Abstract
Spn1 plays essential roles in the regulation of gene expression by RNA Polymerase II (RNAPII), and it is highly conserved in organisms ranging from yeast to humans. Spn1 physically and/or genetically interacts with RNAPII, TBP, TFIIS and a number of chromatin remodeling factors (Swi/Snf and Spt6). The central domain of Spn1 (residues 141-305 out of 410) is necessary and sufficient for performing the essential functions of SPN1 in yeast cells. Here we report the high-resolution (1.85Å) crystal structure of the conserved central domain of Saccharomyces cerevisiae Spn1. The central domain is comprised of eight alpha-helices in a right handed super helical arrangement, and exhibits structural similarity to domain I of TFIIS. A unique structural feature of Spn1 is a highly conserved loop, which defines one side of a pronounced cavity. The loop and the other residues forming the cavity are highly conserved at the amino acid level among all Spn1 family members, suggesting that this is a signature motif for Spn1 orthologs. The locations and the molecular characterization of temperature-sensitive mutations in Spn1 indicate that the cavity is a key attribute of Spn1 that is critical for its regulatory functions during RNAPII-mediated transcriptional activity.
Introduction
Transcription by RNA polymerase II (RNAPII) in vivo is a complex process involving a large number of protein factors. TATA-binding protein (TBP), often in a complex with TBP-associated factors (TAFs), nucleates the pre-initiation complex (PIC) by binding the core promoter and serving as a platform for other general transcription factors, including TFIIA, TFIIB, TFIIE, TFIIF and TFIIH, in addition to RNAPII.1 The transition from the PIC to an actively elongating RNAPII-containing complex also involves a number of additional factors, such as TFIIS, CTD kinases, Spt4/5 (DSIF in humans) etc., each of which can facilitate different aspects of the process (reviewed in2). Since the template for transcription in vivo is packaged into chromatin3, additional factors that modify or remodel chromatin (like Swi/Snf, Spt6, etc.) are also critical to achieve appropriate transcriptional regulation of particular genes (reviewed in2; 4; 5).
For a large number of well-characterized promoters the rate-limiting step in the transcription process is the formation of the pre-initiation complex. Thus, recruitment of TBP and RNAPII to the promoter correlates strongly with transcriptional output.6; 7; 8 However, there are a growing number of genes that are regulated at a step after the recruitment of the general transcription machinery. Such genes include the yeast CYC1 gene, the Drosophila heat shock genes, and mammalian HIV-1 and the proto-oncogene c-Myc.8; 9; 10; 11; 12 TBP and RNAPII are pre-loaded at the promoters of these genes even when transcriptional output is very low, indicating distinct rate-limiting steps from recruitment-regulated genes. Genome-wide studies suggest that a large number of developmental and stress-inducible genes have RNAPII pre-loaded at promoter-proximal regions.13; 14; 15; 16 Thus, an increasing number of promoters appear to be regulated primarily after recruitment of the PIC (for reviews see17; 18; 19). Although our mechanistic understanding of these inactive RNAPII complexes is incomplete, these observations suggest that functions critical for a high level of transcription are either inhibited or absent under non-inducing conditions.
The transcription factor Spn1 was identified in a genetic screen, which was targeted to discover factors important in transcriptional processes after the assembly of the pre-initiation complex.20 SPN1 is an essential gene, and the centrally located 140 amino acid region of the protein, which are highly conserved from yeast to humans, is necessary and sufficient for yeast viability. Spn1 plays an important role in CYC1 expression since a mutation in SPN1 (spn1-K192N; which is lysine at position 192 substituted with asparagine) results in up-regulated transcription from the pre-loaded CYC1 gene. We, and others, have found that Spn1 interacts with RNAPII.21; 22; 23; 24; 25 Spn1 co-localizes with RNAPII along the entire open reading frame of a number of constitutively active genes.22; 26 Spn1 also physically interacts with Spt6, which is a conserved interaction in yeast, plants and humans.21; 22; 23; 24; 27; 28 Spt6 functions during transcription through chromatin in vivo 29; 30; 31; 32; 33 and is important for transcribing long open reading frames.34 Spt6 also plays a role in elongation in vitro on naked DNA.35 Importantly, the spn1-K192N derivative fails to interact with both RNAPII and Spt6.24 Moreover, genetic interactions are observed between spn1 mutants and SPT4, SPT5, and TFIIS21; 24, factors that interact directly with RNAPII and serve to enhance the transition from a paused polymerase to an active elongating complex.36; 37 Taken together, these observations suggest that Spn1 plays a critical role in the transition to a productive elongation complex, which is crucial for full gene activation.
To advance our understanding of how Spn1 functions in the regulation of the pre-loaded CYC1 promoter, we molecularly characterized temperature-sensitive alleles that had been previously described at the genetic level.21 We determined the amino acid substitutions for these spn1 alleles and found that the mutations are contained within the conserved central domain of Spn1. We also found that similar to spn1-K192N, these mutant alleles affect the regulation of CYC1 transcription and disrupt the Spn1-RNAPII interaction. To map the mutations on the tertiary structure of Spn1 and to provide further insight into how this essential gene functions within the cell, we determined the high resolution (1.85 Å) crystal structure of the Spn1 central domain. A unique 467 Å3 cavity was identified on the surface of Spn1. The loop and the residues lining the cavity are nearly invariant among the Spn1 family members at the amino acid level. The position of the three temperature-sensitive mutations in the tertiary structure reveals that the cavity is an important functional aspect of the Spn1 structure. We also found that yeast Spn1 contains a region that is remarkably similar to the structure of mouse TFIIS domain I, another important player in the post-recruitment regulation of RNAPII.
Results
Characterization of two temperature sensitive spn1 mutants
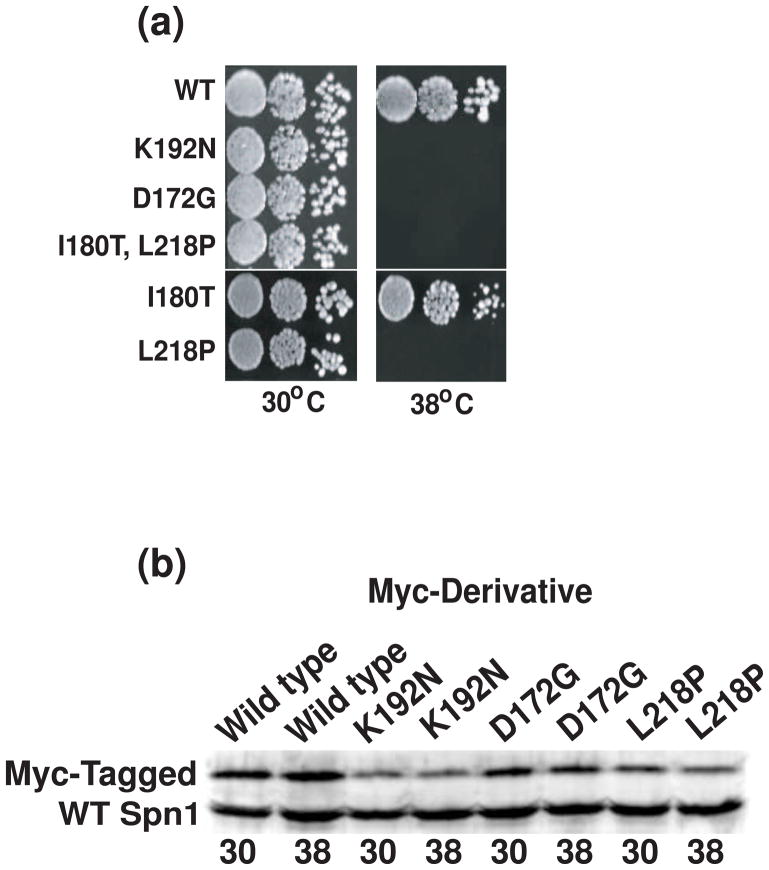
Hartzog and coworkers used random mutagenesis of SPN1 to produce two temperature-sensitive strains of yeast with mutant phenotypes.21 After cloning and sequencing of the spn1 alleles in these strains (graciously provided by Hartzog and colleagues), we found that one of them (iws1-7) has aspartic acid at position 172 changed to glycine (spn1-D172G), and the other (iws1-13) has a double mutation: isoleucine 180 to threonine and leucine 218 to proline (spn1-I180T;L218P). To demonstrate that these mutations in the SPN1 gene are solely responsible for the temperature-sensitive phenotype, plasmid borne copies of the mutant alleles were introduced into cells containing a fully deleted SPN1 gene using plasmid shuffling. Cells harboring wild-type SPN1, the previously identified temperature-sensitive spn1-K192N derivative20; 24, and the two new mutant alleles were assessed for the ability to grow at permissive (30°C) and restrictive (38°C) temperatures. As expected, cells containing spn1-D172G or spn1-I180T;L218P are viable at 30°C and inviable at 38°C (Figure 1A). To determine if one or both substitutions are required for the temperature-sensitive phenotype of spn1-I180T;L218P, the single mutants were generated by site-directed mutagenesis. We found that the proline substitution for leucine at position 218 alone is sufficient to confer the temperature-sensitive phenotype (Figure 1A). Thus, subsequent studies focused on the single substitution mutant spn1-L218P.
