Abstract
Because of adverse health consequences related to smoking, it is important to understand factors that contribute to nicotine dependence. The most replicated genetic finding for nicotine dependence points to variants on chromosome 15, which includes the α5-α3-β4 nicotinic receptor gene cluster. A compelling functional variant is a polymorphism, rs16969968, which alters an amino acid in the α5 nicotinic receptor subunit. Several prominent studies report that the replicated nicotine dependence locus also influences the risk for lung cancer and chronic obstructive pulmonary disease. This represents an exciting convergence of genetic findings and highlights the potential for research on smoking to inform public health.
Introduction
Cigarette smoking is common in both industrialized and developing countries. In the U.S., over 43 million people use tobacco, and worldwide, there are over 1 billion tobacco users [1-2]. Though smoking is on the decline in developed countries because of effective anti-smoking campaigns, in developing countries, tobacco use continues to increase. Despite reductions in current smoking among the U.S. population over the past four decades, each year over 400,000 people die from tobacco-related illnesses [1], and smoking remains the greatest contributor to preventable mortality [3]. Worldwide, the annual death toll from tobacco use is 5 million people, and with increasing tobacco use in developing countries, it is predicted that the worldwide death toll will rise to 8 million per year by 2030 [2].
Lung cancer is the disease most identified with smoking, and its prevalence over time mirrors per capita tobacco consumption (Figure 1). Though there has been a reduction in smoking, and we are beginning to see a decline in the prevalence of lung cancer, more people die from lung cancer each year than from any other cancer [4]. Chronic obstructive pulmonary disease (COPD) is another serious lung disease attributable to smoking, and it is the 4th leading cause of death in the U.S. Smoking causes the irritation, inflammation, and destruction of lung tissue that leads to COPD and difficulty breathing. Because of the large number of people who smoke and the significant health consequences of tobacco use, it is important to understand factors that contribute to dependence on nicotine.
Figure 1.
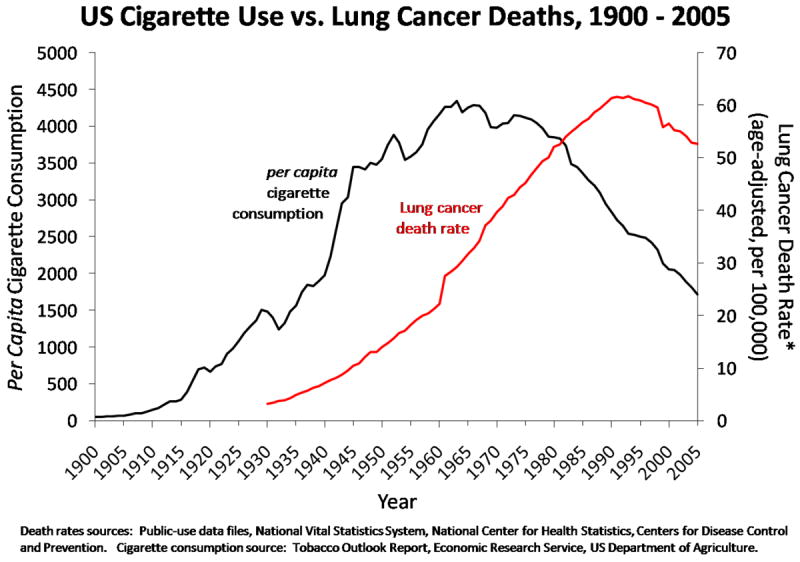
Per Capita cigarette consumption and lung cancer death rate in the U.S. from 1900–2005. Lung cancer prevalence over the 20th century reflects changes in cigarette consumption.
Genetics of nicotine dependence
The development of nicotine dependence requires smoking and results in a sequence of behavioral events that starts with the initiation of cigarette use, the conversion from experimental smoking to the establishment of regular smoking behavior, and then the development of nicotine dependence among smokers [5]. Nicotine dependence, which is characterized by heavier smoking, early morning smoking, tolerance and withdrawal, predicts difficulty quitting.
