Abstract
Kitasatospora setae NBRC 14216T (=KM-6054T) is known to produce setamycin (bafilomycin B1) possessing antitrichomonal activity. The genus Kitasatospora is morphologically similar to the genus Streptomyces, although they are distinguishable from each other on the basis of cell wall composition and the 16S rDNA sequence. We have determined the complete genome sequence of K. setae NBRC 14216T as the first Streptomycetaceae genome other than Streptomyces. The genome is a single linear chromosome of 8 783 278 bp with terminal inverted repeats of 127 148 bp, predicted to encode 7569 protein-coding genes, 9 rRNA operons, 1 tmRNA and 74 tRNA genes. Although these features resemble those of Streptomyces, genome-wide comparison of orthologous genes between K. setae and Streptomyces revealed smaller extent of synteny. Multilocus phylogenetic analysis based on amino acid sequences unequivocally placed K. setae outside the Streptomyces genus. Although many of the genes related to morphological differentiation identified in Streptomyces were highly conserved in K. setae, there were some differences such as the apparent absence of the AmfS (SapB) class of surfactant protein and differences in the copy number and variation of paralogous components involved in cell wall synthesis.
Keywords: Kitasatospora setae, complete genome, Streptomyces
1. Introduction
Among many filamentous bacteria belonging to the phylum Actinobacteria, Streptomyces species have been extensively studied because of the ability to produce various bioactive secondary metabolites and the complex process of morphological differentiation. The life cycle of Streptomyces is initiated by the germination of spores that develop into vegetative mycelia. In response to environmental signals, aerial mycelia emerge from the colony surface and differentiate into chains of spores. Many pioneering works about the regulation of secondary metabolism and differentiation of Streptomyces species were done using model organisms such as S. coelicolor A3(2) and S. griseus IFO 13350.1,2 The complete genome sequences of three Streptomyces species, S. coelicolor A3(2),3 S. avermitilis MA-4680T4 and S. griseus IFO 13350,5 have been reported and further accelerated the studies. Kitasatospora setae is a soil-habiting mycelial bacterium belonging to the same family Streptomycetaceae. All 23 validly published species belonging to the genus Kitasatospora exhibit a similar life style and morphology with Streptomyces species. Kitasatospora may also be comparable with Streptomyces in its capacity to produce bioactive secondary metabolites. The type strain of K. setae, NBRC 14216T, is known to produce setamycin (bafilomycin B1) and bafilomycin A1, specific inhibitors of vacuolar ATPase and commonly used as biochemical reagents for investigation of molecular transport in eukaryotic cells. This genus also includes several other strains reported as producers of bioactive compounds including a proteasome inhibitor and an anti-fungal agent.6,7
After the first proposal of the genus Kitasatospora (originally Kitasatosporia) by Omura et al. in 1982,8 the taxonomic position of Kitasatospora had been under a debate. It was once reclassified as a synonym of Streptomyces based on morphology and partial 16S rDNA analysis reported by Wellington et al.9 and Ochi and Hiranuma.10 Afterward, Zhang et al.11 reported that Streptomyces and Kitasatospora form distinct phyletic groups in the detailed inspection of the 16S rDNA and the 16S–23S rDNA internal spacer region and proposed the revival of the genus Kitasatospora. Such a history made Kitasatospora a suitable model for the development of methods distinguishing closely related species and genera based on the DNA sequence.12,13 Besides the molecular phylogenetic approach, there are clear phenotypic criteria to distinguish Kitasatospora from Streptomyces, of which most notable is the chemical composition of cell wall peptidoglycan.14,15 Cell wall peptidoglycan of streptomycetes contains the ll isomer of diaminopimelic acid (DAP), whereas many other sporoactinomycetes contain the meso isomer. The peptidoglycan of Kitasatospora, in contrast, contains both ll- and meso-DAP.16 In K. setae NBRC 14216T, aerial spores on solid culture and submerged spores in liquid culture both contain exclusively ll-DAP, whereas mycelia in both cultures contain mainly meso-DAP.14,15,17 This suggests that the composition of cell wall peptidoglycan would be regulated in Kitasatospora depending on differentiation stages. Many of the genes involved in the formation of aerial mycelium (bld genes and others) and sporulation (whi genes and others) have been identified by genetic analysis of Streptomyces species,1,2 but the genetic and molecular basis for the differential incorporation of DAP isomers in Kitasatospora raises new questions. Considering the unique taxonomic position of Kitasatospora, as well as the common and distinct features of Kitasatospora and Streptomyces, Kitasatospora would be a key microorganism for further understanding of the evolution of not only actinobacteria within the family Streptomycetaceae but also other mycelial actinobacteria.
We determined the complete genome sequence of K. setae NBRC 14216T (=KM-6054T) and compared it with Streptomyces genomes. Although the overall topology and gene organization of the K. setae NBRC 14216T genome showed close resemblance to Streptomyces genomes, there are discriminative distances between them. We established a robust phylogenetic position of the genus Kitasatospora by multilocus phylogenetic analysis and confirmed the previous results based on 16S rDNA sequences. We also describe the conservation and variation of differentiation-related genes predicted from the annotation of K. setae NBRC 14216T genome, as well as the possible coding capacity of the genome for bioactive secondary metabolites.
2. Materials and methods
2.1. Genome sequencing, assembly and validation
DNA shotgun libraries with average insert sizes of 1.6 and 6.5 kb were constructed in pUC118 (TaKaRa), whereas a fosmid library with average insert size of 37 kb was constructed in pCC1FOS (EPICENTRE) as described previously.18,19 A total of 50 400 clones (34 560, 10 752 and 5088 clones from libraries with 1.6, 6.5 and 37 kb inserts, respectively) were subjected to sequencing from both ends of the inserts on either ABI 3730xl DNA Analyzer (Applied Biosystems) or Base Station DNA Fragment Analyzer BST-0100 (MJ Research, Inc.). Sequence reads were trimmed at a threshold quality value of 20 by Phred and assembled by Phrap and CONSED assembly tools.20,21 For alignment and validation of contigs, Optical Mapping (OpGen) was used. Gaps between contigs were closed by sequencing PCR products, which bridge two neighboring contigs. Finally, each base of K. setae NBRC 14216T genome was ensured to be sequenced from multiple clones and from both directions with Phrap quality score ≥70 or from one direction with Phrap quality score ≥40. Chromosomal terminus was determined after attaching adenine and thymine homopolymers to the naked 3′ ends of the chromosome as described previously.5
2.2. Data analysis and annotation
The prediction of open reading frames (ORFs) was performed using Glimmer3.22 The initial set of ORFs was manually selected from the prediction result in combination with BLASTP23 and FramePlot24 results. Each ORF was annotated manually using in-house genome annotation system OCSS (unpublished). Similarity search results against Uniprot,25 Interpro26 and HAMAP27 databases were used for functional prediction. The KEGG28 database was used for the reconstruction of metabolic pathways. If necessary, annotation was confirmed by molecular phylogenetic analysis using ClustalW, NJplot or GARLI (http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html). Putative transporters and peptidases were independently evaluated using TransportDB29 and MEROPS30 databases, respectively. Non-coding genes were predicted using the Rfam,31 tRNAscan-SE32 and ARAGORN33 programs. Putative oriC region was located using originx34 program. Putative ORFs related to mobile genetic elements were predicted and their boundaries were inferred with the assistance of GenomeMatcher35 software. For accurate assignment of orthologs from actinobacterial genomes, comparative data compiled in the MBGD36 database were used with further molecular phylogenetic evaluation, if necessary.