Figure 1.
Temperature-sensitivity of spn1 mutants is not due to instability of the protein at the restrictive condition. (a) Yeast strains as indicated were serially diluted, plated onto YPD plates and incubated at 30°C or 38°C for two or four days, respectively. (b) Yeast cells containing both untagged wild-type Spn1 and myc-tagged wild-type, myc-tagged Spn1-K192N, myc-tagged Spn1-D172G or myc-tagged Spn1-L218P were grown to mid-log phase and then incubated an additional hour at 30°C or 38°C prior to harvesting. Protein extracts were prepared from the cells and Spn1 levels in the extracts evaluated by immunoblot analysis with polyclonal anti-Spn1 antibodies.
To determine if the temperature-sensitive phenotype of the mutant strains is simply the result of Spn1 instability at the elevated temperature, Spn1 levels were monitored by immunoblot analysis. Protein extracts were prepared from cells containing untagged Spn1 and either myc-tagged wild-type Spn1, myc-tagged Spn1-K192N, myc-tagged Spn1-D172G or myc-tagged Spn1-L218P. Untagged wild-type Spn1 was present in each cell so that the requirement for cellular viability did not artifactually stabilize the mutant Spn1 molecules. Although the expression level of each of the mutated Spn1 proteins is slightly reduced when compared to the wild-type protein (Figure 1B), all of the Spn1 proteins are stably expressed at 38°C with no significant difference in their levels as compared to 30°C. Therefore, the temperature-sensitive phenotype of the mutant strains is not likely to be due to instability of the mutated Spn1 proteins at 38°C.
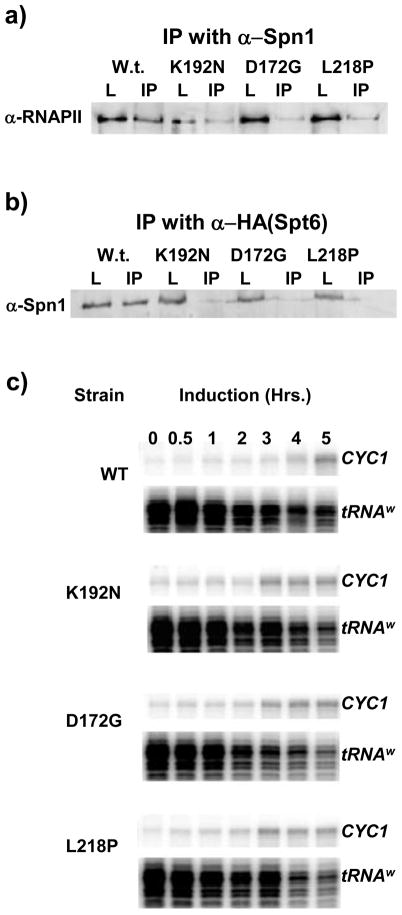
To further characterize the molecular phenotypes of Spn1-D172G and Spn1-L218P we evaluated their ability to associate with RNAPII and Spt6 via co-immunoprecipitation assays. As noted above, we had previously observed that Spn1-K192N is defective for interaction with RNAPII and Spt6.24 Likewise, we observed that the association of the two new mutant derivatives (Spn1-D172G or Spn1-L218P) with either RNAPII or Spt6 is also significantly reduced when compared to wild-type Spn1 (Figure 2A and B).
Figure 2.
Mutations in SPN1 reduce the Spn1-RNAPII and Spn1-Spt6 interactions and confer changes in the regulation of CYC1 transcription. (a) Protein extracts prepared from the indicated strains were immunoprecipitated with protein A-Sepharose beads coupled to polyclonal anti-Spn1 antibodies. RNAPII was detected by immunoblot analysis with antibodies to the largest subunit of RNAPII. The load (L) represents 5% of the input material for the IP of which 50% was loaded. (b) Protein extracts prepared from the indicated SPN1 strains also containing an HA-tagged version of Spt6 were immunoprecipitated with protein A-Sepharose beads coupled to anti-HA antibodies. Spn1 derivatives were detected by immunoblot analysis with polyclonal anti-Spn1 antibodies. The load and IP are as in panel a. (c) The effect of spn1-K192N, spn1-D172G and spn1-L218P on CYC1 transcription was evaluated in an S1 Nuclease protection assay. The indicated strains were grown to mid-log phase in medium containing dextrose (CYC1 partial repression) and then transferred to medium with ethanol (CYC1 activation) for the times indicated (in hours). Total cellular RNA was isolated, hybridized to radioactive CYC1 and tRNAW probes, digested with S1 Nuclease and the products separated on denaturing polyacrylamide gels. Relevant regions of a representative gel are shown. The tRNAW probe was included as a load control and was used to normalize CYC1 expression.
Transcription of the CYC1 gene is regulated by the availability of a fermentable carbon source in the growth medium.38; 39 When yeast cells are grown in the presence of dextrose, CYC1 transcription is partially repressed resulting in very low transcript levels. In the presence of a non-fermentable carbon source (i.g., ethanol) CYC1 transcription is induced approximately ten-fold. We have shown that the spn1-K192N strain has an increase in CYC1 transcription levels under both partially repressed and activated growth conditions.20; 24 We next tested whether Spn1-D172G and Spn1-L218P alter the regulation of CYC1 transcription. Total cellular RNA was isolated from wild-type, spn1-K192N, spn1-D172G and spn1-L218P cells cultured in glucose (partially repressive) or ethanol (inducing). CYC1 transcript levels were measured by a S1 nuclease protection assay. In wild-type cells, CYC1 transcription is barely detectable in glucose, and markedly increases after 5 hours of growth in ethanol. CYC1 transcription prior to induction is higher in the mutant cells when compared to the wild-type strain (Figure 2C and Supplemental Figure 1). Moreover, CYC1 transcription peaks faster (3 hours) compared to wild-type cells (5 hours). Thus, CYC1 transcriptional regulation was altered in a similar manner in all three mutant strains.
To further explore the similarities between the three mutants, we compared the growth phenotypes under an extensive set of conditions, including alternative carbon sources and a variety of stresses (oxidative, osmotic etc.). In all cases, the three mutants behaved indistinguishably (Supplemental Table 1). We also tested for genetic interactions with the Swi/Snf complex, since Spn1-K192N has been shown to suppress the defects associated with mutants in this complex. 24 Similarly, we found that spn1-D172G and spn1-L218P also suppress the defects associated with a deletion in SNF5 (Supplemental Table 2).
Thus, the three mutants share a large number of phenotypic, genetic and molecular properties. Although all three substitutions are located within the conserved central domain of Spn1 (residues 141-305), intriguingly, they are not clustered in the primary sequence of Spn1. To determine if the location of the mutated residues define a structural motif in the tertiary structure of Spn1, we undertook a structural characterization of Spn1.
