum. Alternatively, multiple lines might have been selected as ‘Micro-Tom’ from the breeding population before the complete fixation of the genotypes of each plant.
Though ‘Micro-Tom’ itself, bred as an ornamental plant, has little agricultural value, its genes may be of great value to agriculture. ‘Micro-Tom’ has resistance to several diseases, caused by Alternaria alternata, Corynespora cassiicola, Fusarium oxysporum, and Pseudomonas syringae.47 Moreover, a large number of mutant lines have been developed using ‘Micro-Tom’.31,32 The markers and maps developed in this study may therefore be useful for introgression breeding for disease resistance or targeted genes identified in ‘Micro-Tom’ or its mutant lines. Indeed, ‘Micro-Tom’ mutant lines carry mutated alleles that may confer high agricultural value to tomato, e.g. alleles causing large variations in fruit color, shape, size, and composition. Mutants may also help to decipher the mechanisms controlling specific traits in tomato.
In this study, we demonstrated the validity of the strategy of combining large-scale eSNP discovery with high-throughput SNP genotyping assays. Comparison of sequence data from tomato cultivars has been reported as an efficient strategy for developing a large number of SNP markers for tomato cultivars.24,48,49 Today, extensive amounts of sequence data from crop genomes can be easily collected using massively parallel DNA sequencers.50 In addition, genomic sequences from The International Tomato Genome Sequencing Consortium of SGN will soon become available.51 The accumulating genome sequences can be used to develop custom SNP markers within cultivated tomato. The molecular markers and genetic linkage maps developed in the present study represent one of the initial milestones in the fusion of genomics, genetics, and molecular breeding in cultivated tomato.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Availability
Information on the SNP and SSR markers, the AMF2 and MMF2 linkage maps, and the SNP genotypes for the tomato lines investigated in the current study are available at http://www.kazusa.or.jp/tomato/.
Funding
This work was supported by the Kazusa DNA Research Institute Foundation and the Ministry of Agriculture, Forestry, and Fisheries of Japan with the cooperation of the Genomics for Agricultural Innovation Foundation (DD-4010).
Supplementary Material
Acknowledgements
We are grateful to Dr K. Aoki (Kazusa DNA Research Institute, Japan) for providing the EST data for Micro-Tom. Plant materials were provided by Dr S. D. Tanksley (Cornell University, USA), Dr T. Ariizumi (University of Tsukuba through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan), Dr T. Saito (National Institute of Vegetable and Tea Science, Japan), and Dr S. M. Tam (Tomato Genomic Resource Center, University of California, Davis, USA).
References
- 1.Labate J.A., Grandillo S., Fulton T., et al. Tomato. In: Cole C., editor. Genome Mapping and Molecular Breeding in Plants, vol. 5. New York: Springer; 2007. pp. 1–125. [Google Scholar]
- 2.Paterson A.H., Lander E.S., Hewitt J.D., Peterson S., Lincoln S.E., Tanksley S.D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature. 1988;335:721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- 3.Tanksley S.D., Ganal M.W., Prince J.P., et al. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vicente M.C., Tanksley S.D. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics. 1993;134:585–596. doi: 10.1093/genetics/134.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paran I., Goldman I., Tanksley S.D., Zamir D. Recombinant inbred lines for genetic mapping in tomato. Theor. Appl. Genet. 1995;90:542–548. doi: 10.1007/BF00222001. [DOI] [PubMed] [Google Scholar]
- 6.Fulton T.M., Nelson J.C., Tanksley S.D. Introgression and DNA marker analysis of Lycopersicon peruvianum, a wild relative of the cultivated tomato, into Lycopersicon esculentum, followed through three successive backcross generations. Theor. Appl. Genet. 1997;95:895–902. [Google Scholar]