2.3. Data and strain submission
The nucleotide sequence of the K. setae NBRC 14216T genome has been deposited in the DDBJ/EMBL/GenBank databases under accession number AP010968. The annotated genome sequence is also available at the genome database DOGAN (http://www.bio.nite.go.jp/dogan/Top). The microbial strain and genomic DNA clones used for the sequencing are available through the NBRC (NITE Biological Resource Center, Chiba, Japan, http://www.nbrc.nite.go.jp/e/index.html).
3. Results and discussion
3.1. General features of the K. setae NBRC 14216T genome
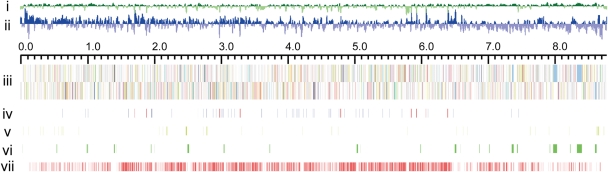
The K. setae genome was composed of a single linear chromosome of 8 783 278 bp with 127 148 bp of terminal inverted repeats (TIRs). These characteristics in the genome topology were similar to those of Streptomyces,3–5 although the microorganism does not harbor a linear or circular plasmid. The general features of the K. setae genome are summarized in Table 1 and Fig. 1. The chromosome was predicted to encode 74 tRNA genes, 9 copies of ribosomal RNA operon and 7569 protein-coding genes. Among the predicted protein-coding genes, 53.5% (4049 ORFs) were assigned putative functions. The average G + C content of the chromosome was 74.2%, which is among the highest in actinobacteria, being nearly equal to that of Kineococcus radiotolerans (http://genome.jgi-psf.org/finished_microbes/kinra/). The putative replication origin containing 20 DnaA box-like sequences was located at the center of the chromosome. This putative origin was flanked by dnaA and dnaN as in the cases of almost all bacteria, including Streptomyces species.37
Table 1.
General features of K. setae NBRC 14216T genome and Streptomyces genomes
| Length (bp) | TIR (bp) | G + C Content (%) | CDS (no.) | rRNA operons (no.) | tRNA genes (no.) | Average CDS length (bp) | Coding density (%) | Reciprocal BLAST best-hit pair (no.)a | |
|---|---|---|---|---|---|---|---|---|---|
| K. setae NBRC 14216T | 8 783 278 | 127 148 | 74.2 | 7569 | 9 | 74 | 1012 | 87.0 | — |
| S. coelicolor A3(2) | 8 667 507 | 21 653 | 72.1 | 7825 | 6 | 63 | 991 | 88.9 | 3550 |
| S. avermitilis MA-4680Tb | 9 025 608 | 49 | 70.7 | 7582 | 6 | 68 | 1027 | 86.3 | 3498 |
| S. griseus IFO 13350 | 8 545 929 | 132 910 | 72.2 | 7138 | 6 | 66 | 1055 | 88.1 | 3513 |
| S. scabies 87.22 | 10 148 695 | 18 488 | 71.5 | 8746 | 6 | 75 | 1005 | 86.2 | 3534 |
aNumbers of best-hit pair between K. setae NBRC 14216T and each Streptomyces are calculated using BLASTP program with a threshold E-value of 1e−20.
bOn the basis of the latest annotation data of S. avermitilis MA-4680T maintained by H. Ikeda (http://avermitilis.ls.kitasato-u.ac.jp).
Figure 1.
Schematic representation of the K. setae NBRC 14216T chromosome. (i) G + C content. (ii) GC skew. (iii) ORFs encoded in forward (upper) and reverse (lower) strand. Each ORF is colored on the basis of the predicted function. (iv) RNA encoding genes. rRNA operons are colored in red and tRNAs are colored in blue. (v) Putative insertion sequences. (vi) Secondary metabolism gene clusters. (vii) Red bars indicate ORFs conserved commonly in all four Streptomyces genomes.
The finished sequence consisted of a big contig representing the major part of the chromosome connected to the same small contig at both termini. As neither sequence variation nor assembly inconsistency was found in the terminal contig, we concluded that the chromosome has inverted identical sequences at both extremities. Terminal sequences of linear chromosomes and plasmids of Streptomyces and Rhodococcus can be classified into at least six groups. The terminal sequence of the K. setae chromosome was distinct from any of these groups; while the first 13 bp sequence exactly matched with that of major Streptomyces groups I and II (Supplementary Fig. S1), the subsequent region containing palindrome structures and loops, which are known in Streptomyces to be required for binding of telomere-associated protein (Tap), was not conserved.38,39 Tap and terminal protein (Tpg) encoding genes (KSE_73020 and KSE_73030) were detected in the K. setae chromosome with lower similarity (42–45% amino acid identity) to those of Streptomyces. Kitasatospora setae thus seems to have similar mechanisms for chromosome maintenance; it possesses a linear chromosome with TIRs, replicates bidirectionally from a centrally located oriC region and maintains terminal sequences by Tap and Tpg. However, the binding specificity of Tap might be different even from those recognizing group I and II replicons of Streptomyces.
No critical difference was found between K. setae and Streptomyces species in the predicted primary metabolism such as carbohydrate metabolism, amino acid metabolism, nucleic acid metabolism and respiration. The numbers of other ubiquitous components, such as membrane transporters, peptidases, transcriptional regulators and sigma factors, were also almost equivalent to those of Streptomyces.