Overall structural features of Spn1
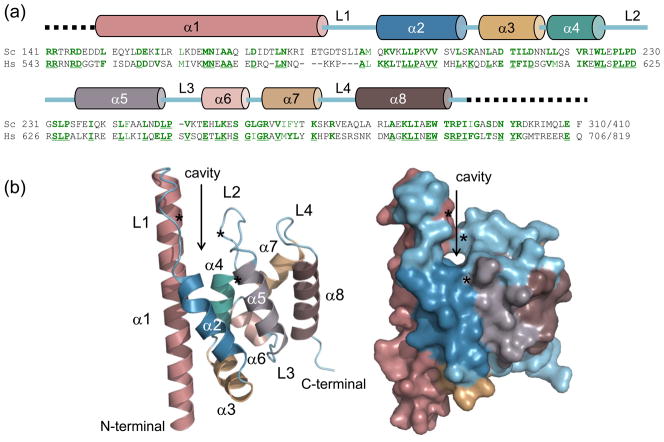
Yeast Spn1 has a highly charged N-terminal domain (pI = 4.6), a central conserved domain, and a highly charged C-terminal domain (pI = 10.4). We previously demonstrated that the nonconserved N-terminal and C-terminal regions of yeast Spn1 are not required to cover a genomic knock-out of the gene.20 Only the conserved central core of Spn1 (referred to here as mini-Spn1; residues 141-305 of yeast Spn1; Figure 3A) is necessary for cellular viability. Cells containing mini-Spn1 grow normally at 30°C and do not exhibit a temperature sensitive phenotype when grown at 38°C (Supplemental Figure 2). Since mini-Spn1 reflects the key properties of the molecule (it contains the highly conserved region, is sufficient for cellular viability, and contains the three residues that were altered in the temperature-sensitive mutations described above), we determined its crystal structure.
Figure 3.
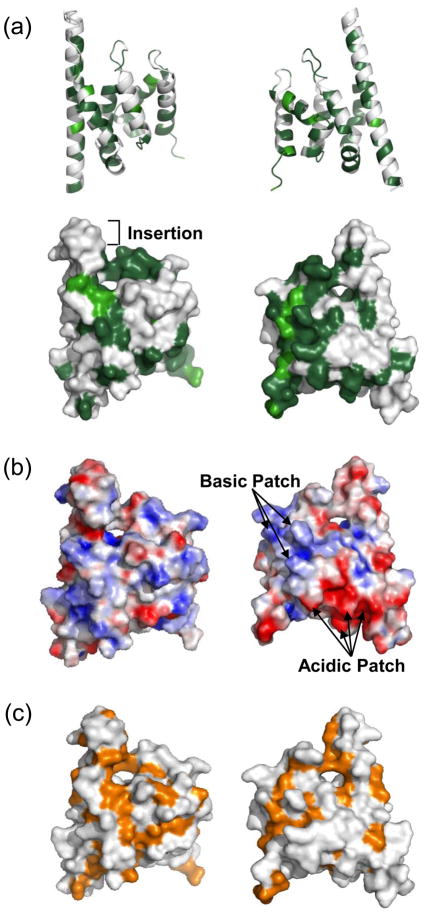
The structure of the conserved central domain of yeast Spn1. (a) primary sequence alignment of Spn1 from Saccharomyces cerevisiae (Sc) and Homo sapiens (Hs) is shown. Identical amino acids are underlined and shown in dark green with similar amino acids in light green. Numbers indicating the starting and ending amino acid positions are shown at the left and right, respectively for each homolog. Secondary structure elements are indicated above the primary amino acid sequence. (b) Overall structure of the highly conserved central domain of yeast Spn1 shown in ribbon format (left) and space-filling mode (right), with the cavity indicated (arrow). Mini-Spn1 consists of eight α-helices (α1 to α8) and four loops (L1 to L4), and contains residues from 149 to 293. From top to bottom, the asterisks denote the locations of L176, L228 and I238 respectively (see text for details).
Spn1 crystallized in the space group P212121 with cell dimensions a=63.6, b=67.8, c=76.0 (Table 1). X-ray diffraction data of Se-Met mini-Spn1 were collected at 3 wavelengths, representing the inflection point, peak and high energy remote. The structure was refined to a resolution of 1.85 Å with a final R/Rfree of 0.203 and 0.265, respectively. Mini-Spn1 is composed of 8 α–helices with 4 loops and 3 turns (Figure 3A and B). The first helix (α1; residues 149-181) is much longer than α2 through α8. A defining structural feature of mini-Spn1 is a surface-exposed cavity, defined by residues contributed from α1, α2, α4 and α5, and completed by the L2 loop (Figure 3B). L176 (α1), L228 (L2) and I238 (α5) form a conserved hydrophobic core lining the cavity between α1 and the L2 loop. This cavity branches into a smaller groove, which is located in the center of helices α1, α2, α4 and α5 (Figure 3B). The volume of the cavity was determined using the 3V Channel Extractor algorithm and found to be 467 Å3.40
Table 1.
Data collection and refinement statistics
| SeMet | |||
|---|---|---|---|
| Peak | Inflection | Remote | |
| Data collection | |||
| Wavelength (Å) | 0.9791 | 0.9793 | 0.9716 |
| Unit cell parameters | |||
| a, b, c (Å) | 63.6, 67.8, 76.0 | 63.6, 67.8, 76.0 | 63.6, 67.8, 76.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Space group | P212121 | P212121 | P212121 |
| Resolution (Å) | 39.6–1.85 | 39.6–1.85 | 33.9–1.85 |
| Total number of reflections | 163,002 | 163340 | 162636 |
| Unique reflections | 53,982 | 54032 | 53955 |
| Redundancya | 3.02 (3.01) | 3.02 (3.01) | 3.01 (3.00) |
| Completenessa (%) | 99.6 (100.0) | 99.6 (100.0) | 99.5 (100.0) |
| R mergea | 0.051 (0.380) | 0.051 (0.396) | 0.061 (0.438) |
| I/σa | 12.1 (2.8) | 11.9 (2.6) | 10.1 (2.4) |
| Refinement | |||
| Resolution (Å) | 33.1–1.85 | ||
| R work (%) | 20.3 | ||
| R free (%) | 26.5 | ||
| Number of reflections in refinement | 28618 | ||
| Model statistics | |||
| Number of atoms (water) | 2304 (414) | ||
| B-average (Å) | |||
| Chain A | 27.37 | ||
| Chain B | 36.46 | ||
| Waters | 50.48 | ||
| RMSD bond length (Å) | 0.018 | ||
| RMSD bond angles (°) | 1.71 | ||
| Ramachandran statisticsb | |||
| Favored (%) | 98.5 | ||
| Allowed (%) | 1.5 | ||
| Generous (%) | 0 | ||
| Disallowed (%) | 0 | ||
Data in parentheses are for the highest resolution shell (1.92–1.85)
Statistics do not include the 8 glycine and 11 proline residues in the protein
The asymmetric unit contains two molecules, with chains designated A or B (rms deviation 0.546 Å for Cα positions). A total of 146 and 142 residues in chain A and B, respectively (of 165 in the construct) could be built into the electron density map. The regions that are disordered include the extreme N-terminus (8 residues, R141-D148 in chain A and B), and the extreme C-terminus (11 residues in chain A; 15 residues in Chain B). There are only minimal contacts between the two chains (Supplemental Figure 3A) with most of their interface covered with water molecules (not shown). At lower concentrations (0.7 mg/ml or less), purified Spn1 eluted from a size exclusion column as a monomer (data not shown). To test the formal possibility that Spn1 functions as a dimer within cells, we performed a co-immunoprecipitation assay using cells that express a myc-tagged and an untagged derivative of Spn1. Immunoprecipitation with anti-myc antibodies precipitated only myc-Spn1 (Supplemental Figure 3B). If Spn1 dimerized efficiently within the cell, untagged Spn1 should have co-immunoprecipitated with the myc-Spn1, and been readily detectable in the immunoprecipitate. Thus, we conclude that Spn1 does not form dimers in vivo.
