Abstract
The c-Myc oncoprotein is overexpressed in many tumors and is essential for maintaining the proliferation of transformed cells. To function as a transcription factor, c-Myc must dimerize with Max via the basic helix-loop-helix leucine zipper protein (bHLH-ZIP) domains in each protein. The small molecule 7-nitro-N-(2-phenylphenyl)-2,1,3-benzoxadiazol-4-amine (10074-G5) binds to and distorts the bHLH-ZIP domain of c-Myc, thereby inhibiting c-Myc/Max heterodimer formation and inhibiting its transcriptional activity. We report in vitro cytotoxicity and in vivo efficacy, pharmacodynamics, pharmacokinetics, and metabolism of 10074-G5 in human xenograft-bearing mice. In vitro, 10074-G5 inhibited the growth of Daudi Burkitt's lymphoma cells and disrupted c-Myc/Max dimerization. 10074-G5 had no effect on the growth of Daudi xenografts in C.B-17 SCID mice that were treated with 20 mg/kg 10074-G5 intravenously for 5 consecutive days. Inhibition of c-Myc/Max dimerization in Daudi xenografts was not seen 2 or 24 h after treatment. Concentrations of 10074-G5 in various matrices were determined by high-performance liquid chromatography-UV, and metabolites of 10074-G5 were identified by liquid chromatography/tandem mass spectrometry. The plasma half-life of 10074-G5 in mice treated with 20 mg/kg i.v. was 37 min, and peak plasma concentration was 58 μM, which was 10-fold higher than peak tumor concentration. The lack of antitumor activity probably was caused by the rapid metabolism of 10074-G5 to inactive metabolites, resulting in tumor concentrations of 10074-G5 insufficient to inhibit c-Myc/Max dimerization. Our identification of 10074-G5 metabolites in mice will help design new, more metabolically stable small-molecule inhibitors of c-Myc.
Introduction
c-Myc is a bHLH-ZIP transcription factor that is involved in cell cycle progression, cellular growth and metabolism, differentiation, and apoptosis (Dang, 1999; Nesbit et al., 1999; Boxer and Dang, 2001; Gardner et al., 2002). c-Myc is overexpressed in numerous cancers, including prostate, gynecological, lung, pancreatic, breast, and colon cancers, B-cell lymphoma, and leukemias. Alterations in c-Myc often are associated with cancer aggressiveness and poor treatment prognosis (Fleming et al., 1986; Nesbit et al., 1999; Pagnano et al., 2001; McNeil et al., 2006). It has been estimated that approximately one-seventh of cancer deaths in the United States per year can be associated with tumors that have changes in the c-Myc gene or its expression (Dang, 1999). Consequently, inhibition of c-Myc is an attractive pharmacological approach in the development of new anticancer treatments. Numerous studies have shown that inactivation of c-Myc can rapidly result in cell-cycle arrest, apoptosis, redifferentiation of tumor cells, tumor vascular degeneration, and ultimately tumor regression (Felsher and Bishop, 1999; Jain et al., 2002; Pelengaris et al., 2002; Russo et al., 2003; Yin et al., 2003; Huang et al., 2006; Mo and Henriksson, 2006; Lin et al., 2007). Our studies demonstrated that depletion of c-Myc, using c-Myc-specific short hairpin RNA, inhibited the growth of 12 various human cancer cell lines, thus suggesting that c-Myc may play a larger role than previously thought in regulating the proliferation and/or survival of transformed cells (Wang et al., 2008). In a preclinical mouse model, inhibition of c-Myc using a dominant-interfering Myc mutant resulted in the regression of established Ras oncogene-induced lung tumors. Effects on normal proliferating tissues were observed in skin, intestine, bone marrow, and testes but were reversed upon stopping treatment (Soucek et al., 2008).
Because c-Myc must heterodimerize with its obligatory bHLH-ZIP partner Max to exhibit transcriptional activity (Walhout et al., 1997), several groups have designed small-molecule inhibitors to interfere with this protein–protein dimerization and thereby inactivate c-Myc function (Berg et al., 2002; Yin et al., 2003; Kiessling et al., 2006; Xu et al., 2006; Wang et al., 2007; Hammoudeh et al., 2009; Shi et al., 2009). Berg et al. (2002) showed that small molecules can interfere with c-Myc/Max dimerization and inhibit c-Myc-dependent transformation of chicken embryo fibroblasts. The inhibitory small molecules disturbed the protein–protein interface, and it was proposed that certain areas of the protein dimer interface contributed disproportionately to the binding affinity between c-Myc/Max (Berg et al., 2002). Yin et al. (2003), through yeast two-hybrid assays, screened chemical libraries and identified several low-molecular-weight compounds that inhibited the heterodimerization of c-Myc and Max, including the thioxothiazolidinone [Z-E]-5-[4-ethylbenzylidene]-2-thioxo-1,3-thiazolidin-4-one (10058-F4) and the compound of interest in this article, 7-nitro-N-(2-phenylphenyl)-2,1,3-benzoxadiazol-4-amine (10074-G5) (Yin et al., 2003). In vitro, 10058-F4 exhibited high binding affinity to the bHLH-ZIP domain of c-Myc, promoted dissociation of the c-Myc/Max dimer, prevented binding of the heterodimer to DNA targets, induced cell cycle arrest, and caused apoptosis (Yin et al., 2003; Gomez-Curet et al., 2006; Huang et al., 2006; Lin et al., 2007; Sampson et al., 2007; Wang et al., 2007; Follis et al., 2008). Hammoudeh et al. (2009) examined in detail the interactions of seven small-molecule inhibitors identified by Yin et al. (2003) and found that 10058-F4 and 10074-G5 inhibited c-Myc/Max heterodimer formation similarly, but bound to different residues in the c-Myc bHLH-ZIP domain. 10058-F4 bound to residues 402 to 409, and 10074-G5 bound to residues 366 to 375. Moreover, 10058-F4 and 10074-G5 could bind simultaneously and independently (Hammoudeh et al., 2009). Based on NMR structures of these small molecules in association with synthetic peptides, it has been postulated that they distort the bHLH-ZIP domain of the unstructured c-Myc monomer, thereby preventing or reversing its heterodimeric association with Max (Follis et al., 2009; Hammoudeh et al., 2009). Based on the activity of 10058-F4 in vitro, Guo et al., (2009) evaluated 10058-F4 in C.B-17 SCID mice bearing PC3 and DU145 human androgen-independent prostate xenografts. In vivo, 10058-F4 was rapidly metabolized, did not reach effective concentrations in the tumor, and showed little or no efficacy against established xenografts.
In the studies presented here, we have assessed the activity of 10074-G5 in vitro and in vivo. Our in vitro objectives were to confirm the growth inhibitory potency of 10074-G5 and its ability to block c-Myc/Max dimerization and to examine the cellular accumulation of 10074-G5. In vivo, we evaluated the efficacy, pharmacokinetics, and metabolism of 10074-G5 in C.B-17 SCID mice bearing Daudi Burkitt's lymphoma xenografts.
Materials and Methods
Drugs and Reagents.
10074-G5 (Fig. 1A) was a gift from Dr. Steven Metallo (Georgetown University, Washington, DC). 10058-F4 (Fig. 1B) was purchased from Chembridge Corporation (San Diego, CA). Doxorubicin (NSC 123127) was provided by the Developmental Therapeutics Program, National Cancer Institute, National Institutes of Health (Rockville, MD). Acetonitrile (HPLC grade), water (HPLC grade), and dimethyl sulfoxide (DMSO) were purchased from Thermo Fisher Scientific (Waltham, MA). Nitrogen gas and liquid nitrogen were purchased from Valley National Gases, Inc. (Pittsburgh, PA). Ammonium acetate, Cremophor EL, and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were purchased from Sigma–Aldrich (St. Louis, MO). Sterile saline (0.154 M NaCl), sterile water, and phosphate-buffered saline (PBS) were purchased from Baxter Healthcare Corporation (Deerfield, IL). Ethanol (200 proof) was purchased from Pharmaco Products (Brookfield, CT). Nitrogen gas for the mass spectrometer was generated with a 75-880 generation system (Parker Balston, Columbus, OH).
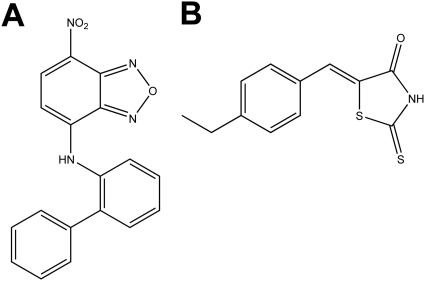
Fig. 1.
Structures of 10074-G5 (A) and 10058-F4 (B).
Dosing Solutions.
