Abstract
The Pit-1 transcription factor (also know as POU1F1) plays a critical role in cell differentiation during organogenesis of the anterior pituitary in mammals and is a transcriptional activator for pituitary gene transcription. Increased expression of Pit-1 has been reported in human tumorigenic breast cells. Here, we found that Pit-1 overexpression or knockdown in human breast cancer cell lines induced profound phenotypic changes in the expression of proteins involved in cell proliferation, apoptosis, and invasion. Some of these protumorigenic effects of Pit-1 were mediated by upregulation of Snai1, an inductor of the epithelial-mesenchymal transition. In immunodeficient mice, Pit-1 overexpression induced tumoral growth and promoted metastasis in lung. In patients with invasive ductal carcinoma of the breast and node-positive tumor, high expression of Pit-1 was significantly correlated with Snai1 positivity. Notably, in these patients elevated expression of Pit-1 was significantly and independently associated with the occurrence of distant metastasis. These findings suggest that Pit-1 could help to make a more accurate prognosis in patients with node-positive breast cancer and may represent a new therapeutic target.
Introduction
Significant clinical consequences from cancer result from the development of metastatic disease. The metastasis of breast cancer is often a multistep event, comprising invasion of mammary carcinoma cells into the adjacent tissues, entry of tumor cells in the systemic circulation, extravasation to distant organs, and finally metastatic colonization, mainly to bones, lungs, and central nervous system (1). To develop metastasis, epithelial mammary cells need to break their intercellular adhesion complexes (i.e., adherent, tight, and gap junctions and desmosomes), as well as their basement membrane that separates the epithelium from other tissues, and acquire motility to invade adjacent tissues. One of the main processes involved in the change from immobile epithelial cells to mobile mesenchymal cells is the epithelial-mesenchymal transition (EMT). EMT is a crucial process that occurs during physiological embryonic development, when epithelial cells acquire a motile morphology appropriate for migration and formation of numerous organs and tissues (2). Mesenchymal cells may again acquire a fully differentiated epithelial phenotype via a mesenchymal-to-epithelial transition (MET). In addition to the physiological role of EMT/MET, it is now known that EMT is a mechanism for carcinoma progression, inducing mammary neoplastic epithelial cells to acquire mesenchymal malignant traits, such as motility, invasiveness, and resistance to apoptosis, thereby contributing to the formation of metastasis (3). Acting as physiological regulators of EMT during the embryonic development, several transcription factors, including Twist, Snai1, Slug, Goosecoid, FOXC2, or SIX-1, have also been shown to be also involved in the metastatic process (4–8). In addition, a recent study by Mani et al. (9) demonstrated that the induction of EMT either by Twist or Snai1 in human mammary epithelial cells results in the acquisition of both mesenchymal and stem cell traits. These authors also found that stem-like cells isolated from human mammary glands had markedly increased expression of EMT markers, including Twist and Snai1, suggesting a strong relation between the EMT process and stem-like cells.
POU class 1 homeobox 1 (Pit-1, also known as POU1F1/GHF-1) belongs to the Pit-Oct-Unc (POU) family of transcription factors that play a key role in inhibition and promotion of cell proliferation and determination of cell lineages as well as in regulation of cell migration, survival, and terminal differentiation (10). Pit-1 is critical for cell differentiation during organogenesis of the anterior pituitary gland in mammals (11) and as a transcriptional activator for pituitary gene transcription (i.e., transcription of prolactin [PRL], growth hormone [GH], and Pit-1 itself) (12–14). Mice with inactivating mutations or deletions of the Pit1 gene fail to generate somatotropes, lactotropes, and thyrotropes and consequently exhibit anterior pituitary hypoplasia and dwarfism (15), demonstrating the importance of Pit-1 in the ontogeny of the pituitary gland. However, Pit-1 is also expressed in nonpituitary cell lines and tissues, such as human placenta, hemapoietic lymphoid tissues, and human breast (16–20). In these extrapituitary tissues it has been suggested that Pit-1 could also be related to cell proliferation and tumorigenesis (21, 22). Specifically in breast, Pit-1 presents higher expression in tumors than in normal breast, increases cell proliferation, and regulates the expression of 2 breast cancer-related hormones, GH and PRL (20, 23, 24).