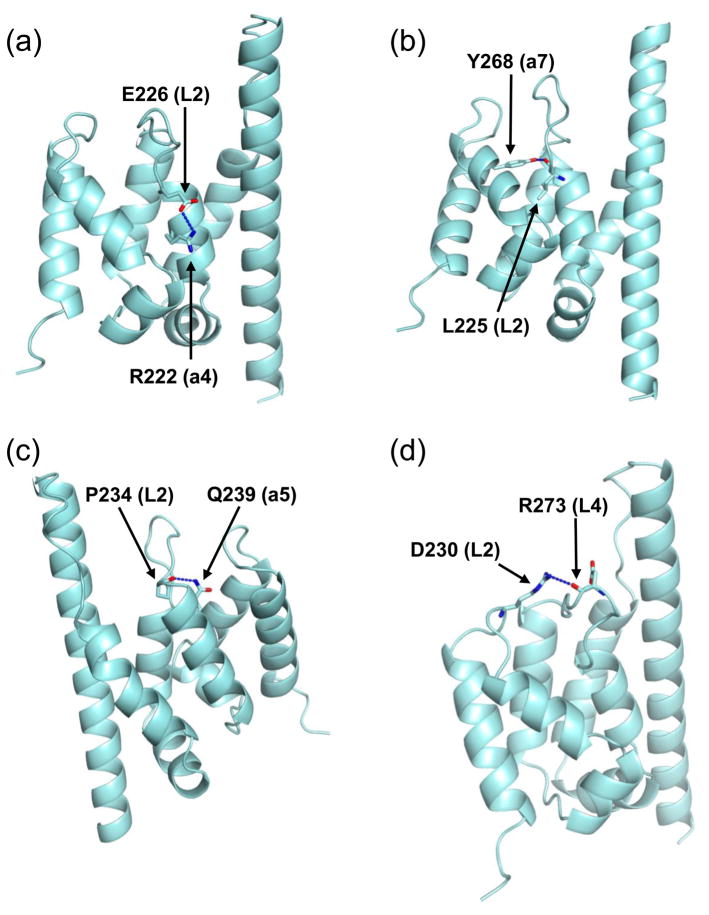
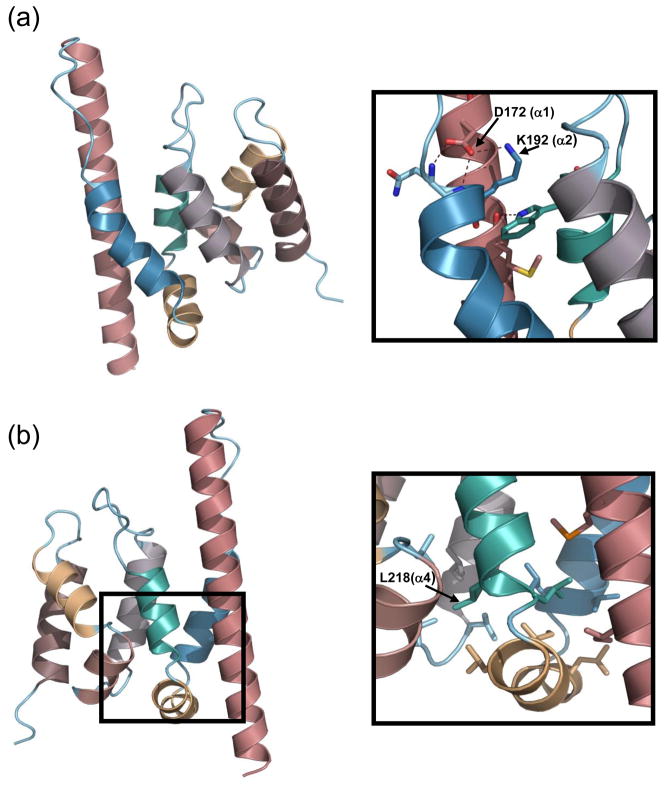
The average B-factors for chain A, chain B and waters are 27.37, 36.46 and 50.48 Å, respectively (Table 1). Two intermolecular hydrogen bonds were formed between mini-Spn1 molecules in the crystal dimer resulting in significantly lower B-factors for L2 and α7 in chain A (Supplemental Figure 4). The difference in B-factors for the L2 loop is especially noteworthy. The hydrogen bond formed between the L2 loop of chain A and α1 helix of chain B holds the loop in a conformation that allows it to form four intramolecular hydrogen bonds (Figure 4). E226 (in L2) forms a hydrogen bond with the side chain of R222 (from α4). The L225 (in L2) carbonyl group forms a hydrogen bond with the Y268 (from α7) hydroxyl group. The P234 (in L2) carbonyl group forms a hydrogen bond with the Q239 (from α5) side chain. The D230 (in L2) carbonyl oxygen forms a hydrogen bond with the R273 (in L4) side chain. In chain B, the top of the L2 loop is not constrained by intermolecular hydrogen bonding and the hydrogen bonds formed between E226 and R222, and D230 and R273 were not observed. This contributes to the higher B-factor observed for L2 in chain B (40.97 Å) compared to L2 in chain A (21.03 Å). The L2 loop is clearly more flexible in chain B and it is possible that this flexibility could lead to transiently more open or closed cavity conformations in solution.
Figure 4.
Four hydrogen bonds stabilize the L2 loop in chain A. (a) E226 forms a hydrogen bond with the side chain of R222. (b) The L225 carbonyl group forms a hydrogen bond with the Y268 hydroxyl group. (c) The P234 carbonyl group forms a hydrogen bond with the Q239 side chain. (d) The D230 carbonyl oxygen forms a hydrogen bond with the R273 side chain. In panels a, b, and d the mini-Spn1 structure has been rotated 180 degrees from the orientation in Figure 3B. To optimize visualization of the hydrogen bonds, panels b and d have been rotated slightly into the plane of the paper.
Spn1 sequence conservation in a structural context
The sequence alignment of Spn1 family members shows the primary sequence conservation is distributed across the molecule (Figure 3A). However, when the sequence conservation is mapped on the 3-dimensional structure of Spn1, highly conserved as well as highly variable residues cluster in particular regions of the molecule (Figure 5A and B). The sequence conservation highlights the importance of the rim surrounding the cavity, part of which is formed by residues in the L2 loop, which is the most highly conserved sequence among the Spn1 family members (Figure 3A). In addition, there are two wide bands of highly conserved residues found on the molecule, one of which extends downward from the cavity on the front surface of the molecule (Figure 5B).
Figure 5.
The surface of mini-Spn1 has a number of conserved and distinctive structural aspects. (a) The sequence conservation as determined from the alignment of Spn1 from Saccharomyces cerevisiae and Homo sapiens is plotted on the front (left) and back (right) sides of Spn1. The top panel shows the structure in a ribbon diagram. The lower panel shows the structure in a space-filling model. Dark green indicates identical residues and light green similar residues. (b) The electrostatic potential of mini-Spn1 is shown on its molecular surface. The molecule is shown in the same orientation as in (a). The basic (blue) and acidic (red) residues are indicated. (c) Hydrophobic residues are shown (orange) on the surface of mini-Spn1. The cavity region of Spn1 is rimmed with hydrophobic amino acids as can be seen on both the front- and back sides. The front surface of mini-Spn1 also has a pronounced region comprised of hydrophobic residues that extends downward from the cavity.
The primary sequence alignment of Spn1 from different species indicates that E181-S186 is an insertion, which is found only in species of Saccharomyces, and is absent in S. pombe and higher eukaryotes (Figure 3A). This insertion falls at the end of α1 and the turn to α2, and results in the projection of this region beyond the top surface of Saccharomyces Spn1. Thus, the end of the α1 helix in other organisms would be predicted to be ‘flush’ with the peripheral edge of the L2 loop.
Localized clusters of charged and hydrophobic residues are apparent on the surface of mini-Spn1. The charged residues are evenly distributed on the front side of the molecular surface of mini-Spn1 while the backside has both a basic and acidic patch. (Figure 5C). Residues K257, R263, K270 (conserved residues) and K272 (not conserved) create the positively charged patch on the back surface (Figure 5C). D155, D210, D214 and E254 (all conserved), and E151 and E258 (not conserved) form the negatively charged patch on the concave surface of the back. There are also two regions on the surface of mini-Spn1 that are highly conserved and comprised principally of hydrophobic amino acids (Figures 5B and 5D). One of these regions includes the cavity, which is rimmed by hydrophobic residues. The other region extends in a band from the cavity along the front surface of mini-Spn1.