Dosing solutions were prepared by dissolving 10074-G5 at a final concentration of 2 mg/ml in Cremophor EL/ethanol/saline (1:1:8, v/v/v). Doxorubicin was dissolved at a final concentration of 0.25 mg/ml in saline. Mice received 0.01 ml/g exact body weight of the dosing solutions or the appropriate vehicle.
Mice.
Specific-pathogen-free, female C.B-17 SCID mice (4–6 weeks of age) were purchased from Charles River Laboratories Inc. (Wilmington, MA) and allowed to acclimate for 1 week to the animal facilities at the University of Pittsburgh Cancer Institute. Mice were maintained in microisolator cages and handled in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and a protocol approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Ventilation and airflow in the animal facility were set to >12 changes/h. Room temperature was regulated at 22 ± 2°C, room humidity was between 30 and 70%, and the rooms were kept on automatic 12-h light/dark cycles. Mice received Prolab ISOPRO RMH 3000, Irradiated Lab Diet (PMI Nutrition International, Brentwood, MO) and water ad libitium except on the evening before dosing for the pharmacokinetic studies, when all food was removed and withheld until 4 h after dosing. Sentinel animals (CD-1 mice in cages with bedding of which 20% was bedding removed from the study animal cages at cage change) were maintained in the room housing the study animals and assayed at monthly intervals for specific murine pathogens by murine antibody profile testing (Charles River Laboratories Inc.). Sentinel animals remained free of specific pathogens throughout the study period, implying that the study animals were free of specific pathogens.
Tumor Cell Lines.
Daudi, human Burkitt's lymphoma cells, and HL-60 human promyelocytic leukemic cells were obtained from American Type Culture Collection (Manassas, VA) and cultured in RPMI medium 1640 with l-glutamine (Lonza Walkersville, Inc., Walkersville, MD), containing 10% heat-inactivated fetal bovine serum, 100 units of penicillin/ml, and 100 μg of streptomycin/ml (Biofluids; Biosource International, Rockville, MD) in an incubator with 95% air, 5% CO2, and 95% humidity at 37°C.
MTT Assay.
Daudi cells (1 × 105 cells) or HL-60 cells (5 × 104 cells) in logarithmic growth were plated into each well of 96-well culture plates and allowed to acclimate for 24 h. Compound was added to wells such that the final concentrations ranged between 1 and 100 μM in medium containing 0.3% DMSO. After 72 h, 50 μl of 1 mg/ml MTT was added to each well and incubated for 4 h. At the end of the incubation, medium containing drug and MTT was removed from each well, and 100 μl of DMSO was added, followed by shaking for 5 min. The absorbance at 570 nm was read on a Dynex NRX Revelation microplate reader (Dynex Technologies, Chantilly, VA). Results were compared with wells containing vehicle-treated cells and expressed as percentage of inhibition. The IC50 was calculated from triplicate experiments by using the Hill equation and the computer program ADAPT II (D'Argenio and Schumitzky, 1997).
Cellular Accumulation of 10074-G5.
Daudi cells (3 × 108 cells in logarithmic growth) were incubated for 0, 1, 3, 6, or 24 h in 3 ml of complete medium containing 10 μM 10074-G5. After incubation, cells were harvested, split into two samples of 1.5 ml each, and overlaid in microcentrifuge tubes containing 0.5 ml of silicon oil (Silicones; General Electric Production Division, Waterford, NY). The tubes were centrifuged at 12,000g for 4 min. After centrifugation, the top 1 ml of medium was removed and stored in cryovials at −70°C until analysis. The remaining medium and silicon oil were carefully removed without disturbing the cell pellets. The sides of the tubes were cleaned with cotton-tipped applicators, and the cell pellets were stored at −70°C until analysis.
Preparation of Cell Lysates and Coimmunoprecipitation Assay.
Daudi cells (1 × 108) were incubated for 4 or 24 h with either 0.3% DMSO in complete medium or 10 μM 10074-G5 in medium. At the end of the incubation, cells were centrifuged at 2000g for 4 min, washed with PBS, pelleted, and resuspended in lysis buffer (200 μl) containing 50 mM Tris-HCl, pH 7.9, 2 mM EDTA, 100 mM NaCl, 1% Nonidet P-40, 10 mM NaF, 10 mM sodium vanadate, and protease inhibitors [1 μg/ml pepstatin, 10 μg/ml aprotin, 5 μg/ml leupeptin, 5 mM phenylmethylsulfonyl fluoride, 0.1 μM microcystin, and 5 mM Na pyrophosphate (BD Biosciences, San Jose, CA)]. Cells were lysed with the tip of a Branson sonifier at the setting of 5 for 30 s (5 s on, 5 s off × 3). Lysate was clarified by centrifugation (12,000g at 4°C for 10 min), and supernatants were collected. The protein concentration in each lysate supernatant was measured by using Bio-Rad protein assay reagents (Bio-Rad Laboratories, Hercules, CA).
For coimmunoprecipitations, equal amounts of protein (400 μg) were adjusted to 500 μl with lysis buffer, and the Max antibody (2 μg; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was added and incubated, with constant agitation, overnight at 4°C. Immobilized protein A-agarose beads (20 μl; Pierce Chemical, Rockford, IL) were then added to the lysate mixture for an additional 4 h at 4°C with agitation. The beads were then washed three times with complete lysis buffer and boiled for 5 min at 95 to 100°C in 80 μl of Laemmli sample buffer (0.4 M Tris-HCL, pH 6.8, 8% SDS, 39% glycerol, 0.04% bromophenyl blue, and 0.4 M dithiothreitol) to which β-mercaptoethanol was added at a final concentration of 5% immediately before use (Bio-Rad Laboratories). The proteins (100 μg, 20 μl) were run on a 7.5% Tris-HCl gel and a 12% Tris-HCl gel at 100 V until the dye front reached the bottom (approximately 90 min). The separated proteins were transferred onto polyvinylidine fluoride membranes and blocked with 5% nonfat milk in PBS + 0.05% Tween for 1 h. The membranes were then incubated with antibodies against c-Myc (Santa Cruz Biotechnology, Inc) on the 7.5% gel or Max (Santa Cruz Biotechnology, Inc) on the 12% gel overnight at 4°C. The blots were probed with a horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG accordingly (Bio-Rad Laboratories), and proteins were detected by ELC detection reagents (PerkinElmer Life and Analytical Sciences, Waltham, MA) following the manufacturer's instructions. The densities of the signal were quantified by densitometry with UN-SCAN-IT (Silk Scientific Inc., Orem, UT). Rabbit serum was used as a control for differentiating the heavy chain IgG and light chain IgG bands from the protein of interest. c-Myc protein expression was normalized to Max protein expression, and then control was set to 1.
Tumor Implantation.
Daudi cells (5 × 106 cells in logarithmic growth) were injected subcutaneously into the right flank of passage C.B-17 SCID mice. When tumors reached approximately 500 mm3, passage mice were euthanized with CO2, and the tumors were harvested aseptically. Tumor cells were isolated by cutting the tumor capsules and flushing the tumors with 10 ml of complete medium. After allowing any clumps to settle by gravity for 5 min, cells were transferred to a 50-ml conical tube and counted on a hemocytometer. The concentration of viable tumor cells, determined by Trypan blue exclusion, was adjusted to 1 × 108 cells/ml in RPMI medium 1640, and 50 μl (5 × 106 cells) was injected subcutaneously into the right flanks of study mice. When the tumors in the study mice were approximately 300 to 400 mm3, the animals were stratified to treatment groups, such that there were no differences in mean body weights or tumor volumes between the groups (analysis of variance and Kruskal-Wallis; p ≥ 0.95).
Efficacy Study.
C.B-17 SCID mice bearing Daudi xenografts were stratified into the following groups (10 mice/group): control; vehicle control (0.01 ml/g body weight, once daily for 5 days); positive control, doxorubicin (2.5 mg/kg/dose, one dose every 4 days for three doses); and 10074-G5 (20 mg/kg/dose, once daily for 5 days). Mice were dosed intravenously on the appropriate schedule, and body weights and tumor volumes were recorded twice weekly. Tumors were measured with digital calipers, and tumor volumes were calculated by using the formula: (L × W2)/2, where L is the longest diameter of the tumor and W is the shortest diameter perpendicular to L. Mice were followed for 10 days after the last 10074-G5 dose (6 days after the last doxorubicin dose) so that tumor growth could be monitored. To calculate doubling time, the tumor volume of each mouse was normalized to day 0 (first day of dosing).
Statistical Analysis.
The mean data were analyzed by using analysis of variance with pairwise comparisons using Dunnett's test or t test. Nonparametic analysis of median data were performed by using Kruskall-Wallis, and pairwise comparisons were conducted by using the Mann–Whitney test. Significance was set at p ≤ 0.05. All statistics were performed with Minitab statistical software (Minitab, Inc., State College, PA).
Pharmacodynamic Study.