3.2. Taxonomic reevaluation and comparative analysis of K. setae NBRC 14216T
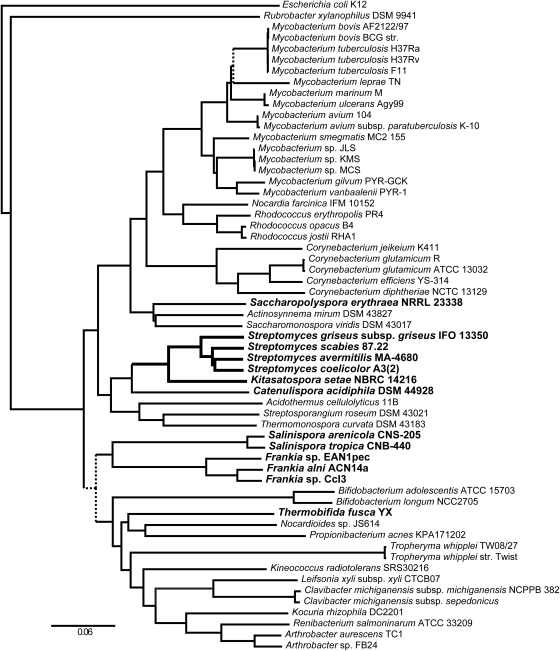
From morphological similarity and rDNA relatedness to Streptomyces species, genus Kitasatospora was once regarded as a synonym of Streptomyces.9 Although phylogenetic analysis based on the 16S rDNA sequence usually separates Streptomyces and Kitasatospora species into distinct sister groups, the results sometimes depend on the choice of the outgroup11 or the region used for the alignment, depicting the difficulties in determining correct taxonomic relationships only from the nucleotide sequences of rDNA. In order to obtain a more robust measure of the taxonomic position of Kitasatospora by taking advantage of its genomic information, we performed a multilocus phylogenetic analysis using 31 conserved amino acid sequences. Ciccarelli et al.40 reported the construction of a tree of life across all three domains using 191 species including 14 actinobacteria. We adopted the same method by utilizing 58 actinobacterial genomes with the Escherichia coli genome as an outgroup (Supplementary Tables S1 and S2). Amino acid sequences were aligned for each of the 31 conserved protein genes, and then all 31 alignments were concatenated and ambiguous portions were deleted before performing phylogenetic reconstruction. Figure 2 shows a phylogenetic tree obtained by the neighbor-joining method (a phylogenetic tree obtained by the maximum-likelihood method is shown in Supplementary Fig. S2). Relationships between major taxonomic groups were broadly consistent with the previous results obtained by 16S rDNA sequences. Within the phylogenetic tree, 82% of all predicted branches were supported by bootstrap proportions greater than 90% (i.e. 900 of 1000). Four Streptomyces species were grouped in the same branch, with S. griseus IFO 13350 branching out at the deepest position. Kitasatospora setae NBRC 14216T was placed within the same clan as Streptomyces creating the outermost branch. All these results were supported by bootstrap proportions of 100%, reinforcing the idea that the genera Kitasatospora and Streptomyces were generated from a common progenitor and have diverged into distinct sister groups. The closest to this group was Catenulispora acidiphila DSM 44928,41 a mycelial actinobacterium with a circular chromosome.
Figure 2.
Phylogenetic tree based on amino acid sequences of 31 protein-coding genes analyzed by the neighbor-joining method. Branches with less than 90% bootstrap support are represented in dashed lines. Lists of organisms and genes used for the analysis are shown in Supplementary Tables S1 and S2, respectively. Names of the organisms mentioned in the text are shown in bold type.
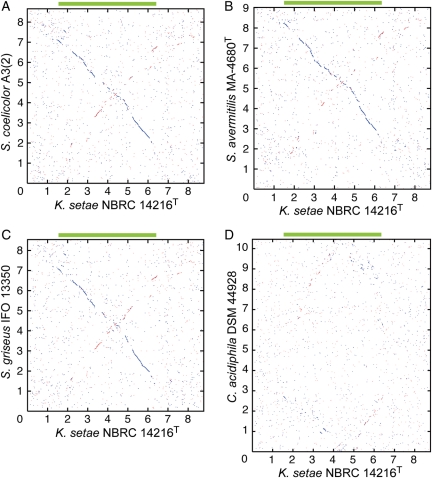
For further analysis of the relationship between K. setae and Streptomyces, we compared all annotated proteins of K. setae and four Streptomyces species. More than half of ORFs predicted in K. setae had orthologs (reciprocal best-hit pairs) in each of four Streptomyces species using a BLASTP threshold of E < 10−20 (Table 1). About 34% of K. setae ORFs had orthologs in all four Streptomyces genomes. The average amino acid identity between orthologous pairs from K. setae and Streptomyces species was around 60%. Despite such high similarities observed between orthologs, genome-wide comparison using ortholog plots demonstrated smaller extent of synteny between K. setae and Streptomyces genome (Fig. 3A–C). Many short synteny blocks were observed along either of the diagonal lines, suggesting that frequent inversions around the replication origin have had occurred. This is in contrast to the higher extent of colinearities with only 2–4 inversions observed in comparative analysis among Streptomyces genomes.3–5 Conserved core region of the K. setae genome predicted by threading major synteny blocks was ∼5 Mb in length ranging from KSE_13770 to KSE_57600, which is ∼1 Mb smaller than the core regions deduced from comparison among Streptomyces genomes.5 Long-range synteny between K. setae and C. acidiphila genomes was much less obvious compared with that between K. setae and Streptomyces genomes (Fig. 3D), although the number of orthologous gene pairs between K. setae and C. acidiphila (3185) was only 10% smaller than that between K. setae and Streptomyces (3498–3550, Table 1).
Figure 3.
Synteny between the genomes of K. setae NBRC 14216T and S. coelicolor A3(2) (A), K. setae NBRC 14216T and S. avermitilis MA-4680T (B), K. setae NBRC 14216T and S. griseus IFO 13350 (C) and K. setae NBRC 14216T and C. acidiphila DSM 44928 (D). Reciprocal BLAST best-hit pairs with a threshold value of E < 10−20 were plotted. The direction of each chromosome was adjusted so that the dnaA gene faces the same direction. Green bar in each panel represents the conserved core region on the K. setae chromosome.
3.3. Conservation and variation of genes related to developmental regulation
Streptomyces is well characterized as a model organism by its complex life cycle; it grows as a thread-like mycelium and forms aerial mycelium in the air under nutrient-limited conditions.1,2,42,43 A series of genes whose mutations cause defects in aerial growth (‘bld’ genes) were characterized in S. coelicolor A3(2). A complex interaction cascade among bld gene products results in the secretion of surfactant proteins, which assist Streptomyces in extending aerial mycelium upward and in differentiating into spores. A number of genes in ‘whi’ loci were also characterized mainly in S. coelicolor A3(2) as causing sporulation deficiency. Most of these classical differentiation genes were found to have orthologs in K. setae. The number of conserved bld and whi genes and their similarity (amino acid identity) to S. coelicolor A3(2) counterparts were higher than those in other mycelial actinobacteria, such as C. acidiphila DSM 44928, Saccharopolyspora erythraea NRRL 2333844 and Salinispora arenicola CNS 205 (Table 2), consistent with the closer morphological similarity of K. setae to Streptomyces. Notably, bld gene pairs orthologous between K. setae and Streptomyces species represented 80–97% amino acid identities, which are much higher than the average (60%) of all orthologous pairs and comparable with those between Streptomyces species. Higher than the average conservation of bld genes was also observed between C. acidiphila and Streptomyces (58–95% amino acid identities). The bldA tRNA gene was also highly conserved in K. setae, with its cognate codon UUA being the rarest in K. setae; used only in 69 predicted ORFs, most of which were either of unknown function or with predicted regulatory functions. In four completely sequenced Streptomyces genomes, only three genes were found to have a conserved UUA codon at the same position in each ortholog.45 One of these, the bldH gene, shared a UUA codon at an equivalent position also in K. setae. The high conservation of these regulatory components may further imply that each component in the regulatory cascade undergoes a higher than the usual number of interactions with other bld gene products or with other cellular components.