The residues altered in spn1-D172G and spn1-K192N are located on the rim of the cavity. These mutations both lead to a loss of interaction between Spn1 and RNAPII, and affect the regulation of CYC1 transcription. D172 and K192, along with the conserved residues M165 and W224 form multiple hydrogen bonds that stabilize helices α1, α2, and α4 in the cavity region (Figure 6A). The side chain of K192 in helix α2, which projects into the cavity, forms a hydrogen bond with D172 from helix α1. The side chain of D172 forms two hydrogen bonds with the Q191 (from loop L1) and K192 back bone nitrogens. The M165 (from helix α1) carbonyl group forms a hydrogen bond with the side chain nitrogen of W224 (from helix α4). The spn1-D172G and Spn1-K192N mutations are predicted to eliminate the hydrogen bond formed between D172 and K192 side chains and could dramatically alter the projection of the K192 side chain into the cavity. This could further affect the surface conformation of the Spn1 cavity.
Figure 6.
The location of D172, K192 and L218 on the mini-Spn1 structure is shown. (a) D172 is located on helix α1 while K 192 is on helix α2. (b) L218 is located at the beginning of helix α4 within the hydrophobic core region. The mini-Spn1 structure shown in Panel B has been rotated 180 degrees relative to Panel a.
The L218 residue, which is altered to proline in spn1-L218P, is invariably a hydrophobic residue in different species, and is located at the beginning of helix α4 (Figure 6B). Helix-helix packing interactions in the hydrophobic core region could significantly stabilize the mini-Spn1 structure. Changing L218 to a proline residue at this position could potentially alter the helical packing in this region or affect the geometry between α3 and α4. However, it is unlikely that the substitution dramatically alters the overall structure of Spn1 as the mutant allele covers the genomic knockout of SPN1 at the permissive temperature. The Spn1-L218P protein was also stably expressed, even at elevated temperatures (Figure 1).
Structural comparison of mini-Spn1 with other proteins
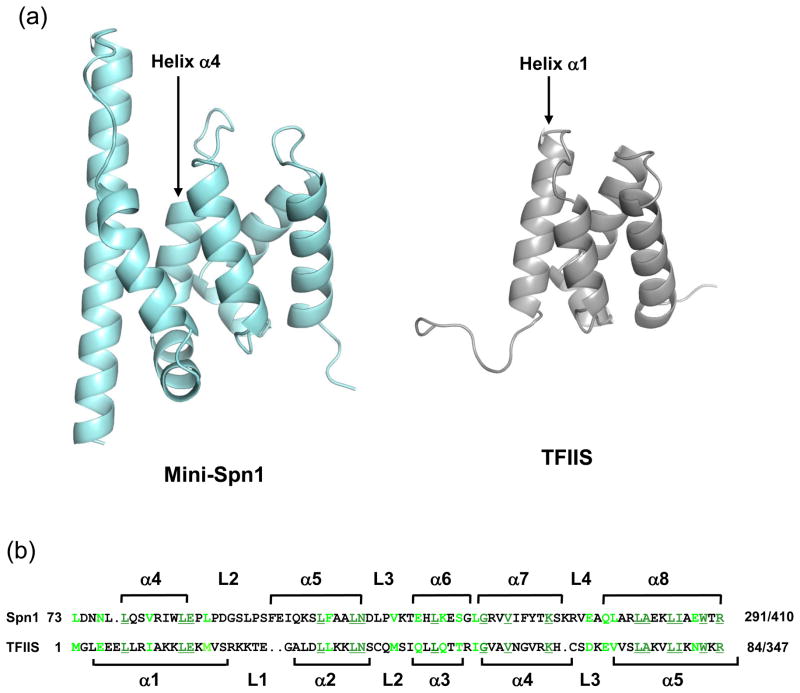
A search for structurally similar proteins was performed using DALI.41 The first five proteins with structural similarity to mini-Spn1 (Table 2) contain heat or armadillo motifs.42; 43 These motifs are comprised of 2 (heat) or 3 (armadillo) α-helices and are repeated 7–13 times within a protein. The mini-Spn1 structure does not contain multiple copies of either motif and the sequence similarity between mini-Spn1 and these proteins is extremely low. Therefore, this structural similarity is most likely due to similar α-helical packing, rather than a conserved structural motif. The DALI alignment also identified structural similarity between mini-Spn1 and domain 1 of mouse Transcription Factor IIS (TFIIS) protein 3, a tissue-specific TFIIS isoform. Structural similarity between the two proteins begins with helix α4 of mini-Spn1 and extends through the remainder of the mini-Spn1 structure (Figure 7). An overlay of the two proteins shows remarkable structural similarity with an rmsd of 2.0 (Table 2). However, the highly conserved sequence corresponding to the Spn1 L2 loop that is present in all Spn1 family members is lacking in TFIIS (Figure 7). In addition, the first three helices (α1-α3) and the L1 loop of mini-Spn1 are not present in the mouse TFIIS structure.
Table 2.
Structural homologues to mini-Spn1 as assessed by DALI
| PDB code | Name | Z-score | RMSD (Å) | Residues total | Residues aligned | Sequence identity (%) |
|---|---|---|---|---|---|---|
| 1UKL | Importin β | 9.1 | 3.7 | 876 | 126 | 13 |
| 1IAL | Importin α | 8.7 | 1.8 | 438 | 68 | 10 |
| 2QIZ | Ubiquitin Conjugation Factor E4 | 8.6 | 3.6 | 943 | 138 | 9 |
| 2BNX | Diaphanous Protein Homolog 1 | 8.5 | 2.9 | 339 | 108 | 14 |
| 1WA5 | GTP-Binding Nuclear Protein RAN | 8.3 | 2.0 | 463 | 70 | 13 |
| 1WJT | Transcription Elongation Factor S-II Protein 3 | 7.9 | 2.0 | 103 | 71 | 23 |
| 2Z6G | β–Catenin | 7.7 | 2.0 | 550 | 70 | 10 |
| 2FV2 | RCD1 Required for Cell Differentiation Homolog | 7.7 | 3.2 | 268 | 125 | 10 |
| 1Z2C | Rho-Related GTP-Binding Protein RHOC | 7.6 | 3.7 | 346 | 114 | 12 |
| 1Z9E | PC4 and SFRS1 Interacting Protein 2 | 7.3 | 2.8 | 83 | 74 | 11 |
Figure 7.
Spn1 and mouse TFIIS protein 3 have primary sequence and structural similarities. (a) The yeast mini-Spn1 and mouse TFIIS (PDB ID: 1WJT) protein structures are shown in ribbon format. The structural similarity begins with helix α4 of Spn1 and helix α1 of TFIIS. (b) The primary sequence alignment of Spn1 (residues 213-291) and mouse TFIIS (residues 1-77) is shown above with conserved amino acids underlined and shown in dark green. Similar residues are shown in light green. Amino acid insertions are indicated with a period in opposite sequence. The secondary structure of mini-Spn1 is shown above the sequence alignment. The secondary structure of domain I of mouse TFIIS protein 3 is shown below the sequence alignment.
It was previously shown that the N-terminal domain of TFIIS and an 80 amino acid region internal to Spn1 (residues 215-295) share primary sequence similarity.44 The region of sequence similarity directly overlaps the structural similarity identified between yeast mini-Spn1 and mouse TFIIS (Figure 7). We mapped the residues conserved between yeast Spn1 and mouse TFIIS on the structure of each protein (Supplemental Figure 5). Approximately one-third of the conserved residues were buried within the structures of the two proteins. The remaining residues were predominantly localized within two patches on the surfaces of the two molecules. The first patch, located on the front of the Spn1 structure is comprised of amino acids that are poorly conserved with human Spn1 (Supplemental Figure 5A). The second patch is located on the side and back of the Spn1 molecule (Supplemental Figure 5B) and is comprised of residues that are highly conserved with human Spn1. These residues form a shallow groove along the surface of both Spn1 and mouse TFIIS.