- 7.Shirasawa K., Asamizu E., Fukuoka H., et al. An interspecific linkage map of SSR and intronic polymorphism markers in tomato. Theor. Appl. Genet. 2010;121:731–739. doi: 10.1007/s00122-010-1344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frary A., Nesbitt T.C., Grandillo S., et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Van Eck J., Cong B., Tanksley S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl Acad. Sci. USA. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong B., Barrero L.S., Tanksley S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- 11.Xiao H., Jiang N., Schaffner E., Stockinger E.J., van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 12.Dixon M.S., Jones D.A., Keddie J.S., Thomas C.M., Harrison K., Jones J.D. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell. 1996;84:451–459. doi: 10.1016/s0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- 13.Martin G.B., Brommonschenkel S.H., Chunwongse J., et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 14.Jones D.A., Thomas C.M., Hammond-Kosack K.E., Balint-Kurti P.J., Jones J.D. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- 15.Thomas C.M., Jones D.A., Parniske M., et al. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant. Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridman E., Pleban T., Zamir D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA. 2000;97:4718–4723. doi: 10.1073/pnas.97.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J C., Tanksley S.D. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- 18.Saliba-Colombani V., Causse M., Gervais L., Philouze J. Efficiency of RFLP, RAPD, and AFLP markers for the construction of an intraspecific map of the tomato genome. Genome. 2000;43:29–40. [PubMed] [Google Scholar]
- 19.Frary A., Xu Y., Liu J., Mitchell S., Tedeschi E., Tanksley S. Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theor. Appl. Genet. 2005;111:291–312. doi: 10.1007/s00122-005-2023-7. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama A., Asamizu E., Negoro S., et al. Characterization of tomato SSR markers developed using BAC-end and cDNA sequences from genome databases. Mol. Breed. 2009;23:685–691. [Google Scholar]
- 21.Tang X., Szinay D., Lang C., et al. Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics. 2008;180:1319–1328. doi: 10.1534/genetics.108.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Tang X., Cheng Z., Mueller L., Giovannoni J., Tanksley S.D. Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics. 2006;172:2529–2540. doi: 10.1534/genetics.106.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta P.K., Rustgi S., Mir R.R. Array-based high-throughput DNA markers for crop improvement. Heredity. 2008;101:5–18. doi: 10.1038/hdy.2008.35. [DOI] [PubMed] [Google Scholar]
- 24.Jimenez-Gomez J.M., Maloof J.N. Sequence diversity in three tomato species: SNPs, markers, and molecular evolution. BMC Plant Biol. 2009;9:85. doi: 10.1186/1471-2229-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labate J.A., Baldo A.M. Tomato SNP discovery by EST mining and resequencing. Mol. Breed. 2005;16:343–349. [Google Scholar]
- 26.Yang W., Bai X., Kabelka E., et al. Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Mol. Breed. 2004;14:21–34. [Google Scholar]
- 27.Yamamoto N., Tsugane T., Watanabe M., et al. Expressed sequence tags from the laboratory-grown miniature tomato (Lycopersicon esculentum) cultivar Micro-Tom and mining for single nucleotide polymorphisms and insertions/deletions in tomato cultivars. Gene. 2005;15:127–134. doi: 10.1016/j.gene.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Ezura H. Tomato is a next-generation model plant for research and development. J. Jpn Soc. Hort. Sci. 2009;78:1–2. [Google Scholar]
- 29.Scott J.W., Harbaugh B.K. Micro-Tom—a miniature dwarf tomato. Florida Agr. Expt. Sta. Circ. 1989;370:1–6. [Google Scholar]
- 30.Meissner R., Jacobson Y., Melamed S., et al. A new model system for tomato genetics. Plant J. 1997;12:1465–1472. [Google Scholar]