Inhibition of c-Myc/Max dimerization in Daudi xenografts was measured by coimmunoprecipitation. C.B-17 SCID mice bearing Daudi xenografts were treated with either vehicle or 20 mg/kg 10074-G5 intravenously consecutively for 5 days, and tumor samples were collected 24 h after the first dose (five mice per treatment group) and 2 h after the fifth dose (five mice per treatment group). Protein extracts were prepared as described previously (Guo et al., 2009). Equal amounts of protein (400 μg) were coimmunoprecipitated by using the same protocol as described above for the cellular lysate.
Pharmacokinetic Study.
C.B-17 SCID mice bearing Daudi xenografts (three mice per time point) were fasted overnight and treated intravenously with 20 mg/kg 10074-G5. Mice were euthanized at 5, 10, 15, 30, 60, 120, 240, 360, 1020, and 1440 min after 10074-G5 administration or 5 and 1440 min after intravenous vehicle administration. The 10074-G5-treated mice to be euthanized at 1020 and 1440 min and vehicle-treated mice to be euthanized at 1440 min were housed in separate metabolism cages for the collection of urine and feces at 0 to 6 h and 6 to 24 h or 0 to 17 h after 10074-G5 or vehicle administration, depending on the time of euthanasia. After the final collection of excreta, cages were washed with 15 ml of sterile water. Mice were euthanized by CO2 inhalation, and blood was collected by cardiac puncture using heparinized syringes. Tumor, liver, kidney, spleen, heart, lung, brain, fat, and skeletal muscle were collected, weighed, and snap-frozen in liquid nitrogen. Blood was centrifuged at 13,000g for 4 min to obtain plasma and packed red blood cells. All samples, including excreta and cage washes, were stored at −70°C until analysis.
Sample Preparation.
Plasma, tissue culture medium, urine, and cage wash samples were extracted directly. Daudi cell pellets, murine tissue samples, and feces were homogenized in five volumes of PBS, pH 7.4. To a 200-μl sample of plasma, urine, red blood cells, tissue homogenate, or fecal homogenate, 20 μl of 10 μg/ml benzanthracene in 90% acetonitrile in water was added as an internal standard. Proteins were precipitated with 1 ml of acetonitrile followed by mixing for 15 s on a Vortex Genie 2 (model G560; Scientific Industries, Bohemia, NY) set at 4. Samples were subsequently centrifuged at 13,000g for 10 min, and the supernatants were transferred to 12 × 75-mm glass tubes and evaporated to dryness under a stream of nitrogen. Each dried residue was suspended in 300 μl of starting mobile phase (30% acetonitrile in water), and 100 μl of each suspended sample was injected onto the HPLC.
HPLC Analysis.
HPLC analysis was performed on a Beckman System Gold HPLC (Beckman Coulter, Fullerton, CA) fitted with a 4.6 × 100-mm, 5-μm Luna C18(2) column (Phenomenex, Torrance, CA) and Brownlee C18 guard column (PerkinElmer Life and Analytical Sciences) that were perfused with a gradient mobile phase that consisted of a linear gradient from acetonitrile containing 1% formic acid/water containing 1% formic acid (30:70, v/v) to 100% acetonitrile containing 1% formic acid over 10 min followed by a 5-min isocratic period before returning to starting mobile phase at 20 min. The mobile phase was pumped at a flow rate of 1 ml/min. Column eluate absorbance at 470 nm was monitored with a Beckman 166 UV-visible absorbance detector (Beckman Coulter). Under these conditions, the retention times of 10074-G5 and the internal standard were approximately 12 and 16 min, respectively. Standard curves of 10074-G5 at concentrations of 0.01 to 3.0 μg/ml (0.02–4.5 μM) in plasma or control tissue homogenates were prepared in triplicate. The 10074-G5-to-internal standard ratio was calculated for each standard by dividing the area of each analyte peak by the area of the respective internal standard peak for that sample. Standard curves of 10074-G5 were constructed by plotting the 10074-G5-to-internal standard ratio versus the known concentration of 10074-G5 in each sample. Standard curves were fitted by linear regression followed by back calculation of concentrations. Concentrations in unknown samples were calculated by comparison with the appropriate standard curve of area ratios of 10074-G5 to internal standard. Concentrations of metabolites present in plasma and tissues are expressed as micromolar equivalents to 10074-G5 based on the assumption that their molar extinction coefficients are the same as 10074-G5 at the wavelength used for quantitation. The lower limit of quantitation of 10074-G5 was 0.01 μg/ml. Coefficients of variation in plasma at a low midrange concentration (0.15 μg/ml, 0.45 μM) and high midrange concentration (1.5 μg/ml, 4.5 μM) were <20 and 8.7%, respectively. Recoveries of 10074-G5 from plasma containing 0.1 μg/ml (0.3 μM) and 1 μg/ml (3.0 μM) were 80 and 85.4%, respectively.
Plasma Protein Binding.
Initial attempts that used Amicon Centrifree ultrafiltration devices (Millipore Corporation, Billerica, MA) to determine plasma protein binding failed, because 10074-G5 bound to the filter of the Centrifree devices. 10074-G5 at concentrations of 0.3 and 3 μg/ml was added to separate 500-μl samples of plasma or PBS containing 100 mM sodium phosphate and 150 mM sodium chloride, and 500 μl of each sample was placed into the sample chamber of a rapid equilibrium dialysis device (Pierce Biotechnology, Inc., Rockford, IL) and dialyzed against 750 μl of PBS for 4 h at 37°C with shaking following the manufacturer's instructions. Complete contents of each chamber were removed and processed for HPLC analysis as described above for plasma samples.
β-Glucuronidase Treatment of Plasma and Urine.
Samples of 15-min plasma and 0- to 6-h urine (200 μl) from mice treated intravenously with 20 mg/kg 10074-G5 were incubated in a shaking water bath for 24 h at 37°C with 10,000 units of β-glucuronidase (10,000 units/mg from bovine liver; Sigma–Aldrich). Samples were then collected, extracted, and processed as described above for plasma or urine.
Pharmacokinetic Analysis.
The terminal half-life, clearance, volume of distribution at steady state, area under the concentration (AUC) versus time curves for plasma, and the AUC and terminal half-life for tissues were estimated by noncompartmental analysis using the Lagrange function (Yeh and Kwan, 1978) as implemented by the LAGRAN program (Rocci and Jusko, 1983) The AUCs were also calculated by using the trapezoidal method to evaluate agreement between the two methods.
Partial Mass Balance.
The percentage of dose in each tissue or organ was estimated by calculating the exact mean dose of 10074-G5 that each mouse received, e.g., 1.305 μmol for an average 21.67-g mouse. For tissues and organs where we have the exact tissue weight, the concentration/g was multiplied by the organ/tissue weight and divided by the dose administered and multiplied by 100% to determine the percentage dose in that tissue at each time point. For tissues or organs for which we did not have total weight, but only the weight that we collected at euthanasia, the percentage dose in that organ was calculated based on Table 21 in Tissue Mass Balance in Physiological Parameter Values for PBPK Models, a report prepared by the International Life Science Institute Risk Science Institute under a cooperative agreement with the U.S. Environmental Protection Agency, Office of Health and Environmental Assessment, December 1994 (Brown et al., 1997). For example, muscle represents 38.4% of body weight for a mouse. Tissues that were not measured, such as skin, gastrointestinal tract and contents, bone, and the rest of the body can account for approximately 39.6% of body weight. We do not know the concentrations of 10074-G5 or metabolites in these tissues or organs.
LC-MS/MS.
Metabolite identifications were performed on plasma samples obtained 15 min after intravenous administration of 20 mg/kg 10074-G5. A control murine plasma was also analyzed to exclude endogenous signals. Sample preparation (200 μl of plasma) consisted of protein precipitation with 1 ml of acetonitrile. The clear supernatant was evaporated to dryness under a gentle stream of nitrogen. The dried residue was reconstituted in 100 μl of 10% acetonitrile and transferred to autosampler vials. A sample volume of 30 μl was injected onto the LC system. The HPLC system consisted of an Agilent Technologies (Santa Clara, CA) 1200 autosampler and binary pump, a Phenomenex (Torrance, CA) hydro Synergi (4 μm, 100 × 2 mm) column kept at ambient temperature, and a gradient mobile phase. Mobile-phase solvent A consisted of 0.1% (v/v) formic acid in methanol, and mobile-phase solvent B consisted of 0.1% (v/v) formic acid in water. Initially, we attempted to use the gradient system described above for the quantitation of parent compound, but that resulted in unsatisfactory ionization of analytes. Therefore, for analyte identification studies, the initial mobile phase composition was 40% solvent A and 60% solvent B pumped at a flow rate of 0.2 ml/min. From 0 to 40 min, solvent A was increased linearly from 40 to 70%. From 40.0 to 40.1 min, the flow was increased to 0.4 ml/min, and the gradient was increased to 100% A. These conditions were maintained until 45 min. From 45.0 to 45.1 min, the flow rate was increased to 0.5 ml/min, and the gradient was decreased to 40% A. These conditions were maintained until 50 min, after which the next sample was injected.