To analyze the function of Pit-1 in mammary carcinogenesis, the present study uses human mammary cell lines and immunodeficient mice to evaluate the effects of overexpression and knockdown of Pit-1 on key features of the carcinogenic and metastatic process. Moreover, we evaluated Pit-1 expression in 110 human breast invasive ductal carcinomas and then correlated expression with prognostic factors.
Results
Pit-1 in the human breast adenocarcinoma cell line MCF-7 regulates cell proliferation and apoptosis.
To analyze the functions of Pit-1 on breast cancer, we first examined the effect of Pit-1 overexpression and knockdown on cell proliferation and apoptosis in the human mammary adenocarcinoma cell line MCF-7. Pit-1 overexpression has been shown to lead to an increase in cell proliferation in pituitary and extra pituitary cell lines (20, 25, 26). Our data demonstrate a significant (P < 0.001) increase in BrdU uptake in MCF-7 cells transfected with hemagglutinin-tagged Pit-1, compared with that of control cells. In addition, a significant (P < 0.001) decrease in cell proliferation was also observed using 2 Pit-1 small interfering RNA (Pit-1 siRNA-1 and Pit-1 siRNA-2), each composed by a pool of 3 pairs of primers (see Supplemental Methods), targeting different sequences in the Pit1 gene (Figure 1, A and B).
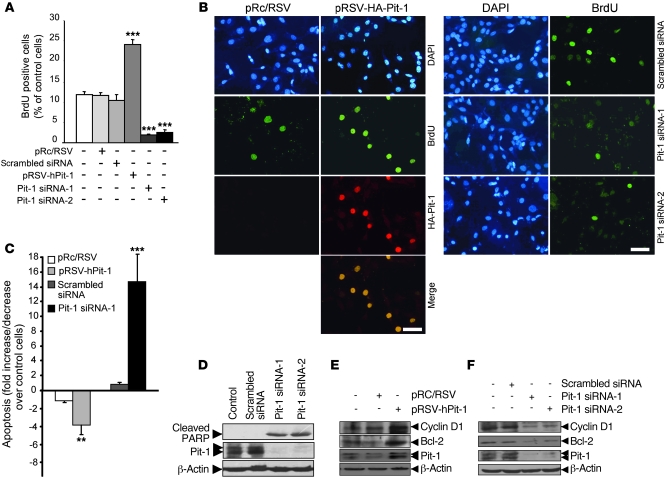
Figure 1. Pit-1 regulates proliferation and cell death in the MCF-7 human breast adenocarcinoma cell line.
(A) Overexpression of Pit-1 in the MCF-7 cell line for 48 hours increases BrdU uptake, while knockdown of Pit-1 has the opposite effect. (B) Representative example of incorporation of BrdU in MCF-7 cells after overexpression or knockdown of Pit-1. For overexpression, the cells were immunostained with an anti-hemagglutinin antibody to localize Pit-1 expression. Scale bar: 50 μm. (C) Flow cytometric analysis of apoptosis in MCF-7 cells 48 hours after Pit-1 overexpression or knockdown. (D) Western blot shows cleavage of poly (ADP-ribose) polymerase (PARP) (89 kDa), which is used as an apoptosis indicator, 48 hours after transfecting MCF-7 cells with Pit-1 siRNA-1 and siRNA-2. (E and F) Western blot of cyclin D1, Bcl-2, Pit-1, and β-actin in MCF-7 cells 48 hours (E) after Pit-1 overexpression or (F) after Pit-1 knockdown. (D–F) The major 31- and 33-kDa immunoreactive bands corresponding to Pit-1 are indicated with double arrowheads. Values are reported as mean ± SD. **P < 0.01, ***P < 0.001.