Many compounds are present in tobacco, but nicotine is the addictive component that confers the risk of developing dependence. Nicotine binds to nicotinic cholinergic receptors, which are widely distributed in the central and peripheral nervous system. The nicotinic cholinergic receptor is composed of 5 subunit proteins arranged around a central pore, and the receptor is involved in physiologic responses related to smoking. There are 16 different nicotinic receptor subunit genes, α 1–7,9,10, β1–4, δ, ε, γ, and different combinations of these subunit proteins result in receptors that vary in biologic function, distribution throughout the body, and other pharmacologic properties. Binding of nicotine to nicotinic cholinergic receptors forms the basis of the molecular pharmacology that leads to dependence.
Not all smokers are nicotine dependent. Among current smokers, approximately 60% are nicotine dependent based on the Fagerström Test for Nicotine Dependence, a well established scale for assessing nicotine dependence [6-7]. Dependence on nicotine has multiple underlying etiologies that include genetic predispositions and environmental risk factors. Evidence for genetic factors contributing to the risk of smoking behaviors and nicotine dependence is shown by the clustering of heavy smoking and nicotine dependence in families and the similarity of smoking behaviors in identical twins [8-9]. In contrast to nicotine dependent smokers, light, non-dependent smokers or “chippers” smoke a few cigarettes per day and can easily quit smoking [10]. These light smokers represent about 15% of the smoking population and can provide a genetic contrast group compared to heavy, nicotine dependent smokers.
It is likely that a large number of genes contribute to nicotine dependence, as well as many environmental factors. This complexity represents a considerable challenge, but it also underscores the potential benefits of genetic studies of nicotine dependence. By understanding genetic factors that contribute to nicotine dependence, we may start to tease apart the different contributions of genes and environments that lead to heavy smoking, which in turn may improve interventions to reduce nicotine dependence and improve smoking cessation treatment.
At this time there are three main pharmacologic approaches to smoking cessation: nicotine replacement therapy (NRT), varenicline, and buproprion. NRT substitutes the nicotine from cigarettes with a sustained nicotine replacement which can then be tapered over several weeks. Varenicline is a partial nicotinic receptor agonist that minimizes nicotine craving and withdrawal. Buproprion acts through a different mechanism via inhibition of catecholamine (noradrenaline and dopamine) reuptake to improve quit rates. It is likely that there are different genetic profiles that identify who will best respond to one pharmacologic treatment or another. A potential promise of the study of the genetics of nicotine dependence is that we may gain insights into these profiles.
Rapid advances in genetic technology now provide a set of tools to identify genetic contributions to common diseases. A map of human genetic variation across major populations and lower cost testing of genetic variants allow large-scale genome-wide association studies (GWAS) that test one million or more genetic variants (single nucleotide polymorphisms or SNPs) in each person. These studies, which include thousands of subjects, test hundreds of thousands of variants across an individual’s complete set of DNA to identify genetic variations associated with a disease. In the last three years this has led to the discovery of new genetic contributions to diabetes, prostate cancer, and many other diseases, including nicotine dependence [11].
The aims of this paper are to review the most robust genetic findings for nicotine dependence, lung cancer, and COPD and to place these in the context of smoking cessation.
Genetic association with chromosome 15 and nicotine dependence
To date, the most compelling evidence for genetic risk variants contributing to nicotine dependence comes from a series of studies that have examined smoking behaviors and nicotine dependence across numerous populations. These genetic studies implicate variants in the chromosome 15q24-25 region in the development of heavy smoking and nicotine dependence, and this region includes the α5-α3-β4 nicotinic receptor gene cluster. This finding was first reported in a study that compared nicotine dependent cases to light smokers who had no symptoms of dependence, which focused the genetic study on the transition from regular smoking to dependence [12]. This genetic association, a correlation between genetic variants and nicotine dependence, is marked by multiple SNPs, including rs16969968 in the α5 cholinergic nicotinic receptor subunit gene CHRNA5. Individuals who have one copy of the risk variant have a 1.3 fold increase in developing nicotine dependence once exposed to smoking, and those with two copies of the risk variant have almost a two-fold increase in risk [13].
Replication of genetic association increases the validity of a finding. Independent replication using nicotine dependence and correlated phenotypes (light versus heavy smokers or cigarettes per day) has been reported repeatedly for rs16969968, either through direct testing or by examining other highly correlated genetic variants [14-19]. Association of this locus for early-onset smokers has also been reported [20]. Community based samples, lung cancer patients, and alcohol dependent subjects all show this same genetic association with smoking behavior and nicotine dependence. Further analysis of the chromosome 15 region identifies at least two statistically distinct variants that contribute to nicotine dependence [12-14, 16, 20].