Discussion
SPN1 is an essential gene in yeast and contains a central domain that has been conserved throughout evolution.20 Genetic interaction of Spn1 with TBP, Swi/Snf, and TFIIS,20; 24 and its co-localization with RNAPII during transcription and association with mRNA capping enzyme,21 Spt5 and Spt621; 22; 23 suggest potential roles for Spn1 in transcription initiation, elongation and mRNA processing. Utilizing the temperature-sensitive spn1-K192N strain we previously analyzed the role of Spn1 in the regulation of CYC1 transcription.20; 24 In contrast to wild-type Spn1, Spn1-K192N does not bind RNAPII or Spt6 and is not present in the PIC formed on the CYC1 promoter. Transcription from the CYC1 gene was also higher in the spn1-K192N strain in both uninduced and induced growth conditions. Collectively these results demonstrate that repression of CYC1 transcription requires the binding of Spn1 to RNAPII.
Two other temperature-sensitive spn1 strains had been previously reported21 although neither mutation was fully characterized. We obtained these strains and have shown that spn1-D172G (iws1-7) and spn1-L218P (iws1-13) confer the temperature-sensitive phenotypes. Similar to spn1-K192N, the spn1-D172G and spn1-L218P mutations both result in reduced binding of Spn1 to RNAPII and Spt6. The regulation of CYC1 transcription in the spn1-D172G and spn1-L218P strains is also clearly altered. Uninduced CYC1 transcription was higher and induction of CYC1 transcription was significantly faster in the mutant cells.
The spn1-K192N, spn1-D172G and spn1-L218P mutants share many properties, and the mutations are all contained within the conserved central domain of yeast Spn1. This region of Spn1 was shown to be sufficient for cellular viability and has been conserved throughout evolution. The 1.85 Å structure of mini-Spn1 reveals eight α–helices in a right-handed superhelical arrangement. A striking and unique feature of the mini-Spn1 structure is a surface-exposed cavity on the “top” of the protein. This cavity is lined with conserved amino acids (Figure 5A and 5B) and is closed off by the highly conserved L2 loop (residues 225–235, Figure 3). In the crystal lattice, the mobility of the L2 loop of one of the two molecules in the asymmetric unit is restricted due to an intermolecular hydrogen bond, whereas the L2 loop in the second molecule has a greater range of movement. This inherent mobility could result in transient opening/closing of the cavity. There is a large patch of conserved hydrophobic residues extending down from the cavity of the “front” surface of mini-Spn1 (Figure 5B and 5D). This hydrophobic patch along with the cavity could serve as a protein-protein interaction site, the access to which may be regulated by loop conformation.
The binding of Spn1 to RNAPII could repress CYC1 transcription by affecting the binding and/or function of several proteins that are involved in productive transcription. Using an in vivo mapping technique, the C-terminus of Spn1 was shown to be within 8 nm of the C-termini of Rpb2 and Rpb5, two subunits of RNAPII.25 The C-termini of Rpb2 and Rpb5 are in close proximity to each other on the RNAPII surface and are located just above Rpb4-Rpb7 subunit heterodimer. Rpb4-Rpb7 border the RNA exit channel and are thought to bind the newly synthesized RNA as it exits the polymerase.45 Rpb4-Rpb7 have also been shown to interact directly with the head module of Mediator.46 Spn1 binding to RNAPII in the region of Rpb4-Rpb7 could repress transcription by blocking the RNA exit channel or by preventing the interaction between Rpb4-Rpb7 and Mediator. We have recently shown that Mediator (especially the head module) is required for activated CYC1 transcription. 47 Unlike Spn1, which is present on the promoter in the pre-loaded state 24, Mediator is recruited only upon activation.47
Structural alignment revealed similarity between mini-Spn1 and the N-terminal domain of mouse TFIIS protein 3, a tissue-specific TFIIS isoform found in humans and mouse. The TFIIS protein is composed of three structurally and functionally distinct domains (domains I, II and III).48; 49; 50; 51 Domains II and III are required for TFIIS binding to RNAPII and for the release of transcription arrest.49 Domain I is required for the interaction of TFIIS with holoenzyme52 and interacts with the MED13 and Spt8 subunits of Mediator and the SAGA complex, respectively.53 The structural similarity between Spn1 and TFIIS domain I is intriguing. As noted above, the N-terminal domain of TFIIS and an 80 amino acid region internal to Spn1 (residues 215-295) share primary sequence similarity.44 Yeast Spn1 and TFIIS interact genetically with each other,24 and physically or genetically with over 30 other proteins (http://thebiogrid.org/), most of which are involved in transcription or mRNA processing and export. Burckin and colleagues have also concluded that Spn1 and TFIIS share functional roles in the cell based on the way both proteins affect pre-mRNA and mRNA levels.54
The structure of the highly conserved central domain of yeast Spn1 provides a strong foundation for future studies on Spn1 function. Although the mechanism of action by Spn1 is not yet understood, it is intriguing to speculate about the similarities between Spn1 and TFIIS. Interference with TFIIS activity, a factor that rescues RNAPII from pausing, backtracking, and arrest by cleavage of nascent transcripts49; 55; 56 could be a likely target. Indeed, recent studies indicate that reactivation of backtracked RNAPII is essential for transcription in vivo.57 Further analysis of the residues near the cavity of Spn1 and in the region conserved with TFIIS may serve to identify and further inform on Spn1 functions at pre-loaded promoters in yeast. The extensive primary amino acid sequence identity between yeast and human Spn1, as well as the existence of homologues for the other transcription players involved, suggests the potential for conservation of Spn1 function in higher eukaryotes. As in yeast, mammalian Spn1 (also known as Iws1) is essential for cell viability.58 Moreover, expression of post-recruitment regulated HIV-1 requires functional Spn1 and Spt6, as knockdown and mutational analysis demonstrate defects in transcript production and processing.27 In addition, human Spn1 is involved in the expression of the proto-oncogene c-Myc27, another gene that is regulated after recruitment of the pre-initiation complex.11; 59 It is also important to note that yeast Spn1 may play a role in activation of transcription at certain promoters 20, which is consistent with the observation that Spn1 is required for hormone-inducible gene expression in plants.28. Perhaps Spn1 plays a negative versus positive role depending on the particular promoter context, or Spn1 may switch between a negative and positive factor depending on interactions with co-factors or modification status. The latter hypothesis is much like the functional role of DSIF, a factor that can both repress and activate transcription36; 60. DSIF also participates in pausing in higher eukaryotes.10; 61 Thus, the exciting and emerging picture is one in which Spn1 plays complex and critical roles in transcription mechanisms across the evolutionary spectrum.
Materials & Methods
Cloning, expression and purification of mini-Spn1
Mini-Spn1 (amino acids 141-305) was subcloned into a pET15b vector, which contains an N-terminal Histidine-tag and expressed using Rosetta pLysS cells at 16°C. Seleno-methionine labeled protein was produced by growing cells in M9 medium under inhibition of methionine biosynthesis.62 In brief: 1 l of M9 medium with 100μg/ml ampicillin and 34μg/ml of chloramphenicol was inoculated with cells from a 20 ml overnight culture. The cells were grown at 37°C to an OD600 of 0.5–0.6, transferred to 16°C and allowed to grow for 30 minutes. During the incubation at 16°C methionine biosynthesis was blocked by the addition of 100mg each of lysine, phenylalanine and threonine, 50 mg each of isoleucine, leucine and valine, and 60 mg of Se-methionine (Acros) per liter of broth. The cells were induced with 1mM IPTG, incubated overnight and collected by centrifugation at 4000 rpm for 15 minutes. The cell pellet from one liter of medium was suspended in 15ml of lysis buffer (20 mM Tris pH 7.5, 200 mM NaCl and 500 μM PMSF). The cells were lysed by sonication and the cellular debris pelleted by centrifugation at 18,000 rpm for 20 mins. Recombinant mini-Spn1 was purified on a Hi-Trap chelating column, followed by thrombin cleavage to remove the histidine-tag. The histidine-tag and mini-Spn1 were separated by fractionation on a Superdex 200 (16/60) column. Mass spectrometric analysis of Se-met mini-Spn1 showed that two selenomethionines were incorporated into the protein and that the histidine-tag was completely removed.