To determine the role of Pit-1 in apoptosis, MCF-7 cells were transfected with the pRSV-hPit-1 expression vector (or the pRc/RSV control vector) or with Pit-1 siRNA-1 (or scramble siRNA). Forty-eight hours later, apoptosis was determined by labeling the cells with propidium iodide (PI) or FITC-conjugated Annexin V or by performing a TUNEL assay and then subjecting cells to flow cytometry. Cells transfected with Pit-1 showed a significant (P < 0.01) decrease in apoptosis, as compared with cells transfected with the control vector (Figure 1C). Conversely, reduction of endogenous levels of Pit-1 by transfection with Pit-1 siRNA-1 induced a significant (P < 0.001) increase in cell death, 14-times greater than that of cells transfected with scramble siRNA (Figure 1C). Western blot analysis of MCF-7 cells transfected with the Pit-1 siRNA-1 and Pit-1 siRNA-2 clearly showed a 89-kDa band corresponding to cleaved poly (ADP-ribose) polymerase, a marker of apoptotic cells (Figure 1D). We found that Pit-1, probably acting at posttranscriptional level (S. Seoane, unpublished observations), also mediated in the expression of cyclin D1 (a critical regulator of the G1/S transition) and Bcl-2 (an antiapoptotic factor), suggesting that these factors could be involved in Pit-1 regulation of cell proliferation and apoptosis (Figure 1, E and F).
Overexpression of Pit-1 in the noninvasive MCF-7 cell line increases invasive potential, while the knockdown of Pit-1 in the highly invasive human adenocarcinoma cell line MDA-MB-231 reduces metastatic features.
To explore the role of Pit-1 in mammary tumor cell invasion and metastasis, we overexpressed Pit-1 transiently in the noninvasive human mammary cell line MCF-7 and stably in MCF-7 Tet-Off cells. On the other hand, we reduced the endogenous expression of Pit-1 by transiently transfecting the highly invasive mammary cell line MDA-MB-231 with Pit-1 siRNA or stably transfecting it with a doxycycline-induced vector (pSingle-tTS-Pit-1shRNA) containing a short hairpin RNA interference (shRNA) (Supplemental Figure 1, A and C; supplemental material available online with this article; doi: 10.1172/JCI42015DS1). Given that the acquisition of anchorage-independent growth is one of the in vitro properties associated with the malignancy of cells, we tested the ability of Pit-1 to induce colony formation in soft agar. We found that noninvasive MCF-7 cells stably transfected with Pit-1 for 8 days significantly (P < 0.001) increased in the number and size of colonies in soft agar, with respect to that of control cultures (Figure 2A). Conversely, after blocking endogenous Pit-1, the ability of the invasive MDA-MB-231 cells to form colonies at 12 days was significantly reduced (P < 0.001; Figure 3A). We next evaluated the motility of cells in the Matrigel invasion assay. MCF-7 cells overexpressing Pit-1 were 5-times more invasive than control cells (Figure 2, B and C). However, when endogenous Pit-1 was blocked by transfecting the MDA-MB-231 cells with Pit-1 siRNA, both migration and invasion were reduced by 90%, as compared with that of control cells (Figure 3, B and C). We also performed wound healing-induced migration assay. Cells were forced to migrate through the space created by scraping the monolayer with a tip. After 24 hours, Pit-1–expressing MCF-7 cells had completely filled the wound (Figure 2D).
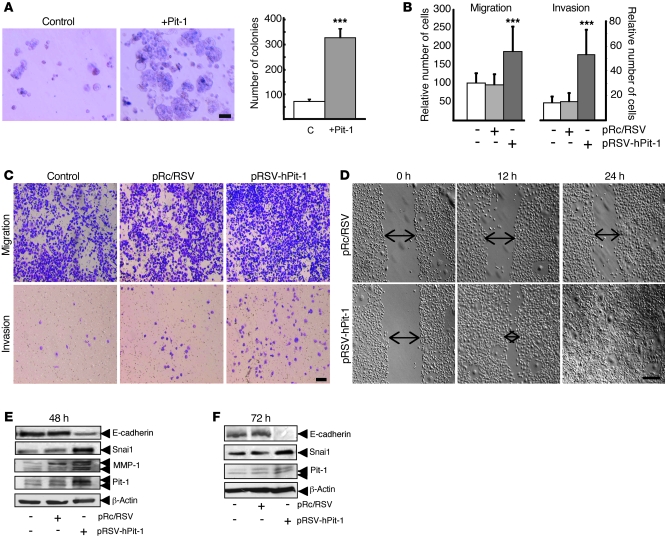
Figure 2. Pit-1 induces invasive and malignant features in the noninvasive MCF-7 human breast carcinoma cell line.