Table 2.
Conservation of aerial mycelium and spore formation-related genes in K. setae NBRC 14216T and other mycelial actinobacteria
| Kitasatospora setae NBRC 14216Ta | Streptomyces coelicolor A3(2) | Streptomyces avermitilis MA-4680T | Streptomyces griseus IFO 13350 | Streptomyces scabies 87.22 | Catenulispora acidiphila DSM 44928 | Saccharopolyspora erythraea NRRL 23338 | Salinispora arenicola CNS 205 | |
|---|---|---|---|---|---|---|---|---|
| Aerial mycelium | ||||||||
| bldA | KSE_t0069 | SCOt24 | SAV_t57 | SGR_tRNA42 | SCAB_t50 | Caci_R0007 | SACE_8016 | Sare_R0014 |
| bldB | (KSE_16220)b (64) | SCO5723 (SCO7246) | SAV_2529 (SAV_1241) | SGR_1796 | SCAB_25271 (SCAB_84941) | ND | SACE_6156 | Sare_3856 |
| bldC | KSE_41620 (97) | SCO4091 | SAV_4130 | SGR_3882 | SCAB_47901 | Caci_0301 | SACE_6926 | Sare_0170 |
| bldD | KSE_13950 (92) | SCO1489 | SAV_6861 | SGR_6045 | SCAB_75171 | Caci_2399 | SACE_2077 | Sare_1844 |
| bldG (=rsbV) | KSE_35840 (96) | SCO3549 | SAV_4614 | SGR_3307 | SCAB_40861 | Caci_8450 | SACE_4194 | Sare_4413 |
| bldH (=adpA) | KSE_26930 (80) | SCO2792 | SAV_5261 | SGR_4742 | SCAB_57831 | Caci_5972 | SACE_4523 | ND |
| bldKA-bldKE | KSE_48250–KSE_48290 (41–78) | SCO5112–SCO5116 | SAV_3152–SAV_3156; SAV_3172–SAV_3176 | SGR_2414–SGR_2418 | SCAB_31501–SCAB_31541 | Caci_7147–Caci_7151 | ND | ND |
| bldM (=whiK) | KSE_31060 (97) | SCO4768 | SAV_4998 | SGR_2759 | SCAB_36231 | Caci_1011 | SACE_6712 | Sare_4229 |
| bldN (=adsA) | KSE_33690 (92) | SCO3323 | SAV_4735 | SGR_4151 | SCAB_39121 | Caci_8330 | SACE_6951 | Sare_0420 |
| amfC | KSE_43740 (54) | SCO4184 | SAV_4026 | SGR_3974 | SCAB_49711 | Caci_8239 | SACE_7115 | ND |
| Lantibiotic-like surfactant | ||||||||
| amfR (=ramR) | ND | SCO6685 | SAV_7499 | SGR_2393 | SCAB_8642c | ND | ND | ND |
| amfS (=ramS) | ND | SCO6682 | SAV_7502 | SGR_2396 | SCAB_8621 | Caci_4240 | SACE_4231 | ND |
| amfB (=ramA) | ND | SCO6683 | SAV_7501 | SGR_2395 | SCAB_8631 | Caci_4242 | SACE_4232 | ND |
| amfA (=ramB) | ND | SCO6684 | SAV_7500 | SGR_2394 | SCAB_8641 | Caci_4243 | SACE_4233 | ND |
| amfT (=ramC) | KSE_63000 (44) | SCO6681 | SAV_7503 | SGR_2397 | SCAB_8611 | Caci_4239 | SACE_4230 | ND |
| Sporulation | ||||||||
| whiA | KSE_55390 (89) | SCO1950 | SAV_6294 | SGR_5572 | SCAB_69681 | Caci_5609 | SACE_2141 | Sare_3326 |
| whiB | KSE_29410 (83) | SCO3034 | SAV_5042 | SGR_4503 | SCAB_55081 | Caci_7609 | SACE_6464 | ND |
| whiD (=wblB) | KSE_31070 (90) | SCO4767 | SAV_4997 | SGR_2760 | SCAB_36241 | Caci_1009 | SACE_5583 | ND |
| whiE | KSE_72410–KSE_72480 (46–67) | SCO5314–SCO5321 | SAV_2837–SAV_2844 | ND | SCAB_43281–SCAB43341 | Caci1083–Caci1090 | ND | Sare_2682–Sare_2690 |
| whiG | KSE_52540 (79) | SCO5621 | SAV_2630 | SGR_1866 | SCAB_26051 | Caci_1526 | SACE_6040 | ND |
| whiH | KSE_54050 (74) | SCO5819 | SAV_2445 | SGR_1702 | SCAB_24461 | Caci_1760 | ND | ND |
| whiI | KSE_55090 (77) | SCO6029 | SAV_2230 | SGR_1475 | SCAB_20881 | Caci_6781 | SACE_1833 | ND |
| sigF | KSE_37310 (69) | SCO4035 | SAV_4185 | SGR_3551 | SCAB_47291 | Caci_7708 | ND | ND |
| sigN | KSE_37320 (80) | SCO4034 | SAV_4186 | SGR_3552 | SCAB_47281 | ND | ND | ND |
| crgA (=whiP) | KSE_39340 (50) | SCO3854 | SAV_4331 | SGR_3718 | SCAB_45631 | Caci_0043 | SACE_0037 | Sare_0058 |
aThe amino acid identity between K. setae NBRC 14216T ORF and S. coelicolor A3(2) ortholog was shown in parentheses. The range of amino acid identity was shown where multiple genes were assigned.
bbldB gene paralog found in S. coelicolor A3(2) and S. avermitilis MA-4680T are also included. KSE_16220 corresponds to an ortholog of the bldB paralog.
cThis ORF tentatively named SCAB_8642 (corresponding to position 975 194–975 802 on the complementary strand) has not yet been incorporated in the submitter annotation.