HPLC eluent was led through an Agilent 1200 DAD series G1315C diode array detector (DAD) set at 470 nm, to allow comparison of the peaks identified by the LC-MS/MS system with those identified by the HPLC-UV system.
Mass spectrometric detection was carried out with an ABI 4000 QTRAP (MDSSciex, Concord, ON, Canada) mass spectrometer with electrospray ionization in positive mode. The settings of the mass spectrometer were as follows: channel electron multiplier, 2500 V; ion source nitrogen gas 1 (GS1), 40 L/h; ion source nitrogen gas 2 (GS2), 30 L/h; collision-activated dissociation, high; curtain gas, 10; ion spray, 5500 V; declustering potential, 60 V; collision energy spread, 10 V; collision energy, 40 V. The mass spectrometer was operated in the enhanced product ion mode, with collision energy operated at 30, 40, and 50 V. The HPLC system and mass spectrometer were controlled by Analyst software (version 1.4.2; MDSSciex), and data were collected with the same software.
Based on the structure of the parent compound, we screened for masses corresponding to expected metabolic transformations, such as reduction of the nitro-moiety, hydroxylation, acetylation, and glucuronidation.
Results
In Vitro Cytotoxicity.
The IC50 values for 10074-G5 and 10058-F4 against Daudi cells after 72 h of incubation were 15.6 ± 1.5 and 17.8 ± 1.7 μM, respectively (Fig. 2A). The IC50 values for 10074-G5 and 10058-F4 against HL-60 cells were 13.5 ± 2.1 and 26.4 ± 1.9 μM, respectively (Supplemental Data).
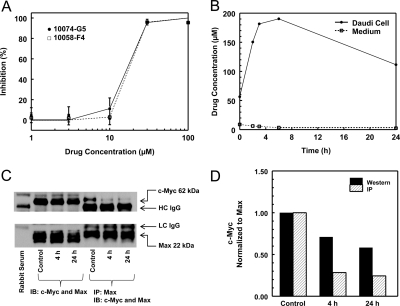
Fig. 2.
In vitro analysis of 10074-G5 against Daudi Burkitt's lymphoma cells. A, in vitro growth inhibition of 10074-G5 and 10058-F4 determined by MTT assay at 72 h. A representative assay of three assays is shown. Data points are mean ± S.D. of three wells/concentration, and curves are the best fit to the data using the Hill equation. B, concentrations of 10074-G5 in medium and Daudi cells in culture. Daudi cells were incubated with 10 μM 10074-G5 for various times between 0 and 24 h, and cell concentrations and medium concentrations were measured by HPLC assay. C, coimmunoprecipitation (IP) and Western blot (IB) analysis of c-Myc and Max protein expression in Daudi cells incubated for 4 or 24 h in either 0.3% DMSO (control) or 10 μM 10074-G5. D, normalized c-Myc protein with control set to 1.
Cellular Accumulation of 10074-G5.
Daudi cells accumulated 10074-G5, and the highest intracellular concentration was observed at 6 h (Fig. 2B). Assuming that the volume of 108 cells is approximately 100 μl, the accumulation in the cells was approximately 50-fold higher than the concentration present in the medium. The concentration of 10074-G5 in the medium decreased with time, but no additional peaks were observed in the HPLC chromatograms. After 24 h of incubation, the amount of 10074-G5 recovered in the medium and cell pellets accounted for only 35% of the initial amount added to the medium. In control samples, where 10 μM 10074-G5 was incubated in medium with no cells for 24 h and processed in the same manner, >95% drug was recovered. This suggested that with continuous incubation 10074-G5 was metabolized by the cells to moieties that could not be detected by using the HPLC assay described.
Coimmunoprecipitation and Western Blot Results.
10074-G5 inhibited c-Myc/Max dimerization in Daudi cells by approximately 75% at 4 h, and this inhibition was maintained through 24 h of incubation (Fig. 2, C and D). Total c-Myc protein expression also decreased, and after 24 h exposure to 10 μM 10074-G5, c-Myc protein expression decreased approximately 40% compared with vehicle-treated control. There were no observed changes in Max expression in 10074-G5-treated cells.
Efficacy Data.
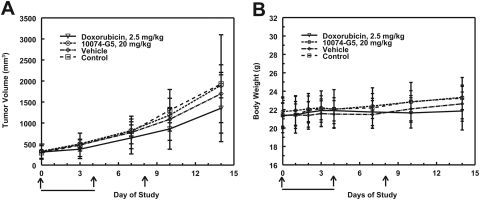
Treatment of C.B-17 SCID mice, bearing Daudi xenografts, with the maximum soluble dose of 10074-G5, 20 mg/kg i.v., for 5 consecutive days did not significantly inhibit tumor growth (Fig. 3A). The maximum mean percentage treated tumor/control tumor occurred on day 10 and was 91% for mice treated with 20 mg/kg 10074-G5 and 66% for mice treated with 2.5 mg/kg doxorubicin on days 0, 4, and 8. Mean time to one doubling for doxorubicin-treated mice was 7.13 days, which was significantly different from the doubling time for the 10074-G5-treated mice (5.05 days) and the control mice (4.50 days). No significant decreases in body weight were observed in any treatment group (Fig. 3B).
Fig. 3.
Efficacy study of 10074-G5 in mice bearing Daudi tumor xenografts. Results are the mean ± S.D. for tumor volumes (A) and body weight (B) of 10 mice per treatment group. The days of treatment with 10074-G5 are indicated by bold lines. Days of doxorubicin treatment are indicated by arrows.
Pharmacodynamics.
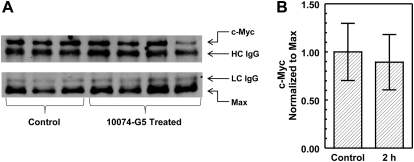
In Daudi tumors harvested at 24 h after the first dose and 2 h after the fifth dose of 20 mg/kg i.v. 10074-G5, there was no difference in c-Myc or Max protein expression compared with controls for either time point. The results for the tumors excised 2 h after the fifth dose are shown in Fig. 4.
Fig. 4.
Coimmunoprecipitation and immunoblot analysis of c-Myc and Max protein expression in Daudi xenografts from mice treated for 5 days with 20 mg/kg i.v. 10074-G5. A, tumors were collected 2 h after the fifth dose. Representative immunoblots of tumors from three control and four 10074-G5-treated mice are shown. c-Myc protein expression was normalized to the heavy chain (HC) IgG (top), and Max protein expression was normalized to the light chain (LC) IgG (bottom). The adjusted c-Myc protein expression was normalized to the adjusted Max protein expression, and control was set to 1. B, the graph represents mean ± S.D. of normalized c-Myc expression of the bands in the blots in A.
Protein Binding.
10074-G5 was >98% plasma protein bound at both 0.3 and 3 μM.
Pharmacokinetics of 10074-G5.