(A) Overexpression of Pit-1 (pTRE2-hPit-1) in MCF-7 cells after 8 days induces a significant (***P < 0.001) increase in cell colony formation in soft agar, with respect to that of control cells. C, control cells. Scale bar: 75 μm. (B and C) Cell motility through uncoated filters (migration) and through Matrigel-coated filters (invasion) at 48 hours in the control cells, pRc/RSV–transfected MCF-7 cells, and pRSV-hPit-1–transfected MCF-7 cells (***P < 0.001 versus control cells). Numbers represent mean ± SD. (C) Representative example of B. Cells were stained with crystal violet, visualized with microscopy, and counted. Scale bar: 100 μm. (D) Wound healing assay. The wounded areas (space indicated by arrows) were examined with microscopy at the indicated times. Scale bar: 250 μm. (E) Protein extracts of MCF-7 cells 48 hours after transfection with the empty vector (pRc/RSV) or with pRSV-hPit-1–expressing vector were prepared and tested using Western blotting for E-cadherin, Snai1, MMP-1, Pit-1, and β-actin. (F) Western blot assay showing a suppression of E-cadherin expression 72 hours after Pit-1 overexpression in MCF-7 cells. (E and F) Double arrowheads in the MMP-1 immunoblot indicate proenzyme form (top arrow) and active form (bottom arrow). Double arrowheads in Pit-1 immunoblots indicate 33-kDa and 31-kDa immunoreactive bands.
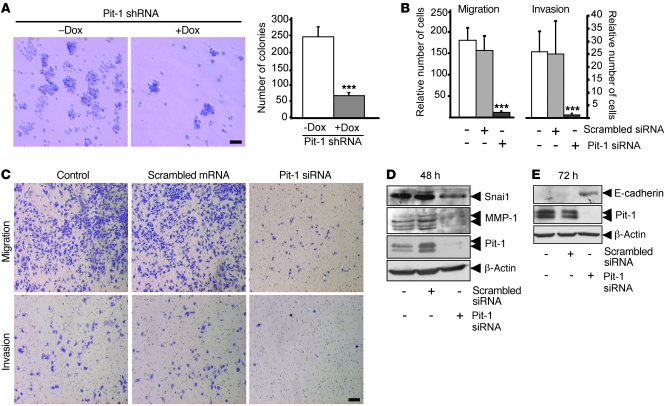
Figure 3. Knockdown of endogenous Pit-1 in the invasive MDA-MB-231 breast carcinoma cell line reduces malignant traits.
(A) Knockdown of Pit-1 in MDA-MB-231 cells (treated with doxycycline [+Dox]) significantly (***P < 0.001) reduces the number of colonies in soft agar at 12 day, as compared with that of controls (not treated with doxycycline [–Dox]). Scale bar: 75 μm. (B and C) Knockdown of Pit-1 for 48 hours in MDA-MB-231 cells significantly inhibits cell motility through uncoated filters (migration) and through Matrigel-coated filters (invasion). Error bars represent SD. ***P < 0.001 versus controls. Cells were stained with crystal violet, visualized with microscopy, and counted. Scale bar: 100 μm. (D) Western blot in MDA-MB-231 cells indicating a decrease of Snai1 and MMP-1 expression 48 hours after endogenous Pit-1 suppression. (E) A band corresponding to the E-cadherin size is readily visible using Western blot 72 hours after blocking endogenous Pit-1 in MDA-MB-231 cells. (D and E) Double arrowheads in the MMP-1 immunoblot indicate proenzyme form (top arrow) and active form (bottom arrow). Double arrowheads in Pit-1 immunoblots indicate 33-kDa and 31-kDa immunoreactive bands.