Crystallization, data collection, and refinement
Recombinant yeast mini-Spn1 was crystallized at 16°C by hanging drop vapor diffusion. Protein (6–10 mg/ml in 20mM Tris pH 7.5, 100mM KCl, 500μM PMSF, 10% glycerol) was combined in a 1:1 ratio with reservoir solution containing 200mM sodium thiocyanate, 20% (w/v) PEG3350. Crystals grew in clusters, usually from a single nucleation site, and were long and thin. Crystals were grown reproducibly from several batches of protein. Crystals were cryoprotected with successive soaks in increasing concentrations of glycerol (10, 20 and 30% v/v) in the mother liquor. The crystals were flash-cooled by plunging the loop in liquid nitrogen.
Data was collected at three wavelength (0.9793 Å Inflection; 0.9791 Å Peak; 0.9716 Å High energy remote) at the Advanced light source (Lawrence Berkeley National Laboratory). Data were reduced by using d*TREK63 (see Table 1 for details).
The location of heavy atom sites and heavy atom refinement was performed with SOLVE.64 The model was built in O 65 after applying density modification. The model was refined using REFMAC5.66 All images were prepared with Pymol.67
Volume determination of the cavity
The volume of the mini-Spn1 cavity was determined using the 3V Channel extractor algorithm.40 A 5 Å probe was rolled over the surface of mini-Spn1 to localize the cavity. A 1.5 Å probe was then placed in the cavity and rolled over the surface of the cavity to measure its’ volume.
Cloning and characterization of spn1 mutants
The plasmid pCR311 was cleaved with PflM1 and BamHI and the resulting DNA fragments separated by agarose gel electrophoresis. The large DNA fragment containing the pRS313 vector, SPN1 promoter, 50 bp of the 5′-end of the SPN1 gene and the SPN1 terminator was excised and purified using a QIAquick gel extraction kit. Genomic DNA was isolated from GHY1199 or GHY1150 cells exactly as described.68 The mutated Spn1 gene was amplified from the genomic DNA by PCR using Pfu Turbo and the manufacturer’s (Stratagene) suggested protocol. Primer pairs used in the PCR amplification were designed to anneal to the 5′ and 3′-ends of the gene and to incorporate a BamHI, site at the 3′-end. The amplified Spn1 gene was cleaved with PflM1 and BamHI and cloned into the purified vector portion of pCR311 (see above) using standard techniques, and sequenced.
Phenotypic analysis of spn1 mutants
To assess the temperature-sensitive phenotype, the wild-type or mutant spn1strains were grown to mid-log phase in YPD (1% yeast extract, 2% bacto-peptone and 2% dextrose) and 10-fold serial dilutions of each spotted onto duplicate YPD plates. The plates were incubated at 30°C or 38°C for two or four days, respectively. To determine protein stability at 38°C, cells were grown to mid-log phase at 30°C, transferred to 38°C for 15 minutes, returned to 30°C for an additional hour and then transferred back to 38°C for one hour. The cells were harvested and whole-cell protein extracts prepared by glass bead lysis in 25 mM Tris-Phosphate (pH 6.7), 2mM phenylmethylsulfonylfluoride. 20 μg of each protein extract were resolved on a 10% polyacrylamide gel and the protein then electroblotted onto nitrocellulose. The Spn1 proteins were detected using a polyclonal antibody to Spn1 and standard techniques. The Spn1 and RNAPII and Spt6 co-immunoprecipitation assays and mutant growth phenotypes were assayed as described previously.24 The S1 nuclease assay was conducted as described.20; 24 For CYC1 induction cells were grown in YPD to mid-log phase, collected by centrifugation, washed three times in YP, suspended in YPE (1% yeast extract, 2% bacto-peptone and 3% ethanol) and cultured at 30°C for the indicated length of time.
Supplementary Material
Supplemental Figure 1. The regulation of CYC1 transcription is altered in cells containing Spn1-K192N, Spn1-D172G or Spn1-L218P. Total RNA was isolated from cells containing wild type Spn1, Spn1-K192N, Spn1-D172G or Spn1-L218P after growth in ethanol for the indicated times. CYC1 transcript levels in the RNA samples were measured via an S1 nuclease protection assay, normalized to a tRNA internal control followed by setting wild-type at time zero to 1. The mean and standard deviation of three independent biological replicates is shown.
Supplemental Figure 2. Cells containing mini-Spn1 grow normally at 30°C and do not exhibit a temperature sensitive phenotype when grown at 38°C. Yeast strains as indicated to the left above were serially diluted, plated onto YPD plates and the plates incubated at 30°C or 38°C for two or three days, respectively.
Supplemental Figure 3. The asymmetric unit contains two mini-Spn1 molecules. (a) The two mini-Spn1 molecules are designated as chain A (brown) or chain B (light blue). For orientation and reference, the L2 loop of both chains is shown in dark blue. The carbonyl group of Pro229 (in the L2 loop) from chain A forms a hydrogen bond with Gln170 (in helix α1) from chain B. Arg263 (in helix α7) in chain A forms a hydrogen bond with Glu156 and Asp163 (in helix α1) in chain B. (b) Spn1 does not dimerize in vivo. A protein extract prepared from cells containing myc-tagged and untagged wild-type Spn1 (L) was immunoprecipitated (IP) with protein A-Sepharose beads coupled to anti-myc antibodies. Spn1 (tagged and untagged) was detected with polyclonal anti-Spn1 antibodies in an immunoblot assay. Untagged Spn1 was not present in the immunoprecipitate indicating it did not dimerize with the myc-tagged Spn1 in vivo. The load (L) represents 10% of the input material for the IP of which 50% was loaded.
Supplemental Figure 4. The L2 loop and α7 helix have lower B-factors in chain A than in chain B. Two intermolecular hydrogen bonds formed between chain A and chain B in the mini-Spn1 crystal. The carbonyl group of P229 on the L2 loop of chain A formed a hydrogen bond with Q170 on helix α1 of chain B. This interaction restricts the movement of the L2 loop in chain A. Similarly, due to the interaction between R263 (from helix α7) in chain A and E156 and D163 (from helix α1) in chain B, the B factors are lower for helix α7 in chain A (Average B factors for α7 in chain A is 20.96 Å and chain B 36.69 Å). Low B-factors are indicated in blue, higher B-factors in orange.
Supplemental Figure 5. The conserved residues between Spn1 and TFIIS are predominantly localized to two regions. The conserved residues were mapped on the surfaces of Spn1 (left panels above) or TFIIS (right panels above). Absolutely conserved residues are shown in dark green and residues that are conservative replacements are shown in light green. The conserved residues are labeled for each molecule. Yeast Spn1 residues that are also conserved with human Spn1 are underlined. The small insets show the sequence conservation between human Spn1 and yeast Spn1 in the ‘front’ orientation for panel (a) and the ‘back’ orientation, before and after rotation to the right for panel (b). The orientation of Spn1 in panel (b) was rotated approximately 60 degrees to the right for optimal viewing of the conserved residues.
Acknowledgments
The yeast strain GHY1199 (iws1-7) and GHY1200 (iws1-13) were generously provided by Grant Hartzog and were previously described.21 We would also like to thank Alexandra Caban for separating the two substitutions found in GHY1200. This work was supported by a grant from the National Institutes of Health to K.L. (GM067777) and by a National Science Foundation grant (MCB-0843073) to L.A.S. Support is also provided by the Howard Hughes Medical Institute to K.L.