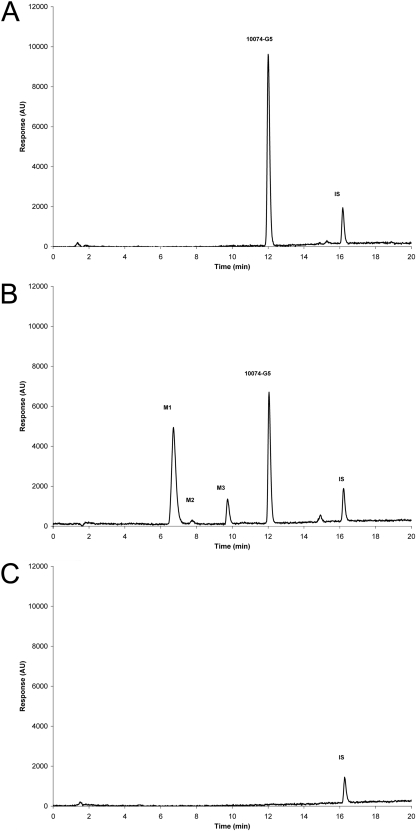
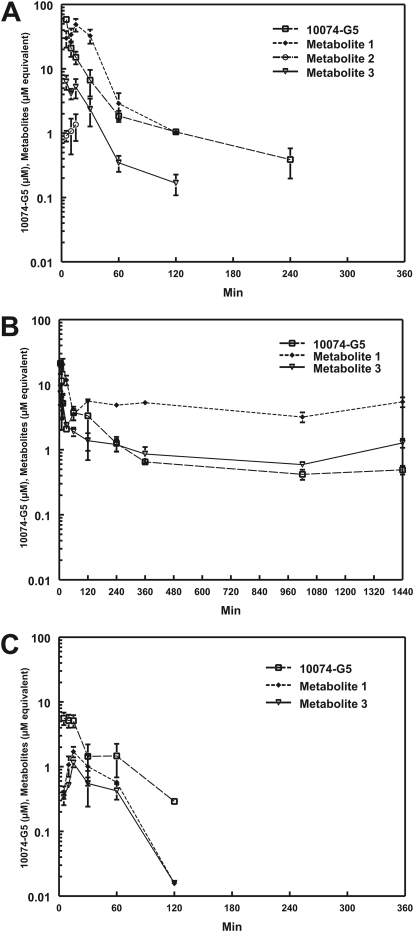
Plasma 10074-G5 peak concentration (Cmax) of 58.5 ± 2.7 nmol/ml was observed at 5 min after intravenous administration of 20 mg/kg to mice bearing Daudi xenografts. 10074-G5 concentration in plasma declined rapidly and was accompanied by the appearance of three more polar metabolites that we labeled M1, M2, and M3, based on their order of elution from the HPLC column (HPLC chromatograms; Fig. 5). Concentrations of M1 increased as concentrations of 10074-G5 and M3 decreased (Fig. 6A). Neither 10074-G5 nor its metabolites were detectable in plasma beyond 240 min. The plasma half-life of 10074-G5 was 37 min, clearance was 69 ml/min/kg, and the volume of distribution was approximately 3000 ml/kg. The highest tissue concentrations of 10074-G5 were observed in liver (40.8 nmol/g) and lung (44.3 nmol/g) and occurred at the earliest time point sampled, 5 min (Table 1). Except for lung, liver, and fat, tissue concentrations of 10074-G5 were lower than those of plasma at all time points. Concentrations of 10074-G5 in lung were higher than those in plasma between 10 and 240 min. Concentrations of 10074-G5 in the liver were higher than those in plasma at 30 min after administration of compound and were still detectable at 1440 min, the last time point sampled. 10074-G5 accumulated in fat more slowly, and peak concentrations of 19.3 nmol/g were not attained until 30 min (Table 1) and remained higher than plasma concentrations until 240 min, after which 10074-G5 was not detectable. Metabolite M2 was detected only in plasma (Fig. 6A) and kidney (data not shown) and was present at much lower concentrations than metabolites M1 or M3. In all other tissues, only M1 and M3 were detected. Based on micromolar equivalents, concentrations of M1 in the liver were higher than those of 10074-G5 at all time points except 5 min (Fig. 6B). Cmax of 10074-G5 in Daudi xenografts (5.6 nmol/g) was observed at the earliest time sampled (5 min) but was only approximately 10% of concomitant peak plasma concentrations (58.5 nmol/ml) (Fig. 6C; Table 1). Cmax correlates with the AUC for all tissues, with the exception of fat, with an R2 of 0.623 for AUC0→∞ versus Cmax.
Fig. 5.
Chromatograms of 10074-G5 in control plasma compared with the plasma of a mouse euthanized 10 min after intravenous administration of 20 mg/kg 10074-G5. A, chromatogram of control plasma to which 1 μg of 10074-G5 and internal standard, benzanthracene, has been added before extraction. B, plasma was collected 10 min after administration of 20 mg/kg 10074-G5 to which internal standard has been added. Three additional peaks were detected and labeled M1, M2, and M3 in the order of their elution from the column. C, chromatogram of plasma from an untreated mouse to which internal standard was added.
Fig. 6.
Plasma (A), liver (B), and Daudi xenograft (C) concentration versus time curves from mice treated intravenously with 20 mg/kg 10074-G5. Potential metabolites were identified as M1, M2, and M3 based on their elution from the HPLC column. The metabolite concentrations are expressed as micromolar equivalents of 10074-G5.
TABLE 1.
Concentrations of 10074-G5 in plasma and tissues after administration of 20 mg/kg 10074-G5 intravenously to C.B-17 SCID female mice bearing Daudi xenografts
| Time | Plasma | Tumor | Liver | Kidney | Lung | Spleen | Heart | Brain | Skeletal Muscles | Red Blood Cells | Fat |
|---|---|---|---|---|---|---|---|---|---|---|---|
| min | nmol/ml | nmol/g | nmol/g | nmol/g | nmol/g | nmol/g | nmol/g | nmol/g | nmol/g | nmol/mlg | nmol/g |
| 5 | 58.46 ± 2.73 | 5.61 ± 1.23 | 40.81 ± 8.71 | 29.26 ± 7.74 | 44.29 ± 8.18 | 7.95 ± 0.60 | 22.00 ± 3.48 | 16.76 ± 3.33 | 8.25 ± 0.83 | 13.64 ± 3.77 | 10.87 ± 1.52 |
| 10 | 20.98 ± 6.54 | 5.16 ± 1.18 | 22.92 ± 18.86 | 17.67 ± 6.31 | 40.37 ± 5.43 | 6.49 ± 1.97 | 11.44 ± 1.86 | 6.83 ± 2.40 | 5.39 ± 2.07 | 3.14 ± 2.07 | 10.09 ± 1.48 |
| 15 | 15.11 ± 3.37 | 5.12 ± 1.11 | 10.36 ± 4.18 | 5.28 ± 0.15 | 26.34 ± 0.10 | 2.88 ± 0.32 | 8.34 ± 2.25 | 3.30 ± 0.35 | 4.73 ± 1.11 | 2.68 ± 0.39 | 18.38 ± 5.01 |
| 30 | 6.66 ± 2.94 | 1.44 ± 0.76 | 6.71 ± 0.12 | 1.77 ± 0.15 | 12.36 ± 0.97 | 1.20 ± 0.12 | 2.53 ± 0.20 | 1.06 ± 0.21 | 2.75 ± 0.81 | 0.48 ± 0.17 | 19.30 ± 7.05 |
| 60 | 1.85 ± 0.37 | 1.47 ± 0.78 | 5.74 ± 0.68 | 1.04 ± 0.27 | 3.59 ± 1.57 | 0.89 ± 0.42 | 0.92 ± 0.22 | 0.52 ± 0.02 | 1.02 ± 0.45 | 0.19 ± 0.11 | 6.83 ± 4.03 |
| 120 | 1.05 ± 0.11 | 0.29 ± 0.03 | 3.33 ± 2.63 | 0.24 ± 0.04 | 3.08 ± 1.11 | ND | 0.52 ± 0.14 | 0.31 ± 0.02 | 0.68 ± 0.27 | 0.06 ± 0.00 | 4.59 ± 0.29 |
| 240 | 0.39 ± 0.19 | ND | 1.25 ± 0.32 | 0.21 ± 0.03 | 0.67 ± 0.12 | ND | ND | 0.04 ± 0.00 | ND | ND | 0.91 ± 0.46 |
| 360 | ND | ND | 0.65 ± 0.03 | ND | ND | ND | ND | ND | ND | ND | ND |
| 1020 | ND | ND | 0.42 ± 0.07 | ND | ND | ND | ND | ND | ND | ND | ND |
| 1440 | ND | ND | 0.49 ± 0.08 | ND | ND | ND | ND | ND | ND | ND | ND |
| t 1/2 | 37 | 29 | — | — | — | — | — | — | — | — | — |
| AUC0→∞ a | 864 (899) | 223 (212) | 1819 (1864) | 621 (681) | 1063 (1232) | 147 (141) | 349 (380) | 216 (228) | 255 (244) | 131 (132) | 1541 (1584) |
ND, not determined.
AUCs were calculated noncompartmentally using both the Lagrange function and the Trapezoidal method (in parentheses). AUC units are nanomoles per milliliter per minute for plasma and nanomoles per gram per minute for tissues.
Urinary and Fecal Excretion of 10074-G5.
Less than 0.02% of the dose of 10074-G5 was excreted in the urine and feces as parent compound. Based on micromolar equivalents, M1 accounted for approximately 1.5% of the dose excreted in the urine and 0.08% excreted in the feces. M2 accounted for 0.06% of the dose of 10074-G5 and was detected only in the urine. M3 accounted for only 0.03% of the dose.
Treatment of Plasma and Urine with β-Glucuronidase.