The Snai1 transcription factor is an important regulator of invasiveness during tumor progression by repressing E-cadherin (27, 28). E-cadherin is essential for the maintenance of cell-cell adhesion, and functional loss of E-cadherin in epithelial cells has been considered a hallmark of EMT (2). Thus, the next step in our evaluation of the effect of Pit-1 focused on Snai1 and E-cadherin expression. It has been shown that Snai1 is more strongly expressed in MDA-MB-231 cells compared with epithelial MCF-7 cells (29). On the other hand, E-cadherin expression is present in MCF-7 cells but not in the invasive MDA-MB-231 cells (28). This is corroborated by our results. At 48 hours, Snai1 presented a strong band both in controls and scrambled siRNA-transfected MDA-MB-231 cells. In addition, we found a visible decrease after Pit-1 siRNA transfection. Furthermore, at 72 hours, a band corresponding to E-cadherin became readily visible (Figure 3, D and E). However, in MCF-7 cells, Pit-1 overexpression at 48 hours increased Snai1 transcription and reduced E-cadherin expression, which was completely repressed at 72 hours (Figure 2, E and F). We also evaluated the expression of the MMP-1. This protein has recently been related to both breast tumorigenicity and basal lung metastagenicity (30). As shown in Figure 2E and Figure 3D, overexpression of Pit-1 increased MMP-1 expression in MCF-7 cells, but Pit-1 siRNA transfection decreased MMP-1 expression in the MDA-MB-231 cells.
Overexpression of Pit-1 in immunodeficient mice increases tumor growth and induces EMT.
We next evaluated the effect of Pit-1 overexpression or depletion in vivo using the severe combined immunodeficient (SCID) mouse tumor xenograft model. Tumor growth at day 12 and 20 was significantly (P < 0.001) increased in mice subcutaneously injected with Pit-1–overexpressing MCF-7 cells, as compared with mice injected with control cells (Figure 4, A and B, and Supplemental Figure 1A). As expected, at day 20, Pit-1 expression was greater in mice injected with Pit-1–overexpressing MCF-7 cells, both by Western blot (Figure 4C) and immunohistochemistry (Figure 4D). Immunohistochemical analysis indicated that mouse tumors with MCF-7 cells overexpressing Pit-1 are involved in EMT, as demonstrated by decreased epithelial markers (e.g., cytokeratin AE1-AE3 [CKAE1-AE3], CK19, E-cadherin, and β-catenin) and increased mesenchymal markers (e.g., vimentin). We also found increased cell proliferation (e.g., Ki-67) (Figure 4E). Interestingly, E-cadherin expression levels seem to be related with tumor size (Supplemental Figure 1B).
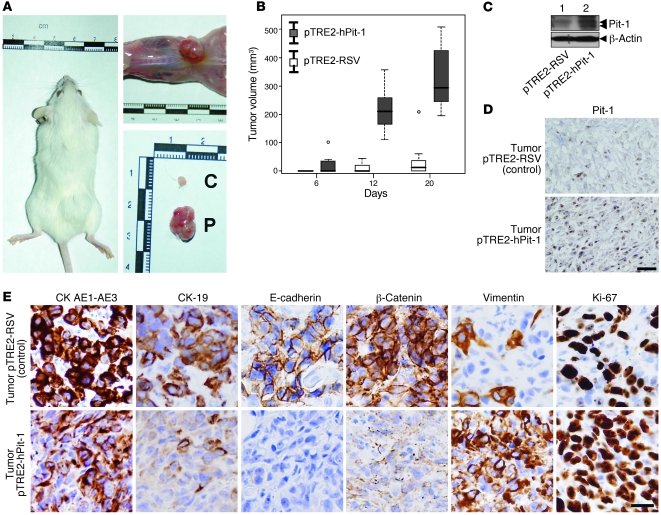
Figure 4. Subcutaneous injection of Pit-1–overexpressing MCF-7 cells in SCID mice increases tumor growth and induces EMT.