Convergence of smoking, lung cancer, and chronic obstructive pulmonary disease genetics
In parallel with the studies of smoking, large scale genetic studies of lung cancer and COPD have been undertaken, and the same variants on chromosome 15 are associated with these smoking related diseases [21-24]. This represents an exciting convergence of genetic findings for nicotine dependence, smoking quantity, lung cancer, and COPD risk. The identification of this common genetic region that is associated with smoking behavior and smoking related illnesses raises the question of whether this locus has a direct effect on lung cancer and COPD vulnerability, or whether this increased genetic risk of lung cancer and COPD can be explained solely through the genetic influence on heavy smoking. A direct effect of this locus on lung cancer and COPD may represent pleiotropy where a single genetic region influences multiple diseases.
In these genetic association studies of lung cancer and COPD, the amount of cigarettes smoked per day was accounted for in analyses. After including number of cigarettes smoked and other measures of smoking exposure such as pack years smoked, the genetic association with lung cancer and COPD remained. This supports the potential of a direct genetic influence of this region on multiple distinct disorders, nicotine dependence, lung cancer, and COPD, or pleiotropy.
There is also evidence that the genetic effect for this region and lung cancer and smoking may be only through the genetic effects of smoking. The association of the chromosome 15 region and lung cancer is not seen in non-smokers [18]. Secondly, though cigarettes smoked per day is a reasonable proxy for exposure, it does not fully capture the carcinogenic and toxic exposure risk associated with smoking. For example, among smokers who report smoking the same number of cigarettes per day, the quantity of toxin and carcinogen exposure varies, and individuals who have this risk variant rs16969968 appear to ingest more toxins, which is likely due to more intensive smoking such as inhaling more deeply and frequently while smoking [25]. Because the vast majority lung cancer and COPD cases are attributable to smoking, and carcinogenic exposure varies between smokers even when smoking the same number of cigarettes per day, it is unlikely that further statistical tests will be able to dissect whether this genetic association with smoking-related diseases is acting only through smoking genetic risk or whether there remains an added direct genetic disease risk. To untangle the genetic relationship of smoking to lung cancer and COPD, animal studies will need to be undertaken which can directly test this lung cancer and COPD risk independent from smoking behaviors.
Animal studies can directly examine the effects of genetic variants given a constant carcinogenic exposure. For example, the variant rs16969968, which causes the amino acid change in the α5 nicotinic receptor, can be inserted into a mouse model (a “knock in” mouse) so that the only difference between the two groups of mice is this one nucleotide change. Animals with and without this human variant can be exposed to a carcinogenic agent, and the frequency of subsequent lung cancer can be measured. If there is a difference in incidence of lung cancer development between mice with the variant and those without, the polymorphism likely has a direct role in the etiology of lung cancer. If no difference is seen, it is more likely that this variant alters smoking behavior in subtle ways that are not captured in smoking measures such as cigarettes per day, which subsequently changes the risk for lung cancer. In other words, the genetic risk is indirect and mediated through smoking. Mouse models are now being developed.
Importance of genetic studies of smoking in other populations
Although cigarette smoking and other tobacco use is common in other world populations, the majority of the large-scale genetic studies to date have been performed in populations of European descent. The chromosome 15 region that is associated with smoking behaviors in populations of European descent shows great genetic diversity in populations of Asian and African origin. This varying allele frequency results in different patterns of correlated SNPs, or linkage disequilibrium, in populations of different origin. While the frequency of disease causing genetic markers may vary across populations, their biological impact on the risk for common diseases is typically consistent across different populations [26]. This expected biological consistency, along with different patterns of genetic correlation between SNPs, can be leveraged to refine association signals by narrowing down the most likely biologically causative variants.
The strongest genetic association finding for smoking, the rs16969968 variant in the α5 nicotinic receptor subunit that causes an amino acid change, is a rare variant in populations of African and Asian descent [15]. Two recent genetic studies of African Americans, one examining nicotine dependence and the other lung cancer, implicated the chromosome 15 region as associated with disease even though the risk allele was rare, which adds further support to the hypothesis that this region contributes to both smoking and lung cancer [27-28]. This chromosome 15 region was also identified in a Chinese population though a different group of correlated variants was reported [29]. Additional genetic studies of individuals of non-European descent will be important to narrow down the search for biologically causal genetic variants that explains the statistical evidence for disease associations. We await these future studies.