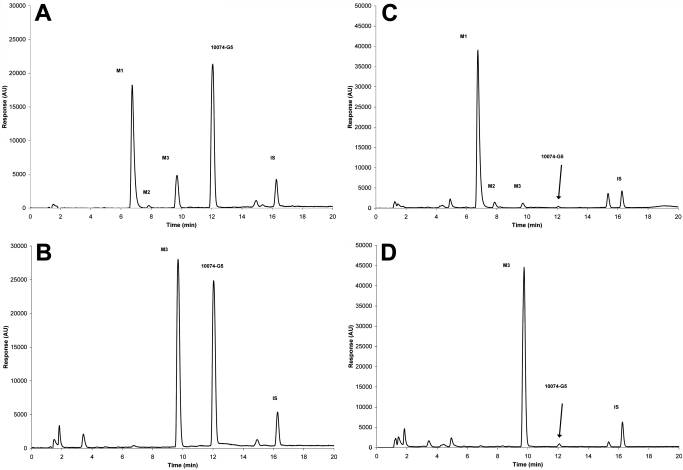
Because concentrations of M1, the major and most polar of the three metabolites, increased as concentrations of 10074-G5 and M3 deceased, we speculated that it represented a glucuronide conjugate of either 10074-G5 or M3. As shown in the chromatograms displayed in Fig. 7, treatment of plasma and 0- to 6-h urine samples with β-glucuronidase resulted in a large decrease in M1 and a comparable increase in M3 with no apparent increase in the parent compound peak, suggesting that M1 was indeed the glucuronide conjugate of M3 and not of 10074-G5. M2 disappeared when incubated with β-glucuronidase (Fig. 7).
Fig. 7.
Effect of treatment with β-glucuronidase on plasma and urine from mice treated with 20 mg/kg i.v. 10074-G5. The metabolites M1, M2, M3, and the parent compound (10074-G5) are shown before (A) and after (B) treatment with β-glucuronidase at 37°C for 24 h for plasma and before (C) and after (D) β-glucuronidase treatment of a 0- to 6-h urine sample.
Partial Mass Balance.
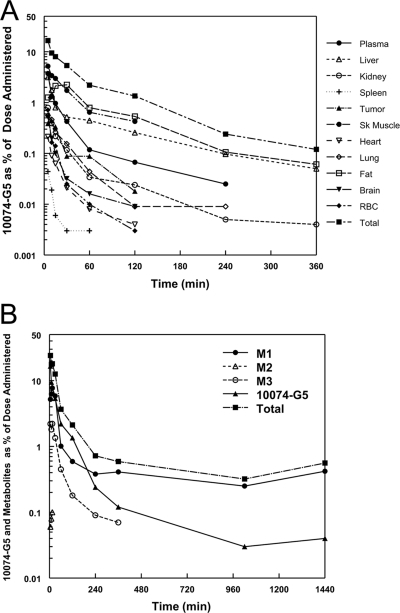
The total amount of the administered dose that was detectable at the earliest time point sampled, 5 min, was 24% of the dose: 16.5% as the parent compound, 10074-G5 (the breakdown of percentage dose of 10074-G5 per tissue is shown in Fig. 8A), 5.2% as M1, 0.06% as M2, and 2.2% as M3 as measured by HPLC (Fig. 8B). Several of the metabolites that are detected by LC-MS/MS in the plasma at 15 min (Table 2; Fig. 9) are not detectable by HPLC-UV and therefore are not accounted for in this partial mass balance. By 1440 min, there was less than 0.04% of parent compound detected and this was localized in liver, the primary site of metabolism. At this same time point, M1, the proposed glucuronide conjugate of the hydroxylated metabolite, represented 0.42% of the dose using calculations based on micromolar equivalents with 10074-G5. Tissues and organs not measured consisted of skin, bone, and gastrointestinal tract as well as the remaining carcass and these organs can account for as much as 40% of the body weight of the mouse.
Fig. 8.
Percentage of mean dose accounted for in plasma and tissues as a function of time after administration. A, percentage of mean dose represented by 10074-G5 in plasma and tissues as a function of time after intravenous administration of 10074-G5. B, total percentage of mean dose accounted for as 10074-G5 and metabolites as a function of time after administration of 10074-G5. Metabolites were determined as micromolar equivalents based on the assumption of the same extinction coefficient at the absorption wavelength used to quantitate 10074-G5 by HPLC.
TABLE 2.
Overview of metabolites of 10074-G5 analyzed by HPLC-UV and LC-MS/MS
| Peak | LC-MS/MS DAD | [M+H]+ (m/z)/Cps | Product Ions | HPLC-UV | Postulated Designation a |
|---|---|---|---|---|---|
| min | min | ||||
| MS1 | 8.9 | 495/2.8 × 106 | 371, 283, 239, 195, 189, 177, 145, 133, 101, 89, 73 | Amino-O-Gluc | |
| MS2 | 9.0 | 345/1.3 × 105 | 333, 289, 277, 263, 233, 219, 189, 174, 167, 145, 101 | Amino-Acetyl | |
| MS3 | 11.1 | 495/1.5 × 106 | 302, 301, 285, 184, 177, 133, 101, 89, 73 | 6.9 (M1) b | Amino-O-Gluc |
| MS4 | 11.6 | 525/2.5 × 106 | 481, 221, 177, 133, 89, 73 | 6.9 (M1) b | Nitro-O-Gluc |
| MS5 | 12.8 | 525/1.4 × 107 | 481, 349, 332, 315, 298, 284, 273, 255, 221, 203, 133, 89, 73 | 6.9 (M1) b | Nitro-O-Gluc |
| 12.6 | |||||
| MS6 | 13.8 | 495/1.6 × 105 | 370, 327, 319, 302, 301, 285, 274, 247, 189, 184, 182, 156, 145, 113, 89 | Amino-O-Gluc | |
| MS7 | 17.2 | 495/4.0 × 105 | 469, 425, 392, 381, 343, 337, 319, 302, 301, 285, 274, 263, 249, 219, 205, 189, 182, 161, 138, 99, 89 | 7.8 (M2) c,d | Amino-O-Gluc |
| MS8 | 17.4 | 479/5.4 × 106 | 449, 437, 405, 393, 361, 349, 317, 305, 261, 243, 217, 199, 177, 133, 101, 89, 87, 73 | Amino-Gluc | |
| MS9 | 21.1 | 495/7.0 × 105 | 453, 393, 374, 343, 319, 302, 301, 288, 284, 273, 256, 242, 225, 189, 178, 169, 145 | Amino-O-Gluc | |
| MS10 | 22.0 | 525/1.5 × 106 | 349, 332, 315, 284, 273, 257, 205, 178, 151, 117, 85 | Nitro-O-Gluc | |
| MS11 | 23.7 | 525/1.5 × 106 | 493, 481, 437, 393, 349, 305, 293, 277, 261, 233, 219, 189, 175, 145, 133, 89 | Nitro-O-Gluc | |
| MS12 | 24.6 | 349/1.1 × 107 | 331, 315, 298, 284, 272, 255, 229, 197, 185, 167, 139, 128, 115, 91, 79 | 9.8 (M3) b | Nitro-OH |
| 24.3 | |||||
| MS13 | 28.0 | 303/2.9 × 106 | 286, 273, 258, 225, 209, 197, 189, 159, 153, 135, 113, 107, 99, 89, 79, 77 | Amino | |
| MS14 | 31.7 | 479/1.2 × 106 | 437, 405, 397, 355, 349, 307, 293, 263, 215, 197, 155, 135, 107, 99, 89 | Amino-Gluc | |
| MS15 | 32.2 | 303/8.0 × 105 | 285, 272, 257, 232, 205, 179, 161, 137, 133, 119, 105, 95, 91, 81, 67, 54 | Amino | |
| MS16 | 33.0 | 319/1.6 × 105 | 301, 284, 272, 257, 245, 233, 167, 145, 119, 105, 91 | Amino-OH | |
| MS17 | 34.2 | 333/6.5 × 107 | 315, 299, 282, 268, 256, 254, 242, 229, 204, 181, 167, 152, 140, 105, 78 | 12.1 (10074-G5) | Parent |
| 34.0 | |||||
| MS18 | 35.7 | 479/7.0 × 105 | 461, 429, 417, 379, 353, 321, 305, 283, 261, 239, 223, 179, 161, 158, 133, 95, 89 | Amino-Gluc | |
| MS19 | 36.8 | 345/4.8 × 105 | 303, 285, 272, 258, 255, 245, 167, 85 | Amino-Acetyl |
Designation includes: the fate of the nitro moiety, either intact as nitro or reduced to an amino moiety; and any oxidations or conjugations inferred from the data; Gluc, glucuronide.
Incubation of urine sample with β-glucuronidase results in disappearance of M1 and appearance of M3, suggesting that M1 contains a glucuronide of M3.
Incubation of urine sample with β-glucuronidase results in disappearance of M2.
M2 could not be detected by the DAD in the LC-MS/MS system. The retention time was estimated by correlating the retention times of M1, M3, and parent compound on both systems, and interpolating the expected LC-MS/MS retention time of M2 based on its retention time on the HPLC-UV system.
Fig. 9.
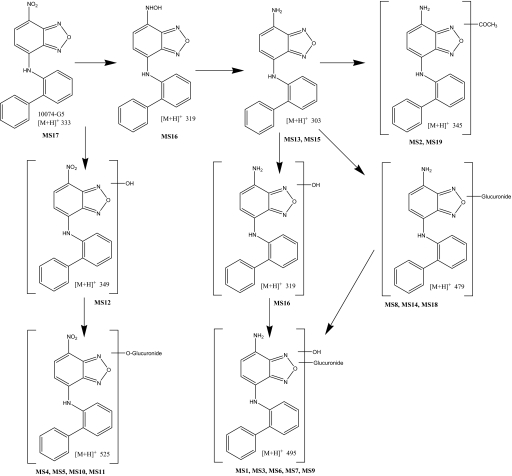
Proposed metabolism of 10074-G5 in mice. Bold MS numbers correspond to LC-MS/MS peaks in Table 2.