(A) Seven SCID mice were subcutaneously injected with control MCF-7 cells (left flank) or MCF-7 cells stably transfected with Pit-1 expression vector (right flank). C, control cells; P, Pit-1–overexpressing MCF-7 cells. (B) Box plot of tumor growth in SCID mice, as described in A. The bottom and the top of the boxes represent the first and third quartiles (i.e., the 25th and 75th percentile, respectively), and the bands near the middle of each box represent the 50th percentile (the median). The ends of the whiskers represent the lowest and the highest values still within 1.5 interquartile range of the lower and upper quartiles, respectively. (C and D) Western blot and immunohistochemistry of Pit-1 in tumors of SCID mice injected with MCF-7 control cells stably transfected either with the pTRE2-hPit-1 expression vector or the control vector (pTRE2) at day 20. Double arrowheads in the Pit-1 immunoblot indicate 33-kDa and 31-kDa immunoreactive bands. Scale bar: 40 μm. (E) Immunohistochemistry analysis of epithelial (CK AE1-AE3, CK-19, E-cadherin, and β-catenin), mesenchymal (vimentin), and proliferation (Ki-67) markers in tumors of SCID mice injected with control or Pit-1–overexpressing MCF-7 cells. Scale bar: 25 μm.
Knockdown of Pit-1 in MDA-MB-231 cells injected into SCID mice reduces tumor growth but does not modify EMT-associated proteins.
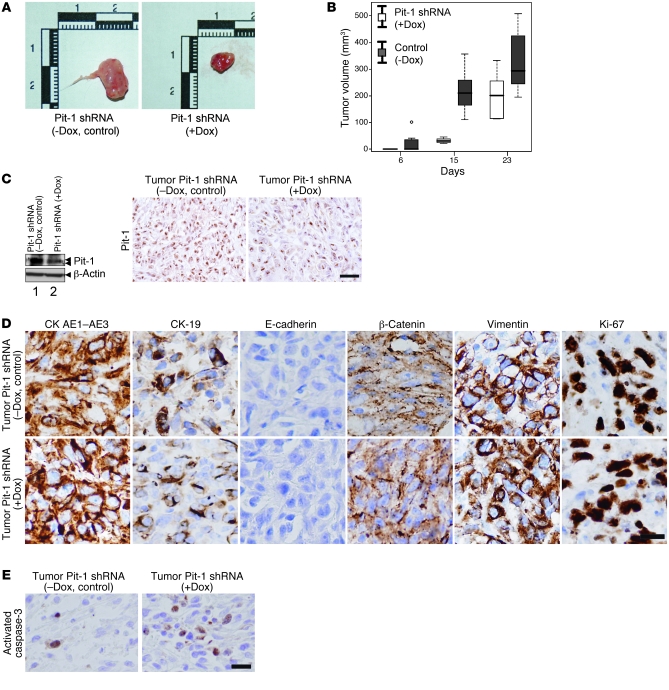
Mice injected with MDA-MB-231 cells transfected with the pSingle-tTS-Pit-1shRNA vector and fed doxycycline (to knockdown Pit-1 expression) showed a significant reduction in tumor growth, without significant weight loss at 15 and 23 days after injection, as compared with mice not fed doxycycline (Figure 5, A and B, and Supplemental Figure 1C). In mouse tumors with Pit-1 knocked down, we observed a decreased Pit-1 protein content (Figure 5C). However, surprisingly, no changes in the expression of proteins involved in epithelial-to-mesenchymal transition (i.e., CKAE1-AE3, CK19, E-cadherin, β-catenin, and vimentin) were observed in either group of mice (Figure 5D). Using immunohistochemistry, we also evaluated activated caspase-3 as an indicator of apoptosis. As shown in Figure 5E, a significant (P < 0.001) increase in immunopositivity was seen in tumors of mice with Pit-1 knocked down (5.49% ± 0.72%), with respect to the control mice (0.31% ± 0.18%), suggesting that the lower tumor growth observed after Pit-1 knockdown could be the result of increased apoptosis (see also Figure 1C).
Figure 5. Knockdown of Pit-1 levels in MDA-MB-231 cells reduces tumoral growth when injected into SCID mice.