On the contrary, we could not identify most of the amf genes in K. setae (Table 2). The amf genes are highly conserved among Streptomyces species and are necessary for the synthesis of AmfS [SapB in S. coelicolor A3(2)] surfactant protein, which is known in Streptomyces to be secreted before the initiation of aerial growth. The presence of amfAB homologs in C. acidiphila41 and S. erythraea,44 mycelial actinobacteria more distantly related to Streptomyces, may suggest that the pre-existing AmfS system was eliminated in the Kitasatospora lineage, although other possibilities such as the horizontal acquisition of these genes in each lineage cannot be excluded. In addition to the lack of the amf gene cluster, differences from Streptomyces were also suggested in the variation of paralogous components (Supplementary Table S3) such as SsgA-like proteins involved in peptidoglycan synthesis in sporogenic cell division, WhiB family transcriptional regulators and chaplins, another class of surfactant protein shown in S. coelicolor to play a part in aerial mycelium formation.46,47
A number of whi genes control the process of sporulation septation and spore maturation in Streptomyces. Of the classical whi genes, whiA, whiB and whiD are commonly found in actinobacteria, and their exact functions in simpler (non-sporulating) actinobacteria need to be elucidated.1 All other whi genes, which had been considered specific to Streptomyces species,1,42 were also conserved in K. setae with remarkably high amino acid identities (69–90%). Despite such close similarities in genetic background, preliminary experimental results suggest that K. setae produces seemingly less mature spores compared with Streptomyces; (i) although K. setae genome encodes full set of whiE gene cluster (encoding biosynthetic enzymes for polyketide spore pigment), no expression of whiE genes nor the production of spore pigment was observed in any culture conditions for sporulation, (ii) both aerial and submerged spores produced by K. setae are much more sensitive to freeze-thaw cycles than those of Streptomyces (S. Kitani and H. Ikeda, unpublished observations). Further studies are needed to elucidate mechanisms underlying such differences.
The initiation of secondary metabolite synthesis is known to be linked with morphological differentiation in S. griseus via the gamma-butyrolactone autoregulator cascade. In this regard, it would be noteworthy that K. setae possesses three homologs of the autoregulator receptor: KsbA (KSE_58650), KsbB (KSE_01050t and KSE_75690t; identical genes encoded in the TIR) and KsbC (KSE_44580). However, KsbA in K. setae was experimentally confirmed to be involved only in secondary metabolism,48 but not in morphological differentiation, whereas the involvement of other two remains to be clarified. In addition, the AfsA family protein, which contains two A-factor biosynthesis repeat motifs, has been reported to be a crucial enzyme to synthesize the gamma-butyrolactone autoregulator in Streptomyces.49 Interestingly, three AfsA family proteins were encoded on the genome of K. setae: KsbS2 (KSE_01060t and KSE_75680t; identical and present in the TIR, similar to the case of KsbB), KsbS3 (KSE_22970) and KsbS4 (KSE_44600). These findings suggested that there might be a more complicated signaling network for secondary metabolism and/or morphological differentiation in K. setae.
3.4. Genes involved in peptidoglycan biosynthesis
One of the most important features of the genus Kitasatospora from the chemotaxonomic viewpoint is that the cell wall peptidoglycan contains both ll-DAP and meso-DAP.14,15 DAP analysis of Kitasatospora strains showed that spores contain only ll-DAP, whereas mycelia contain mostly meso-DAP.16,17 This observation suggests that Kitasatospora incorporates different DAP isomers into the cell wall depending on differentiation stage. We can speculate two steps which might be responsible for the differential incorporation of DAP isomers. (i) DAP biosynthesis and isomerization: ll-DAP is isomerized to meso-DAP by dapF gene product in the course of lysine biosynthesis.50 (ii) Incorporation of DAP into peptidoglycan: the addition of DAP to UDP-N-acetylmuramyl-l-alanyl-d-glutamate precursor is catalyzed by an enzyme encoded by murE.51
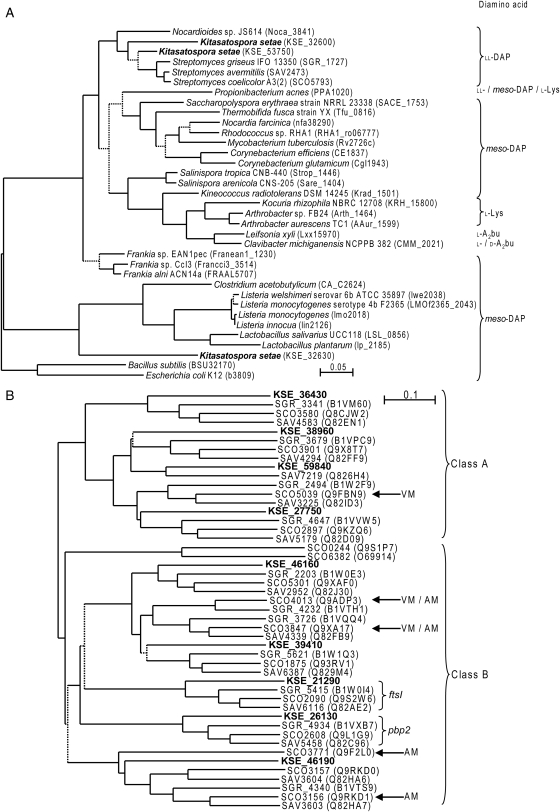
As in the case of Streptomyces species whose peptidoglycan is composed of ll-DAP, K. setae had only one murE gene (KSE_21280), which is located in the center of a long conserved dcw (division cell wall) cluster. Amino acid residues known to be responsible for substrate recognition in the enzymes from meso-DAP containing bacteria are also conserved in the enzymes from ll-DAP containing bacteria including Streptomyces. The MurE protein purified from E. coli can incorporate ll-DAP in addition to meso-DAP, the natural substrate in this bacterium.52 Thus, the MurE protein may not be responsible for the differentiation-dependent alterations in peptidoglycan synthesis, unless the substrate specificity is regulated by unknown factor(s). On the other hand, K. setae had three dapF genes (KSE_32600, KSE_32630 and KSE_53750) whose products share highly conserved active site residues. Phylogenetic analysis based on amino acid sequences showed that two of the DapF proteins (encoded by KSE_32600 and KSE_53750) were closely related to those from ll-DAP containing actinobacteria such as Streptomyces species and Nocardioides sp. BAA-499/JS614 (Fig. 4A). In contrast, the third DapF protein (KSE_32630) was positioned apart from the actinobacterial groups, but in the vicinity of proteins from meso-DAP containing Listeria, Lactobacillus and Clostridium. In the dapF mutant strain of E. coli, an increase in ll-DAP containing peptidoglycan was observed,52 suggesting that the relative size of ll- and meso-DAP pools would affect the composition of peptidoglycan. Differential regulation of the multiple dapF genes, or their protein products, in the course of morphological differentiation might play an important role in the change in peptidoglycan composition.