LC-MS/MS Analysis of Metabolites of 10074-G5.
Because the amount of 10074-G5 and the three metabolites detected with the HPLC assay in urine and feces accounted for so little of the dose of 10074-G5 administered, we used LC-MS/MS methodology to examine plasma for other potential metabolites of 10074-G5 (Table 2). Because the product ions for each [M+H]+ mass were obtained by individual injections and mass spectrometric analyses, the retention times listed in Table 2 have a variability of ≤0.7 min.
LC-MS/MS analysis indicated that the peak identified by HPLC-UV as M1 actually corresponded to three metabolites (MS3, MS4, and MS5) that were chromatographically resolved on the longer LC-MS/MS system. MS5 is quantitatively the most important contributor to M1 and could be detected by the DAD system. MS3, MS4, and MS5 all are glucuronides of hydroxylated parent compound, which is in agreement with the results of the β-glucuronidase experiment.
The peak identified by HPLC-UV as M2 could not be detected by the DAD in the LC-MS/MS system. The retention time of M2 was estimated by correlating the retention times of M1, M3, and parent compound on HPLC-UV and LC-MS/MS systems and interpolating the expected LC-MS/MS retention time of M2 based on its retention time on the HPLC-UV system. M2 seems to be the glucuronide of the proposed amino metabolite of G5. This is supported by the results of the β-glucuronidase experiment.
The peak identified by HPLC-UV as M3 corresponded to metabolite MS12, the hydroxyl metabolite of 10074-G5. This was in agreement with the results of the β-glucuronidase experiment, which resulted in conversion of M1 to M3. Other metabolites detected by LC-MS/MS confirm the major metabolic pathways indicated by M1, M2, and M3: oxidation, glucuronidation, and reduction of the nitro moiety to an amino function. These other metabolites suggested various glucuronides of hydroxylated parent compound (MS4, MS5, MS10, and MS11) and similar metabolites with an amino instead of a nitro function (MS1, MS3, MS6, and MS9). Apart from the MS6–MS10 pair, these metabolites pair up in such a way that reduction of the nitro to an amino moiety results in a decrease in retention time of approximately 1.7 to 2.7 min; a similar retention time difference was observed between parent compound 10074-G5 (MS17) and its amino metabolite (MS15). The existence of the various isomeric metabolites is reflective of the various positions on the molecule that are available for possible hydroxylation. Among the glucuronides of the parent compound, MS5 and MS10 had virtually identical enhanced product ion spectra, whereas MS4 and MS11 were each different. The fragment at m/z 349 represents the molecular ion after loss of the glucuronide moiety (−176), and the base peak at m/z 315 represents an additional loss of H20 and O (−34), a loss also observed in the enhanced product ion spectrum of 10074-G5, resulting in the fragment of m/z 299. We were unable to assign the positions of the conjugates, although metabolism of the exposed phenyl rings is most likely.
Furthermore, there were indications of direct glucuronidation of the amino metabolite (MS8, MS14, and MS18) and acetylation of the amino metabolite (MS2 and MS19). Most likely, MS18 and MS19 are conjugated on the primary amino (reduced nitro) moiety, reducing the polarity and possibly explaining their more prolonged retention. Likewise, MS16 might be a hydroxylamine (the intermediate between the parent compound and its amino metabolite). The hydroxylamine metabolite would be expected to have a lower pKa than the amino metabolite, resulting in its longer retention time. Based on the masses observed, we propose the 10074-G5 metabolic scheme as depicted in Fig. 9.
Discussion
Although the development of small-molecule inhibitors of protein–protein interactions has proven to be challenging, significant progress is being made. Several groups, including our own, have identified small-molecule inhibitors that target c-Myc protein–protein complexes and thereby inhibit the growth of c-Myc-dependent cell lines in vitro (Yin et al., 2003; Kiessling et al., 2006; Berg, 2008; Shi et al., 2009). These studies provide evidence that bHLH-ZIP proteins are druggable targets, although translating these in vitro observations into antitumor activity in animals has been met with limited success (Guo et al., 2009).
Several groups have shown that inactivation of MYC in transgenic mouse models greatly reduces cellular proliferation, promotes cellular differentiation and apoptosis, and ultimately leads to tumor regression (Felsher and Bishop, 1999; Yin et al., 2003; Soucek et al., 2008). However, a previously studied small-molecule c-Myc/Max inhibitor, 10058-F4, had limited in vivo efficacy because of its rapid metabolism and consequent failure to reach inhibitory concentrations in the tumor (Guo et al., 2009). Additional work is required to develop a metabolically stable, small-molecule c-Myc/Max inhibitor with activity in animal xenograft models.
In the present study, we have demonstrated that 10074-G5 is cytotoxic in vitro against Daudi and HL-60 cells, which overexpress c-Myc (Dalla-Favera et al., 1982; Li et al., 2003; Wang et al., 2007). This verifies the original study by Yin et al. (2003), which demonstrated an IC50 for 10074-G5 of 32 μM in a c-Myc-overexpressing mammalian cell line. We found 10074-G5 to be highly (50-fold) concentrated in Daudi cells. This large intracellular accumulation suggests that the plasma concentrations required to inhibit in vivo tumor growth will need to be higher than the concentration suggested by in vitro IC50 values, because the plasma protein binding (>98%) and the rapid metabolism (t1/2 37 min) of 10074-G5 limit tumor accumulation.
After 4 h of in vitro incubation with 10 μM 10074-G5, there was a substantial inhibition of c-Myc/Max dimerization and a further decrease in c-Myc levels after longer incubations. These results parallel previous studies demonstrating reductions in endogenous c-Myc protein levels in HepG2, HL-60, and Burkitt's lymphoma cells when exposed to 10058-F4 (Gomez-Curet et al., 2006; Huang et al., 2006; Lin et al., 2007; Sampson et al., 2007; Wang et al., 2007). These results indicate that toxicity may depend on both the ability of the drug to inhibit the c-Myc/Max heterodimer formation and the consequent decrease in intracellular c-Myc protein. c-Myc has a very short half-life (∼20–30 min) and, if unable to heterodimerize with Max, c-Myc is ubiquitinated and subsequently degraded (Gregory and Hann, 2000). It is also possible that much of the decline in overall c-Myc levels simply reflects a secondary reduction in its synthesis as a result of cell cycle arrest in G0/G1 (Yin et al., 2003). On the other hand, the invariant levels of Max probably reflect its much greater stability (t1/2; >24 h), its lack of interaction with small molecules such as 10074-G5 or 10058-F4 (Wang et al., 2007; Follis et al., 2008; Hammoudeh et al., 2009), and its ability to interact with numerous other bHLH-ZIP factors (Baudino and Cleveland, 2001). 10074-G5 was nontoxic to Daudi tumor-bearing mice at 20 mg/kg i.v., its maximum soluble dose. When treated for 5 consecutive days, 10074-G5 had no effect on the growth of the Daudi xenografts in C.B-17 SCID mice. To determine the pharmacokinetics and tissue distribution of 10074-G5, we developed an HPLC method that was used with LC-MS/MS to quantitate parent compound and structurally elucidate potential metabolites. We found that most of the drug-related peaks elute early in the HPLC run and these early eluting metabolites seem to be glucuronide conjugates of hydroxylated or nitro-reduced parent, as is supported by the treatment of the 15-min plasma sample with β-gluronidase. This suggests rapid metabolism by phase I and phase II enzymes localized primarily in the liver. The fact that the same metabolite pattern by HPLC is obtained in the 0- to 6-h urine collection suggests that these metabolites are rapidly formed and cleared in the urine. Furthermore, at least two (M1 and M3) of the early eluting metabolites of 10074-G5 are present in all tissue samples at the earliest time point sampled (5 min after dosing). Because the concentrations of 10074-G5 in the liver, kidney, and lung all are at Cmax at the earliest sample obtained in the mice (5 min), it is clear that these tissues are well perfused and metabolism occurs rapidly. Likewise, the highest concentration of compound and metabolites in the tumor occurs at 5 min, but is a magnitude lower than the highly perfused tissues. Fat is the only tissue with Cmax >5 min. Because of the lipophilic nature of 10074-G5 the compound accumulates longer in fat, resulting in a Cmax of 30 min. As was observed with 10058-F4, 10074-G5 was metabolically labile and rapidly metabolized to more polar compounds.