- 31.Matsukura C., Yamaguchi I., Inamura M., et al. Generation of gamma irradiation-induced mutant lines of the miniature tomato (Solanum lycopersicum L.) cultivar ‘Micro-Tom. Plant Biotechnol. 2007;24:39–44. [Google Scholar]
- 32.Watanabe S., Mizoguchi T., Aoki K., et al. Ethylmethanesulfonate (EMS) mutagenesis of Solanum lycopersicum cv. Micro-Tom for large-scale mutant screens. Plant Biotechnol. 2007;24:33–38. [Google Scholar]
- 33.Dan Y., Yan H., Munyikwa T., Dong J., Zhang Y., Armstrong C.L. MicroTom—a high-throughput model transformation system for functional genomics. Plant Cell Rep. 2006;25:432–441. doi: 10.1007/s00299-005-0084-3. [DOI] [PubMed] [Google Scholar]
- 34.Sun H.J., Uchii S., Watanabe S., Ezura H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006;47:426–431. doi: 10.1093/pcp/pci251. [DOI] [PubMed] [Google Scholar]
- 35.Iijima Y., Nakamura Y., Ogata Y., et al. Metabolite annotations based on the integration of mass spectral information. Plant J. 2008;54:949–962. doi: 10.1111/j.1365-313X.2008.03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aoki K., Yano K., Suzuki A., et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics. 2010;11:210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chevreux B., Pfisterer T., Drescher B., et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 39.Sato S., Isobe S., Asamizu E., et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.) DNA Res. 2005;12:301–364. doi: 10.1093/dnares/dsi018. [DOI] [PubMed] [Google Scholar]
- 40.Van Ooijen J.W. JoinMap®4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen, Netherlands: Kyazma. BV; 2006. [Google Scholar]
- 4!Marti E., Gisbert C., Bishop G.J., Dixon M.S., Garcia-Martinez J.L. Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 2006;57:2037–2047. doi: 10.1093/jxb/erj154. [DOI] [PubMed] [Google Scholar]
- 42.Hyten D.L., Song Q., Choi I.-K., et al. High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor. Appl. Genet. 2008;116:945–952. doi: 10.1007/s00122-008-0726-2. [DOI] [PubMed] [Google Scholar]
- 43.Muchero W., Diop N.N., Bhat P.R., et al. A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc. Natl Acad. Sci. USA. 2009;106:18159–18164. doi: 10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyten D.L., Song Q., Fickus E.W., et al. High-throughput SNP discovery and assay development in common bean. BMC Genomics. 2010;11:475. doi: 10.1186/1471-2164-11-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deulvot C., Charrel H., Marty A., et al. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics. 2010;11:468. doi: 10.1186/1471-2164-11-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rick C.M., Kesicki E., Fobes J.F., Holle M. Genetic and biosystematic studies on two new sibling species of Lycopersicon from interandean Peru. Theor. Appl. Genet. 1976;47:55–68. doi: 10.1007/BF00281917. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi H., Shimizu A., Arie T., et al. Catalog of Micro-Tom tomato responses to common fungal, bacterial, and viral pathogens. J. Gen. Plant Pathol. 2005;71:8–22. [Google Scholar]
- 48.Labate J.A., Robertson L.D., Wu F., Tanksley S.D., Baldo A.M. EST, COSII, and arbitrary gene markers give similar estimates of nucleotide diversity in cultivated tomato (Solanum lycopersicum L.) Theor. Appl. Genet. 2009;118:1005–1014. doi: 10.1007/s00122-008-0957-2. [DOI] [PubMed] [Google Scholar]
- 49.Van Deynze A., Stoffel K., Buell C.R., et al. Diversity in conserved genes in tomato. BMC Genomics. 2007;8:465. doi: 10.1186/1471-2164-8-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varshney R.K., Hoisington D.A., Tyagi A.K. Advances in cereal genomics and applications in crop breeding. Trends. Biotechnol. 2006;24:490–499. doi: 10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Mueller L.A., Lankhorst R.K., Tanksley S.D., et al. A snapshot of the emerging tomato genome sequence. Plant Genome. 2009;2:78–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.