From association to biologic function
When a genetic association is found, it represents not only an association with the tested genetic variants, but also an association with untested, highly correlated SNPs that can span across many genes on the same chromosome. The replication of genetic association justifies the continued search for the specific associated genetic variants that change biological function. First, the association marked by rs16969968 is correlated with genetic variants that extend across 6 genes on chromosome 15 in populations of European descent. See Figure 2. These genes include the α5-α3-β4 nicotinic receptor gene cluster (CHRNA5-CHRNA3-CHRNB4), and correlated SNPs are also located in the following genes: iron-responsive element binding protein 2 (IREB2), an iron regulatory protein; LOC123688, a putative protein of unknown function; and α4 proteasome subunit protein (PSMA4), a protein that cleaves other proteins. The most plausible genes that may influence smoking behavior in this region are the family of nicotinic receptor genes. To understand the biological relationship of the genetic association with nicotine dependence, lung cancer and COPD, we must determine which of these correlated SNPs alter biological function.
Figure 2.
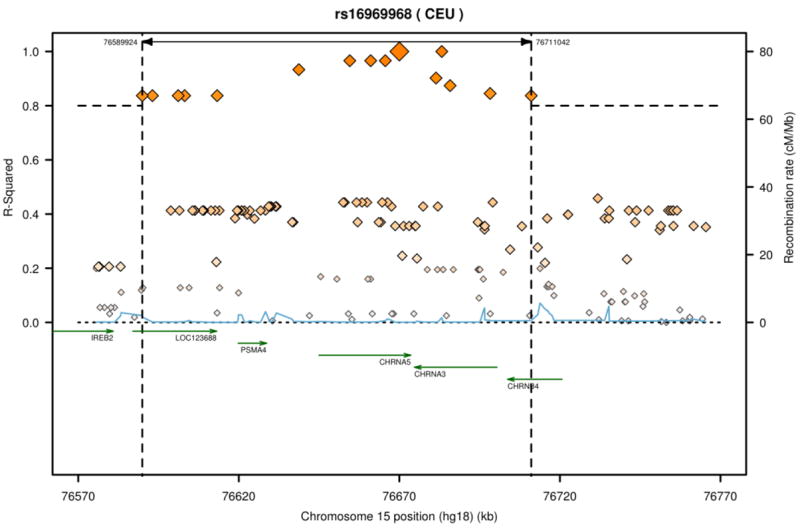
SNPs correlated with rs16969968 on chromosome 15. The SNP rs16969968 is marked by the largest diamond. This SNP is highly correlated (r2 > 0.8) with SNPs that span approximately 120 kilobases across multiple genes including the α5-α3-β4 nicotinic receptor gene cluster. Figure generated by SNAP [42].
A variant that appears most likely to be a biological contributor to nicotine dependence is rs16969968 because this polymorphism changes an amino acid from aspartate to asparagine at position 398 (D398N) in the α5 nicotinic receptor protein. This amino acid is highly conserved across species, and an in vitro model that inserts this single amino acid change into an α5 nicotinic receptor subunit alters nicotinic receptor function [15]. This work supports the hypothesis that the genetic variant rs16969968 causes biological changes that alter the risk of developing nicotine dependence and heavy smoking. Though this amino acid change is the most compelling candidate for the underlying biologic risk responsible for this genetic association, we cannot rule out the role of other correlated genetic variants. A second biologic mechanism that is associated with heavy smoking and nicotine dependence includes different levels of expression for CHRNA5 messenger RNA (mRNA) in the brain [30]. Similar changes in mRNA expression are seen in lung tissue [31-32].
Joint statistical analyses of the two loci, or haplotype analyses, that mark both these biologic mechanisms demonstrate that the amino acid change and varying levels of RNA expression independently contribute to nicotine dependence. The variant CHRNA5 (D398N), which greatly increases the risk for nicotine dependence, lung cancer, and COPD, primarily occurs on the background of low mRNA expression of CHRNA5. The lower mRNA expression of CHRNA5 along with the non-risk allele of rs16969968 is protective for nicotine dependence and lung cancer. These functional receptor data, gene expression results, and genetic analyses point to the nicotinic receptor genes, and specifically the α5 nicotinic receptor subunit gene, as the most likely gene altering the risk for nicotine dependence, lung cancer, and COPD.