(A) Six SCID mice were subcutaneously injected in the left and right flanks with MDA-MB-231 cells stably transfected with the pSingle-tTS-Pit-1shRNA doxycycline-induced vector and treated (+Dox) or not treated (–Dox, control) with doxycycline to induce Pit-1shRNA. A sample tumor from each group is shown. (B) Box plot of tumor growth in SCID mice, as described in A. The box plot is defined as in Figure 4B. (C) Western blots and immunohistochemical detection of Pit-1 in tumors of SCID mice, as described in A. Double arrowheads in the Pit-1 immunoblot indicate 33-kDa and 31-kDa immunoreactive bands. Scale bar: 40 μm. (D) Immunohistochemical detection of epithelial, mesenchymal, and proliferation markers, as described in Figure 4E. Scale bar: 25 μm. (E) Immunohistochemical detection of activated caspase-3 in tumors of SCID mice, as described in A. Scale bar: 40 μm.
Pit-1 transcriptionally regulates Snai1 gene expression.
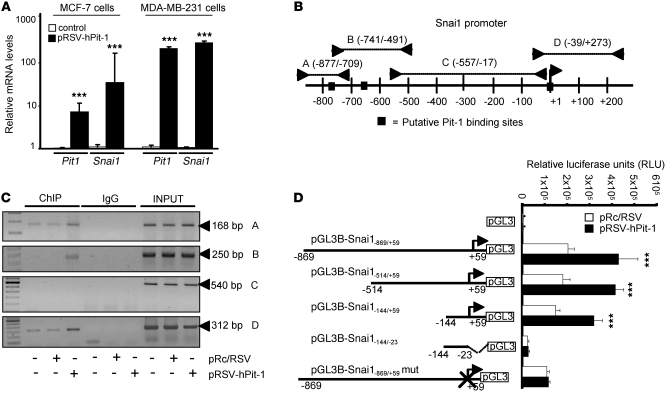
Given that overexpression of Pit-1 in MCF-7 cells increases Snai1 protein and induces EMT in mouse tumors, we evaluated the regulation of Snai1 by Pit-1 in more depth. MCF-7 and MDA-MB-231 cells were transfected with the pRSV-hPit-1 vector or the pRc/RSV control vector, and 48 hours later quantitative PCR was carried out to evaluate Pit1 and Snai1 mRNA expression. As expected, a significant increase in Pit1 mRNA expression was observed after Pit-1 overexpression in both MCF-7 and MDA-MD-231 breast cancer cell lines. In addition, Snai1 mRNA levels rose significantly in pRSV-hPit-1–overexpressed cells in relation to that of control cells (Figure 6A). To determine whether Pit-1 binds to the Snai1 gene, a ChIP assay was carried out using Snai1 gene-specific primers (see Supplemental Methods). Using the Transcription Element Search Software program (University of Pennsylvania, Philadelphia, Pennsylvania, USA; http://www.cbil.upenn.edu/tess), 3 putative Pit-1 binding sites were found in the Snai1 promoter (Figure 6B). Specific binding of Pit-1 to the A, B, and D sites of the Snai1 promoter was observed in both control and pRSV-hPit-1–overexpressed cells, indicating that Pit-1 binds to the Snai1 promoter gene in vivo (Figure 6C). Transfection reporter assays (Figure 6D) indicated that Pit-1 transcriptionally regulates the Snai1 gene. Specific deletions of the Snai1 promoter demonstrate that Pit-1 binds to a region comprised between –23/+59 bp from the start transcription site, and this region is necessary for Pit-1–induced Snai1 transcription. The functionality of the proximal Pit-1 response element was tested by mutating it in the context of the pGL3B-Snai1–869/+59 construct. Specifically, 5 different mutations were introduced into the Snai1–869 sequence at position –5/+4 from the transcription start site (pGL3B-Snai1–869/+59mut). This construct was cotransfected into MCF-7 cells with the pRc/RSV or with the pRSV-hPit-1 vector. As shown in Figure 6D, the response to Pit-1 in cells transfected with the mutant construct was completely abolished.
Figure 6. Pit-1 transcriptionally regulates Snai1 expression.