Figure 4.
Amino acid phylogenetic trees of DapF (A) and HMW-PBPs (B). Branches with less than 70% bootstrap support are represented in dotted line. The distribution of peptidoglycan amino acid component is also presented in (A). Streptomyces coelicolor A3(2) PBPs and their orthologs were grouped and expression patterns were assigned according to Noens et al.55 in (B); those known to be expressed in aerial mycelium and vegetative mycelium are shown by AM and VM, respectively. Amino acid sequences were derived from the HAMAP database (MF_00197).
Streptomyces, a representative of mycelial bacteria, is known to have two distinct modes of cell division. During vegetative growth, cell division occurs occasionally resulting in widely and irregularly spaced cross-walls. In sporulating mycelia, in contrast, sporulating septa are deposited in a coordinated manner resulting in regularly spaced spores.53 The latter suggests a regulated synthesis of peptidoglycan at septa and spore walls. High-molecular weight PBPs (HMW-PBPs) catalyze the cross-linking of peptidoglycan peptides. Streptomyces contain a number of HMW-PBP genes of which some were shown to be expressed at specific stages of differentiation.54,55 Kitasatospora setae seems to have nine HMW-PBP genes, one of which (KSE_46190) being highly homologous to S. coelicolor A3(2) genes (SCO3771 and SCO3156) known to be expressed during aerial growth and sporulation. On the other hand, the gene (SCO5039) specifically expressed in vegetative mycelium in S. coelicolor seemed not conserved in K. setae (Fig. 4B). Such differences in stage-specific PBPs could also be reflected in differentiation specific incorporation of peptidoglycan units in K. setae. In this context, it is also interesting to note that SsgA-like family proteins (SALPs), likely involved in the regulation of peptidoglycan synthesis in sporogenic cell division, are much diversified in K. setae compared with Streptomyces. Kitasatospora setae encodes at least 12 SALPs, about twice as many as those found in Streptomyces species (Supplementary Table S3).
3.5. Gene clusters for secondary metabolite biosyntheses
Bacteria belonging to the genus Kitasatospora have been explored as potential new sources of various bioactive metabolites.56 Kitasatospora setae NBRC 14216T is known to produce setamycin (bafilomycin B1), which bears antitrichomonal activity.57 A total of 24 genes or gene clusters in the K. setae genome were predicted to be involved in the biosynthesis of secondary metabolites (Table 3). Of the 24 clusters, more than 60% (16 clusters) were located in subtelomeric regions; 11 were in the right subtelomeric region, whereas 5 were in the left subtelomeric region. The number of predicted gene clusters for secondary metabolism was slightly lower than that predicted in Streptomyces species (36 in S. griseus IFO 13350, 37 in S. avermitilis MA-4680T, 30 in S. coelicolor A3(2) and at least 20 in S. scabies 87.22), but apparently higher than that in other prokaryotes,58 underscoring the importance of the genus Kitasatospora as the source of bioactive compounds.
Table 3.
Biosynthetic genes of secondary metabolites in K. setae NBRC 14216T
| Orfid | Note |
|---|---|
| Terpene | |
| KSE_00200t (=KSE_76540t)a | Sesquiterpene |
| KSE_12950 | Sesquiterpene |
| KSE_17590-KSE_17630 | Squalene/hopanoid |
| KSE_46080 | Germacradienol/geosmin |
| KSE_70210-KSE_70220 | 2-Methylisoborneol |
| NRPS and PKS | |
| KSE_18000 | Type III PKS |
| KSE_22630-KSE_22810 | Discrete NRPS |
| KSE_27200-KSE_27290 | Type I PKS |
| KSE_33340-KSE_33360 | Discrete NRPS |
| KSE_58150-KSE_58200 | NRPS |
| KSE_61120 | NRPS (incomplete) |
| KSE_65510-KSE_65560 | NRPS and type I PKS |
| KSE_65960-KSE_66030 | NRPS and type II PKS |
| KSE_70570-KSE_70620 | Type I PKS and NRPS (factumycin) |
| KSE_72410-KSE_72480 | Type II PKS (spore pigment) |
| KSE_73410-KSE_73580 | Type I PKS (bafilomycins) |
| KSE_75420-KSE_75430 | NRPS |
| Other | |
| KSE_04750-KSE_04770 | Lantibiotic |
| KSE_09030-KSE_09170 | Similar to valanimycin biosynthetic genes |
| KSE_12660-KSE_12700 | Siderophore |
| KSE_27300-KSE_27440 | Similar to valanimycin biosynthetic genes |
| KSE_45610-KSE_45680 | Lantibiotic |
| KSE_58810-KSE_58830 | Lantibiotic |
| KSE_53800-KSE_53830 | Siderophore |
aKSE_00200t was embedded in TIR and identical to KSE_76540t.