Smoking cessation
Smoking encompasses many different behaviors, from the initiation of smoking to the development of nicotine dependence, and finally cessation. Many variables influence smoking cessation. As people age, more quit smoking, and high education and income are strong predictors of successful quitting. The genetic risk factor on chromosome 15 that is associated with nicotine dependence, smoking quantity, lung cancer, and COPD appears to be a relatively specific risk factor for a smoker to become a heavy, nicotine dependent smoker. This chromosome 15 variant is not related to smoking initiation and experimentation, and it does not distinguish between never smokers and smokers [21].
Whether this chromosome 15 region plays a role in smoking cessation is less clear. Two genetic studies of smoking cessation found no evidence that variants in this region influence successful quitting [33-34]. However, two other studies have demonstrated the role of this locus in smoking cessation [21, 35]. In addition, a recent study of pregnant smokers showed that this chromosome 15 variant predicted who continued smoking during pregnancy [36]. Though the genetic association with nicotine dependence and smoking quantity is very robust, this region appears to play a smaller role in cessation.
Summary
The prevalence of smoking in the U.S. has decreased primarily through smokers’ successful efforts to quit, yet cigarette smoking continues to impose a substantial health burden. Most smokers want to quit, and over 40% give up smoking for at least one day; however, few smokers successfully stop smoking each year [37]. Because there are many causal pathways that lead to nicotine dependence, it is unlikely that any single preventative intervention or pharmacological treatment to improve smoking cessation will be optimal for all individuals. Hopefully an improved understanding of the development of nicotine dependence will allow for tailored treatments to help smokers quit.
In concert with the large-scale genome wide association studies, an extensive and growing range of genomic annotation and biological information is now available, which presents both an opportunity and a challenge in genetic studies. Incorporation of known biology to prioritize and test hypotheses about which genetic variants alter biological processes is the next important step to understand the physiologic mechanisms of nicotine dependence. Though these studies have demonstrated the robust genetic findings with heavy smoking, nicotine dependence, lung cancer and COPD, this chromosome 15 region explains only a small proportion of variance of the disease and the majority of the genetic contributions await discovery. These resources hold the possibility of explaining more of the risk for nicotine dependence and potentially inform better smoking cessation.
These overlapping genetic findings for nicotine dependence, lung cancer, and COPD demonstrate that genetic research on smoking is very important to medical research. Though smoking contributes to many illnesses, such as heart disease, hypertension, and stroke, the clearest genetic convergence is with smoking, lung cancer, and COPD. There have been questions about whether smoking should be a priority for genomic research from the perspective of public health [38-41], it is now clear that the strongest genetic determinants of lung cancer risk and COPD are the underlying genetic risk variants for nicotine addiction. Further insights into the genetic basis of nicotine dependence and smoking thus have strong potential to inform ongoing studies of lung disease and COPD and improve our ability to help smokers quit.
Acknowledgments
Funding was provided by grant P01 CA089392 from the National Cancer Institute (NCI) and K02 DA021237 from the National Institute on Drug Abuse (NIDA).
Glossary
- Allele
one of a number of genetic variants at a specific location (or at a SNP) on a chromosome.
- Genome-wide association study
a study of genetic variation across the complete sets of DNA, or genomes, of many people in order to identify genetic associations with observable traits (e.g., height), health conditions (e.g., hypertension) or diseases (e.g., lung cancer).
- Haplotype
a combination of alleles that are physically close and are inherited together on the same chromosome.
- Linkage disequilibrium
the non-random distribution of alleles so that certain combinations of alleles are more likely to occur together on a chromosome than other combinations of alleles. The correlation between alleles is one measure of linkage disequilibrium.
- Pleiotropy
occurs when a single gene influences multiple diseases or traits.
- Single nucleotide polymorphism (SNP)
a change in a single nucleotide or base in the DNA in a specific location on the chromosome. Millions of SNPs have been catalogued in the human genome.