Footnotes
Accession numbers: Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3NFQ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 3.Svejstrup JD. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim Biophys Acta. 2004;1677:64–73. doi: 10.1016/j.bbaexp.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochim Biophys Acta. 2009;1789:26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24:747–57. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 7.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 9.Martens C, Krett B, Laybourn P. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Molecular Microbiology. 2001;40:1009–1019. doi: 10.1046/j.1365-2958.2001.02445.x. [DOI] [PubMed] [Google Scholar]
- 10.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes & Development. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–13. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 12.Stevens M, De Clercq E, Balzarini J. The regulation of HIV-1 transcription: molecular targets for chemotherapeutic intervention. Med Res Rev. 2006;26:595–625. doi: 10.1002/med.20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelechano V, Jimeno-Gonzalez S, Rodriguez-Gil A, Garcia-Martinez J, Perez-Ortin JE, Chavez S. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 2009;5:e1000614. doi: 10.1371/journal.pgen.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–2. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price DH. Poised polymerases: on your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–4. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 20.Fischbeck JA, Kraemer SM, Stargell LA. SPN1, a conserved yeast gene identified by suppression of a post-recruitment defective yeast TATA-binding protein mutant. Genetics. 2002;162:1605–1616. doi: 10.1093/genetics/162.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JRr, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003:1368–78. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogan NJM, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–92. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavin AC, Bosche M, Krause R, Grandli P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–146. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Fletcher A, Cheung V, Winston F, Stargell LA. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol. 2008;28:1393–403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 26.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. Embo J. 2004;23:354–64. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–74. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci U S A. 2010;107:3918–23. doi: 10.1073/pnas.0909198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–6. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–9. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 31.Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell. 2006;21:405–16. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–34. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. Embo J. 2009;28:1067–77. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morillo-Huesca M, Vanti M, Chavez S. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. Febs J. 2006;273:756–69. doi: 10.1111/j.1742-4658.2005.05108.x. [DOI] [PubMed] [Google Scholar]
- 35.Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, Sato H, Yamaguchi Y, Mandal SS, Reinberg D, Wada T, Handa H. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol. 2004;24:3324–36. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–56. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–92. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 38.Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 39.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 40.Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–38. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 42.Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–91. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 43.Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 44.Ling Y, Smith AJ, Morgan GT. A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS. Nucleic Acids Res. 2006;34:2219–29. doi: 10.1093/nar/gkl239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CY, Chang CC, Yen CF, Chiu MT, Chang WH. Mapping RNA exit channel on transcribing RNA polymerase II by FRET analysis. Proc Natl Acad Sci U S A. 2009;106:127–32. doi: 10.1073/pnas.0811689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator Head module structure and functional interactions. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SK, Fletcher AGL, Zhang L, Chen X, Fischbeck JA, Stargell LA. Activation of a poised RNAPII-dependent promoter requires both SAGA and Mediator. 2010 doi: 10.1534/genetics.109.113464. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morin PE, Awrey DE, Edwards AM, Arrowsmith CH. Elongation factor TFIIS contains three structural domains: solution structure of domain II. Proc Natl Acad Sci U S A. 1996;93:10604–8. doi: 10.1073/pnas.93.20.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Awrey DE, Shimasaki N, Koth C, Weilbaecher R, Olmsted V, Kazanis S, Shan X, Arellano J, Arrowsmith CH, Kane CM, Edwards AM. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem. 1998;273:22595–605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
- 50.Olmsted VK, Awrey DE, Koth C, Shan X, Morin PE, Kazanis S, Edwards AM, Arrowsmith CH. Yeast transcript elongation factor (TFIIS), structure and function. I: NMR structural analysis of the minimal transcriptionally active region. J Biol Chem. 1998;273:22589–94. doi: 10.1074/jbc.273.35.22589. [DOI] [PubMed] [Google Scholar]
- 51.Booth V, Koth CM, Edwards AM, Arrowsmith CH. Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. J Biol Chem. 2000;275:31266–8. doi: 10.1074/jbc.M002595200. [DOI] [PubMed] [Google Scholar]
- 52.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–71. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 53.Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. Embo J. 2004;23:4232–42. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–82. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 55.Kulish D, Struhl K. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol Cell Biol. 2001;21:4162–8. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–12. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 57.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–10. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z, Zhou Z, Chen G, Bao S. A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem Biophys Res Commun. 2007;353:47–53. doi: 10.1016/j.bbrc.2006.11.133. [DOI] [PubMed] [Google Scholar]
- 59.Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem. 1997;272:14747–54. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 61.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & Development. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–24. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 63.Kuret J, Pflugrath JW. Crystallization and preliminary X-ray analysis of the cAMP-dependent protein kinase catalytic subunit from Saccharomyces cerevisiae. Biochemistry. 1991;30:10595–600. doi: 10.1021/bi00107a031. [DOI] [PubMed] [Google Scholar]
- 64.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 66.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 67.DeLano WL. Th PyMOL Molecular Graphics System 2002 [Google Scholar]
- 68.Davis RW, Thomas M, Cameron J, St John TP, Scherer S, Padgett RA. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65:404–11. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. The regulation of CYC1 transcription is altered in cells containing Spn1-K192N, Spn1-D172G or Spn1-L218P. Total RNA was isolated from cells containing wild type Spn1, Spn1-K192N, Spn1-D172G or Spn1-L218P after growth in ethanol for the indicated times. CYC1 transcript levels in the RNA samples were measured via an S1 nuclease protection assay, normalized to a tRNA internal control followed by setting wild-type at time zero to 1. The mean and standard deviation of three independent biological replicates is shown.
Supplemental Figure 2. Cells containing mini-Spn1 grow normally at 30°C and do not exhibit a temperature sensitive phenotype when grown at 38°C. Yeast strains as indicated to the left above were serially diluted, plated onto YPD plates and the plates incubated at 30°C or 38°C for two or three days, respectively.
Supplemental Figure 3. The asymmetric unit contains two mini-Spn1 molecules. (a) The two mini-Spn1 molecules are designated as chain A (brown) or chain B (light blue). For orientation and reference, the L2 loop of both chains is shown in dark blue. The carbonyl group of Pro229 (in the L2 loop) from chain A forms a hydrogen bond with Gln170 (in helix α1) from chain B. Arg263 (in helix α7) in chain A forms a hydrogen bond with Glu156 and Asp163 (in helix α1) in chain B. (b) Spn1 does not dimerize in vivo. A protein extract prepared from cells containing myc-tagged and untagged wild-type Spn1 (L) was immunoprecipitated (IP) with protein A-Sepharose beads coupled to anti-myc antibodies. Spn1 (tagged and untagged) was detected with polyclonal anti-Spn1 antibodies in an immunoblot assay. Untagged Spn1 was not present in the immunoprecipitate indicating it did not dimerize with the myc-tagged Spn1 in vivo. The load (L) represents 10% of the input material for the IP of which 50% was loaded.
Supplemental Figure 4. The L2 loop and α7 helix have lower B-factors in chain A than in chain B. Two intermolecular hydrogen bonds formed between chain A and chain B in the mini-Spn1 crystal. The carbonyl group of P229 on the L2 loop of chain A formed a hydrogen bond with Q170 on helix α1 of chain B. This interaction restricts the movement of the L2 loop in chain A. Similarly, due to the interaction between R263 (from helix α7) in chain A and E156 and D163 (from helix α1) in chain B, the B factors are lower for helix α7 in chain A (Average B factors for α7 in chain A is 20.96 Å and chain B 36.69 Å). Low B-factors are indicated in blue, higher B-factors in orange.
Supplemental Figure 5. The conserved residues between Spn1 and TFIIS are predominantly localized to two regions. The conserved residues were mapped on the surfaces of Spn1 (left panels above) or TFIIS (right panels above). Absolutely conserved residues are shown in dark green and residues that are conservative replacements are shown in light green. The conserved residues are labeled for each molecule. Yeast Spn1 residues that are also conserved with human Spn1 are underlined. The small insets show the sequence conservation between human Spn1 and yeast Spn1 in the ‘front’ orientation for panel (a) and the ‘back’ orientation, before and after rotation to the right for panel (b). The orientation of Spn1 in panel (b) was rotated approximately 60 degrees to the right for optimal viewing of the conserved residues.