Of the 24 clusters, 5 were estimated for terpene biosynthesis, 12 for polyketides or non-ribosomal peptides, 2 for siderophores and 5 for lantibiotics and others. Five ORFs containing the terpene synthase domain (IPR005630) were classified by the phylogenetic analysis described by Komatsu et al.,59 indicating that KSE_46080 was in the group of germacradienol/geosmin synthase60,61 and KSE_70210 in the group of 2-methylisoborneol synthase. Consistently, geosmin and 2-methylisoborneol were identified from the culture of K. setae. Of the 12 clusters for polyketides or non-ribosomal peptides, setamycin (bafilomycin B1) cluster was estimated and experimentally proved to be KSE_73410–KSE_73580 (H. Ikeda, unpublished results). An 84 kb region (KSE_70410–KSE_70650) containing the cluster KSE_70570–KSE_70620 showed striking resemblance in features of gene organization and in deduced amino acid sequences of each ORF to the kirromycin biosynthetic gene cluster, the first characterized combined cis–trans-PKS cluster in Streptomyces collinus Tu 365.62 This region was experimentally confirmed to be responsible for the biosynthesis of factumycin, an antibiotic structurally related to kirromycin (H. Ikeda, unpublished results). Regarding the two siderophore clusters, the one (KSE_12660–KSE_12700) showed good similarity in gene arrangement and deduced amino sequence of each ORF to a cluster in S. avermitilis (SAV_7320–SAV_7323, lacking a homolog corresponding to KSE_12680), both of which were similar to vibrioferrin cluster identified in Vibrio parahaemolyticus.63 Another one (KSE_53800–KSE_53830) was similar in gene arrangement to the rhizobactin cluster in Sinorhizobium meliloti,64 but differed from any of the known siderophore biosynthesis clusters of Streptomyces. Interestingly, K. setae lacks a gene cluster for nocardamin (desferrioxamine) biosynthesis that was commonly found in genome-sequenced Streptomyces species.58,65
3.6. Conclusion
Streptomycetes are thought to have emerged ∼440 million years ago. At present, Streptomyces species are some of the most highly differentiated microorganisms with a complex life cycle. The cell wall peptidoglycan composed of ll-DAP is another feature to be considered to distinguish Streptomyces from other sporoactinomycetes. Here, we analyzed the first genome sequence of a Streptomycetaceae bacterium other than Streptomyces species. Phylogenetic analysis based on amino acid sequences, together with a genome-wide comparison of the predicted genes such as those related to morphological differentiation and cell wall biosynthesis, suggests that the genera Streptomyces and Kitasatospora were diverged directly from their last common ancestor. The chromosomal linearity and the presence of the TIR sequence would also be the features inherited from the common ancestor. Many differentiation-related genes highly conserved in K. setae and Streptomyces species might have been acquired sequentially in the evolution, as previously suggested from the comparison of Streptomyces with other groups of mycelial actinobacteria, Thermobifida fusca and Frankia species.1,42 If additional genomic sequences determined more recently such as C. acidiphila, S. erythraea, Salinispora species and K. setae were taken into consideration, however, the evolutionary pathway of differentiation-related genes seemed not necessarily be straightforward, and gene losses and horizontal acquisitions, in particular lineage, might also have to be considered (Table 2). Phylogenetic data presented in Fig. 2 and Supplementary Fig. S2 also suggest that mycelial actinobacteria are not monophyletic; some organisms such as S. erythraea and T. fusca belong to sub-branches that also contain bacteria without most of the developmental genes. Extraordinary high similarity observed in bld regulatory genes, which are dispersed in the genome, might also reflect recent horizontal transfer. Although the highly regulated differentiation system of Streptomyces has attracted much attention, reasons for the unique abundance of ll-DAP in peptidoglycan may have been overlooked. We provided the possible background of the relative abundance of meso- and ll-DAP in K. setae. Further analysis and comparison of two genera, including genome sequencing of another Kitasatospora species, will provide deeper understanding of the evolution of actinobacteria.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Supplementary Material
Acknowledgements
We thank S. Omata, S. Gomi, S. Fujinami, A. Kawakoshi, K. Koga and C. Koga for the assistance in annotation and A. Hosoyama, T. Harada, R. Igarashi and H. Horikawa for draft sequencing. We also thank T. Tamura, K. Mori and K. Suzuki for the help and fruitful discussions about phylogenetic analysis and H. Komaki for valuable discussions about secondary metabolism-related genes.
References
- 1.Chater K.F., Chandra G. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 2006;30:651–72. doi: 10.1111/j.1574-6976.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 2.Horinouchi S. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 2007;71:283–99. doi: 10.1271/bbb.60627. doi:10.1016/S0168-583X(01)00748-0. [DOI] [PubMed] [Google Scholar]
- 3.Bentley S.D., Chater K.F., Cerdeno-Tarraga A.M., et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–7. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda H., Ishikawa J., Hanamoto A., et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003;21:526–31. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 5.Ohnishi Y., Ishikawa J., Hara H., et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008;190:4050–60. doi: 10.1128/JB.00204-08. doi:10.1016/j.radmeas.2003.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momose I., Sekizawa R., Hirosawa S., et al. Tyropeptins A and B, new proteasome inhibitors produced by Kitasatospora sp. MK993-dF2. II. Structure determination and synthesis. J. Antibiot. (Tokyo) 2001;54:1004–12. doi: 10.7164/antibiotics.54.1004. doi:10.1016/S1350-4487(96)00085-6. [DOI] [PubMed] [Google Scholar]
- 7.Yoon T.M., Kim J.W., Kim J.G., Kim W.G., Suh J.W. Talosins A and B: new isoflavonol glycosides with potent antifungal activity from Kitasatospora kifunensis MJM341. I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. (Tokyo) 2006;59:633–9. doi: 10.1038/ja.2006.84. doi:10.1016/1350-4487(94)00084-E. [DOI] [PubMed] [Google Scholar]
- 8.Omura S., Takahashi Y., Iwai Y., Tanaka H. Kitasatospora, a new genus of the order Actinomycetales. J. Antibiot. (Tokyo) 1982;35:1013–9. doi: 10.7164/antibiotics.35.1013. [DOI] [PubMed] [Google Scholar]
- 9.Wellington E.M., Stackebrandt E., Sanders D., Wolstrup J., Jorgensen N.O. Taxonomic status of Kitasatospora, and proposed unification with Streptomyces on the basis of phenotypic and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int. J. Syst. Bacteriol. 1992;42:156–60. doi: 10.1099/00207713-42-1-156. doi:10.1097/00004032-197108000-00016. [DOI] [PubMed] [Google Scholar]
- 10.Ochi K., Hiranuma H. A taxonomic review of the genera Kitasatospora and Streptoverticillium by analysis of ribosomal protein AT-L30. Int. J. Syst. Bacteriol. 1994;44:285–92. doi: 10.1099/00207713-44-2-285. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Wang Y., Ruan J. A proposal to revive the genus Kitasatospora (Omura, Takahashi, Iwai, and Tanaka 1982) Int. J. Syst. Bacteriol. 1997;47:1048–54. doi: 10.1099/00207713-47-4-1048. [DOI] [PubMed] [Google Scholar]
- 12.Gunther S., Groth I., Grabley S., Munder T. Design and evaluation of an oligonucleotide-microarray for the detection of different species of the genus Kitasatospora. J. Microbiol. Methods. 2006;65:226–36. doi: 10.1016/j.mimet.2005.07.012. doi:10.1016/j.radmeas.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Moller R., Schuler T., Gunther S., Carlsohn M.R., Munder T., Fritzsche W. Electrical DNA-chip-based identification of different species of the genus Kitasatospora. Appl. Microbiol. Biotechnol. 2008;77:1181–8. doi: 10.1007/s00253-007-1241-0. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y., Iwai Y., Omura S. Relationship between cell morphology and the types of diaminopimelic acid in Kitasatospora setalba. J. Gen. Appl. Microbiol. 1983;29:459–65. doi:10.1016/j.radmeas.2008.04.015. [Google Scholar]
- 15.Takahashi Y., Kuwana T., Iwai Y., Omura S. Some characteristics of aerial and submerged spores of Kitasatospora setalba. J. Gen. Appl. Microbiol. 1984;30:223–9. doi:10.1016/j.radmeas.2009.10.089. [Google Scholar]