Footnotes
Conflicts of interest L.J. Bierut is an inventor on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs, including rs16969968, in determining the diagnosis, prognosis, and treatment of addiction. Dr. Bierut served as a consultant for Pfizer Inc. in 2008.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Centers for Disease Control. Cigarette smoking among adults--United States, 2007. Morbidity & Mortality Weekly Report. 2008;57:1221–1226. [PubMed] [Google Scholar]
- 2.World Health Organization. Tobacco key facts. 2009 http://www.who.int/topics/tobacco/facts/en/index.html.
- 3.Mokdad AH, et al. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 4.American Cancer Society. Cancer facts & figures 2009. American Cancer Society; 2009. [Google Scholar]
- 5.Bierut LJ. Nicotine dependence and genetic variation in the nicotinic receptors. Drug Alcohol Depend. 2009;104(Suppl. 1):S64–69. doi: 10.1016/j.drugalcdep.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Substance Abuse and Mental Health Services Administration. Overview of findings from the 2003 National Survey on Drug Use and Health. NSDUH Series H-24 (Studies, O.o.A., ed) 2004 [PubMed] [Google Scholar]
- 7.Heatherton TF, et al. The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 8.Bierut LJ, et al. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 9.Li MD. The genetics of nicotine dependence. Curr Psychiatry Rep. 2006;8:158–164. doi: 10.1007/s11920-006-0016-0. [DOI] [PubMed] [Google Scholar]
- 10.Shiffman S. Light and intermittent smokers: background and perspective. Nicotine Tob Res. 2009;11:122–125. doi: 10.1093/ntr/ntn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saccone NL, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berrettini W, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bierut LJ, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens VL, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grucza RA, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–929. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitz MR, et al. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss RB, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 23.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Marchand L, et al. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis JP, et al. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 27.Saccone NL, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz AG, et al. Racial differences in the association between SNPs on 15q25.1, smoking behavior, and risk of non-small cell lung cancer. J Thorac Oncol. 2009 doi: 10.1097/JTO.0b013e3181b244ef. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, et al. Association of human aryl hydrocarbon receptor gene polymorphisms with risk of lung cancer among cigarette smokers in a Chinese population. Pharmacogenet Genomics. 2009;19:25–34. doi: 10.1097/FPC.0b013e328316d8d8. [DOI] [PubMed] [Google Scholar]
- 30.Wang JC, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falvella FS, et al. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res. 2009;15:1837–1842. doi: 10.1158/1078-0432.CCR-08-2107. [DOI] [PubMed] [Google Scholar]
- 32.Falvella FS, Galvan A, Frullanti E, Dragani TA. Reply to the letter to the editor from Wang. Clin Cancer Res. 2009;15:5599. [Google Scholar]
- 33.Breitling LP, et al. Smoking cessation and variations in nicotinic acetylcholine receptor subunits alpha-5, alpha-3, and beta-4 genes. Biol Psychiatry. 2009;65:691–695. doi: 10.1016/j.biopsych.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Conti DV, et al. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker TB, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine Tob Res. 2009;11:785–796. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freathy RM, et al. A common genetic variant in the 15q24 nicotinic acetylcholine receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) is associated with a reduced ability of women to quit smoking in pregnancy. Hum Mol Genet. 2009;18:2922–2927. doi: 10.1093/hmg/ddp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Centers for Disease Control. Tobacco use among adults--United States, 2005. Morbidity & Mortality Weekly Report. 2006;55:1145–1148. [PubMed] [Google Scholar]
- 38.Carlsten C, Burke W. Potential for genetics to promote public health: genetics research on smoking suggests caution about expectations. JAMA. 2006;296:2480–2482. doi: 10.1001/jama.296.20.2480. [DOI] [PubMed] [Google Scholar]
- 39.Merikangas KR, Risch N. Genomic priorities and public health. Science. 2003;302:599–601. doi: 10.1126/science.1091468. [DOI] [PubMed] [Google Scholar]
- 40.Bierut LJ, et al. Genetic research and smoking behavior. JAMA. 2007;297:809. doi: 10.1001/jama.297.8.809. author reply 810. [DOI] [PubMed] [Google Scholar]
- 41.Berrettini W, et al. Setting priorities for genomic research. Science. 2004:1445–1447. doi: 10.1126/science.304.5676.1445c. author reply 1445-1447. [DOI] [PubMed] [Google Scholar]
- 42.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]