From previous studies with other analogs of 10074-G5, it is clear that the loss of the nitro group or replacement of this group with an amine results in the analogs being inactive in cell culture and in binding assays with c-Myc (Steven Metallo, personal communication). Addition of a hydroxyl group on the phenyl rings results in decreased activity of the analog as well. Glucuronides of compounds generally are accepted as inactive. Therefore, the most likely reason for lack of antitumor activity is the rapid metabolism of 10074-G5 to inactive metabolites, resulting in tumor concentrations of parent compound that are too low to inhibit c-Myc/Max dimerization.
We have identified three discrete binding sites within the 85-residue bHLH-ZIP domain of c-Myc, and NMR experiments have demonstrated that binding at one site on c-Myc does not affect the affinity or binding at other sites, which indicates that small-molecule inhibitors can bind simultaneously and independently on all three sites (Follis et al., 2008). By linking the two compounds, it may be possible to target two sites on c-Myc bHLH-ZIP domain simultaneously and increase the potency of these small-molecule inhibitors. Binding affinity of 10058-F4 to recombinant c-Myc bHLH-ZIP is 5.7 μM, and the binding affinity of 10074-G5 is 2.8 μM, whereas the linked compounds, link N1 and link N3, have a binding affinity of 0.002 and 0.0006 μM, respectively (Steven Metallo, personal communication). Furthermore, the structure of these link compounds may block some of the active sites of metabolism available on the individual compounds. Therefore, the combination of two or more compounds may be more effective than any one alone (Hammoudeh et al., 2009).
Currently, we are using pharmacophore modeling to generate future structures and compounds that have binding affinity similar, or superior, to 10074-G5 and 10058-F4 (Mustata et al., 2009). These compounds will be evaluated in cell culture followed by selection of the most promising candidates for advancement into animal studies. Based on the structural variability of compounds already proven to bind and inhibit c-Myc, the development of newer analogs that are active in vivo and are metabolically stable should be possible.
Supplementary Material
Acknowledgments
We thank Diane Mazzei and the University of Pittsburgh Division of Laboratory Animal Resources staff for the excellent care of the animals and members of the University of Pittsburgh Cancer Institute Writing Group for their helpful suggestions and input regarding this manuscript.
This work was supported by the National Institutes of Health National Cancer Institute [Grants P30-CA47904, CA078039, CA140624]. J.H.B. is the recipient of a Hillman Fellows for Innovative Cancer Research Award. M.J.E. is the recipient of an American Society of Clinical Oncology Cancer Foundation Translational Research Professorship.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.170555.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- bHLH-ZIP
- basic helix-loop-helix leucine zipper protein
- PBS
- phosphate-buffered saline
- HPLC
- high-performance liquid chromatography
- LC-MS/MS
- liquid chromatography/tandem mass spectrometry
- MTT
- methylthiazolyldiphenyl-tetrazolium bromide
- 10074-G5
- 7-nitro-N-(2-phenylphenyl)-2,1,3-benzoxadiazol-4-amine
- 10058-F4
- [Z-E]-5-[4-ethylbenzylidene]-2-thioxo-1,3-thiazolidin-4-one
- AUC
- area under the concentration
- C max
- maximum peak concentration
- DAD
- diode array detector
- DMSO
- dimethyl sulfoxide.
References
- Baudino TA, Cleveland JL. (2001) The Max network gone mad. Mol Cell Biol 21:691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T. (2008) Inhibition of transcription factors with small organic molecules. Curr Opin Chem Biol 12:464–471 [DOI] [PubMed] [Google Scholar]
- Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. (2002) Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci USA 99:3830–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer LM, Dang CV. (2001) Translocations involving c-myc and c-myc function. Oncogene 20:5595–5610 [DOI] [PubMed] [Google Scholar]
- Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP. (1997) Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health 13:407–484 [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. (1982) Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA 79:7824–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. (1999) c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio DZ, Schumitzky A. (eds) (1997) ADAPT II User's Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. University of Southern California, Los Angeles [Google Scholar]
- Felsher DW, Bishop JM. (1999) Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4:199–207 [DOI] [PubMed] [Google Scholar]
- Fleming WH, Hamel A, MacDonald R, Ramsey E, Pettigrew NM, Johnston B, Dodd JG, Matusik RJ. (1986) Expression of the c-myc protooncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer Res 46:1535–1538 [PubMed] [Google Scholar]
- Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. (2009) Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg Med Chem Lett 19:807–810 [DOI] [PubMed] [Google Scholar]
- Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. (2008) Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol 15:1149–1155 [DOI] [PubMed] [Google Scholar]
- Gardner LB, Lee LA, Dang CV. (2002) c-myc protooncogene, in Encyclopedia of Cancer, 2nd Ed (Bertino J. eds) pp 555–561, Elsevier Science, Burlington, MA [Google Scholar]
- Gomez-Curet I, Perkins RS, Bennett R, Feidler KL, Dunn SP, Krueger LJ. (2006) c-Myc inhibition negatively impacts lymphoma growth. J Pediatr Surg 41:207–211 [DOI] [PubMed] [Google Scholar]
- Gregory MA, Hann SR. (2000) c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 20:2423–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Parise RA, Joseph E, Egorin MJ, Lazo JS, Prochownik EV, Eiseman JL. (2009) Efficacy, pharmacokinetics, tissue distribution, and metabolism of the Myc-Max disruptor, 10058-F4 [Z,E]-5-[4-ethylbenzylidine]-2-thioxothiazolidin-4-one, in mice. Cancer Chemother Pharmacol 63:615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. (2009) Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J Am Chem Soc 131:7390–7401 [DOI] [PubMed] [Google Scholar]
- Huang MJ, Cheng YC, Liu CR, Lin S, Liu HE. (2006) A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol 34:1480–1489 [DOI] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. (2002) Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297:102–104 [DOI] [PubMed] [Google Scholar]
- Kiessling A, Sperl B, Hollis A, Eick D, Berg T. (2006) Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem Biol 13:745–751 [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. (2003) A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA 100:8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Liu JD, Chow JM, Liu CR, Liu HE. (2007) Small-molecule c-Myc inhibitor, 10058-F4, inhibits proliferation, down-regulates human telomerase reverse transcriptase and enhances chemosensitivity in human hepatocellular carcinoma cells. Anticancer Drugs 18:161–170 [DOI] [PubMed] [Google Scholar]
- McNeil CM, Sergio CM, Anderson LR, Inman CK, Eggleton SA, Murphy NC, Millar EK, Crea P, Kench JG, Alles MC, et al. (2006) c-Myc overexpression and endocrine resistance in breast cancer. J Steroid Biochem Mol Biol 102:147–155 [DOI] [PubMed] [Google Scholar]
- Mo H, Henriksson M. (2006) Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc Natl Acad Sci USA 103:6344–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustata G, Follis AV, Hammoudeh DI, Metallo SJ, Wang H, Prochownik EV, Lazo JS, Bahar I. (2009) Discovery of novel myc-max heterodimer disruptors with a three-dimensional pharmacophore model. J Med Chem 52:1247–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. (1999) MYC oncogenes and human neoplastic disease. Oncogene 18:3004–3016 [DOI] [PubMed] [Google Scholar]
- Pagnano KB, Vassallo J, Lorand-Metze I, Costa FF, Saad ST. (2001) p53, Mdm2, and c-Myc overexpression is associated with a poor prognosis in aggressive non-Hodgkin's lymphomas. Am J Hematol 67:84–92 [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. (2002) Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109:321–334 [DOI] [PubMed] [Google Scholar]
- Rocci ML, Jr, Jusko WJ. (1983) LAGRAN program for area and moments in pharmacokinetic analysis. Comput Programs Biomed 16:203–216 [DOI] [PubMed] [Google Scholar]
- Russo P, Arzani D, Trombino S, Falugi C. (2003) c-myc down-regulation induces apoptosis in human cancer cell lines exposed to RPR-115135 (C31H29NO4), a non-peptidomimetic farnesyltransferase inhibitor. J Pharmacol Exp Ther 304: 37–47 [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. (2007) MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67:9762–9770 [DOI] [PubMed] [Google Scholar]
- Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. (2009) Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg Med Chem Lett 19:6038–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. (2008) Modelling Myc inhibition as a cancer therapy. Nature 455:679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT. (1997) c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res 25:1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV. (2007) Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther 6:2399–2408 [DOI] [PubMed] [Google Scholar]
- Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. (2008) c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene 27:1905–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. (2006) A credit-card library approach for disrupting protein-protein interactions. Bioorg Med Chem 14:2660–2673 [DOI] [PubMed] [Google Scholar]
- Yeh KC, Kwan KC. (1978) A comparison of numerical integrating algorithms by trapezoidal, Lagrange, and spline approximation. J Pharmacokinet Biopharm 6:79–98 [DOI] [PubMed] [Google Scholar]
- Yin X, Giap C, Lazo JS, Prochownik EV. (2003) Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22:6151–6159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.