- 16.Omura S., Iwai Y., Takahashi Y., Kojima K., Otoguro K., Oiwa R. Type of diaminopimelic acid different in aerial and vegetative mycelia of setamycin-producing actinomycete KM-6054. J. Antibiot. (Tokyo) 1981;34:1633–4. doi: 10.7164/antibiotics.34.1633. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi Y., Seino A., Iwai Y., Omura S. Taxonomic study and morphological differentiation of an actinomycete genus, Kitasatospora. Zentralbl. Bakteriol. 1999;289:265–84. doi: 10.1016/s0934-8840(99)80065-6. doi:10.1016/j.radmeas.2009.10.075. [DOI] [PubMed] [Google Scholar]
- 18.Sekine M., Tanikawa S., Omata S., et al. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 2006;8:334–46. doi: 10.1111/j.1462-2920.2005.00899.x. [DOI] [PubMed] [Google Scholar]
- 19.Takarada H., Sekine M., Kosugi H., et al. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J. Bacteriol. 2008;190:4139–46. doi: 10.1128/JB.01853-07. doi:10.1016/j.radmeas.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- 21.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. doi:10.1016/j.asr.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 22.Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. doi:10.1093/rpd/ncp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa J., Hotta K. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 1999;174:251–3. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- 25.UniProt, Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–5. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder N.J., Apweiler R., Attwood T.K., et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–8. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima T., Auchincloss A.H., Coudert E., et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2008;37:D471–8. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., Araki M., Goto S., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Q., Chen K., Paulsen I.T. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–9. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlings N.D., Morton F.R., Kok C.Y., Kong J., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–5. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–4. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worning P., Jensen L.J., Hallin P.F., Staerfeldt H.H., Ussery D.W. Origin of replication in circular prokaryotic chromosomes. Environ. Microbiol. 2006;8:353–61. doi: 10.1111/j.1462-2920.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 35.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35:D343–6. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakimowicz D., Majka J., Messer W., et al. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology. 1998;144(Pt 5):1281–90. doi: 10.1099/00221287-144-5-1281. [DOI] [PubMed] [Google Scholar]
- 38.Bao K., Cohen S.N. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 2003;17:774–85. doi: 10.1101/gad.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang C.H., Lin Y.S., Yang Y.L., Huang S.W., Chen C.W. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 1998;28:905–16. doi: 10.1046/j.1365-2958.1998.00856.x. [DOI] [PubMed] [Google Scholar]
- 40.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 41.Copeland A., Lapidus A., Rio T.G.D., et al. Complete genome sequence of Catenulispora acidiphila type strain (ID 139908T) Stand. Genomic Sci. 2009;1:119–25. doi: 10.4056/sigs.17259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura M., Canchaya C., Tauch A., et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willey J.M., Willems A., Kodani S., Nodwell J.R. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 2006;59:731–42. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- 44.Oliynyk M., Samborskyy M., Lester J.B., et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007;25:447–53. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 45.Chandra G., Chater K.F. Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie Van Leeuwenhoek. 2008;94:111–26. doi: 10.1007/s10482-008-9231-5. [DOI] [PubMed] [Google Scholar]
- 46.Capstick D.S., Willey J.M., Buttner M.J., Elliot M.A. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol. Microbiol. 2007;64:602–13. doi: 10.1111/j.1365-2958.2007.05674.x. [DOI] [PubMed] [Google Scholar]
- 47.Claessen D., Rink R., de Jong W., et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–26. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi S.U., Lee C.K., Hwang Y.I., Kinoshita H., Nihira T. Cloning and functional analysis by gene disruption of a gene encoding a γ-butyrolactone autoregulator receptor from Kitasatospora setae. J. Bacteriol. 2004;186:3423–30. doi: 10.1128/JB.186.11.3423-3430.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato J.Y., Funa N., Watanabe H., Ohnishi Y., Horinouchi S. Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl Acad. Sci. USA. 2007;104:2378–83. doi: 10.1073/pnas.0607472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richaud C., Higgins W., Mengin-Lecreulx D., Stragier P. Molecular cloning, characterization, and chromosomal localization of dapF, the Escherichia coli gene for diaminopimelate epimerase. J. Bacteriol. 1987;169:1454–59. doi: 10.1128/jb.169.4.1454-1459.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abo-Ghalia M., Michaud C., Blanot D., van Heijenoort J. Specificity of the uridine-diphosphate-N-acetylmuramyl-L-alanyl-D-glutamate: meso-2,6-diaminopimelate synthetase from Escherichia coli. Eur. J. Biochem. 1985;153:81–7. doi: 10.1111/j.1432-1033.1985.tb09269.x. [DOI] [PubMed] [Google Scholar]
- 52.Mengin-Lecreulx D., Michaud C., Richaud C., Blanot D., van Heijenoort J. Incorporation of LL-diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diaminopimelate epimerase encoded by dapF. J. Bacteriol. 1988;170:2031–9. doi: 10.1128/jb.170.5.2031-2039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flardh K., van Wezel G.P. Cell division during growth and development of Streptomyces. Recent Res. Devel. Bacteriol. 2003;1:71–90. [Google Scholar]
- 54.Hao J., Kendrick K.E. Visualization of penicillin-binding proteins during sporulation of Streptomyces griseus. J. Bacteriol. 1998;180:2125–32. doi: 10.1128/jb.180.8.2125-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noens E.E., Mersinias V., Traag B.A., Smith C.P., Koerten H.K., van Wezel G.P. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 2005;58:929–44. doi: 10.1111/j.1365-2958.2005.04883.x. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi Y., Omura S. Isolation of new actinomycete strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol. 2003;49:141–54. doi: 10.2323/jgam.49.141. [DOI] [PubMed] [Google Scholar]
- 57.Omura S., Otoguro K., Nishikiori T., Oiwa R., Iwai Y. Setamycin, a new antibiotic. J. Antibiot. (Tokyo) 1981;34:1253–56. doi: 10.7164/antibiotics.34.1253. [DOI] [PubMed] [Google Scholar]
- 58.Nett M., Ikeda H., Moore B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009;26:1362–84. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komatsu M., Tsuda M., Omura S., Oikawa H., Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc. Natl Acad. Sci. USA. 2008;105:7422–7. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cane D.E., He X., Kobayashi S., Omura S., Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J. Antibiot. (Tokyo) 2006;59:471–9. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- 61.Jiang J., He X., Cane D.E. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat. Chem. Biol. 2007;3:711–5. doi: 10.1038/nchembio.2007.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber T., Laiple K.J., Pross E.K., et al. Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chem. Biol. 2008;15:175–88. doi: 10.1016/j.chembiol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 63.Tanabe T., Funahashi T., Nakao H., Miyoshi S., Shinoda S., Yamamoto S. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 2003;185:6938–49. doi: 10.1128/JB.185.23.6938-6949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lynch D., O'Brien J., Welch T., et al. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 2001;183:2576–85. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueki M., Suzuki R., Takamatsu S., et al. Nocardamin production by Streptomyces avermitilis. Actinomycetologica. 2009;23:34–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.