(A) Relative expression of the mRNAs encoding Pit-1 and Snai1 in MCF-7 and MDA-MB-231 cells before and after Pit-1 overexpression, as determined by real-time PCR. Rn18s mRNA was used to normalize the variability in template loading. Values are expressed as mean ± SD. ***P < 0.001 versus controls. (B) The diagram shows the Snai1 promoter with the putative Pit-1 binding sites (identified by filled squares) and the regions amplified using specific primers (see Supplemental Methods) in the ChIP assay (identified by lines with arrowheads as A, B, C, and D). The initiation transcription site is indicated by a single arrowhead at position +1. (C) Soluble chromatin obtained from control, pRc/RSV–transfected MCF-7 cells, and pRSV-hPit-1–transfected MCF-7 cells was immunoprecipitated with anti–Pit-1 antibody or control IgG. The immunoprecipitated DNA was amplified by PCR using primers (see Supplemental Methods) A, B, C, and D, as shown in B. (D) Deletion analysis and selective mutations at position –5/+4 bp from the start transcription site identified a Pit-1–responsive region in the human Snai1 promoter. MCF-7 cells were transfected with several Snai1 promoter fragments fused to pGL3Basic (pGL3B) vector. Normalized relative luciferase units (RLU) were calculated as the ratio of luciferase activity in Pit-1–overexpressing cells to the activity in the corresponding control cells. *** P < 0.001 versus controls. Data are expressed as mean ± SD.
Pit-1, independently and also through Snai1, modifies expression of proteins involved in EMT and cellular motility.
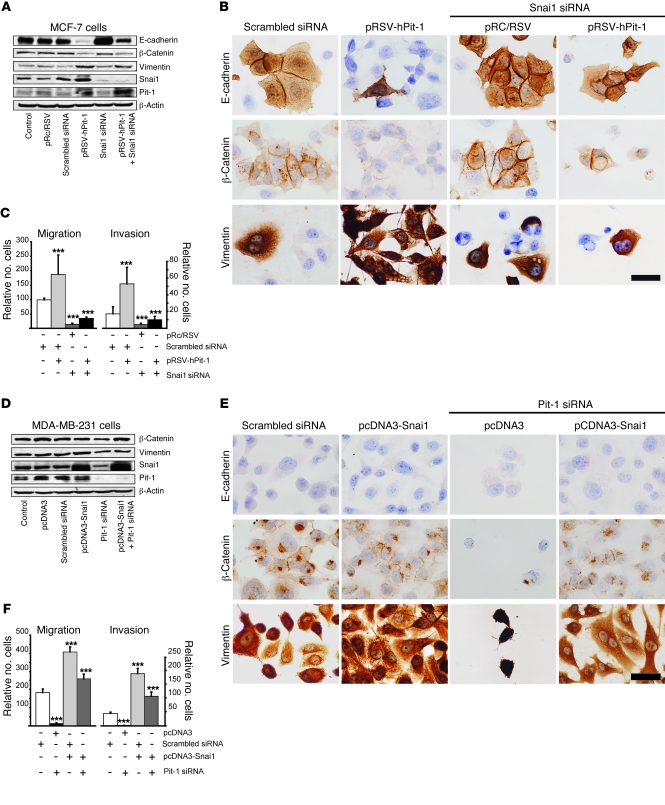
To evaluate whether Pit-1 regulates proteins involved in EMT in some way other than through Snai1 induction, Snai1 expression was knocked down in Pit-1–overexpressing MCF-7 cells, and 48 hours later E-cadherin, β-catenin, and vimentin were evaluated using Western blot and immunohistochemistry. With respect to E-cadherin, our data showed a decreased expression after Pit-1 overexpression and an increased expression after Snai1 knockdown, in relation to that of control cells. Endogenous knockdown of Snai1 in Pit-1–overexpressed MCF-7 cells restored the E-cadherin to basal levels (Figure 7, A and B), suggesting a Snai1-dependent action of Pit-1. On the other hand, although Pit-1 overexpression causes both a decrease in β-catenin and a strong increase in vimentin, unlike the effect of Pit-1 on E-cadherin, neither of these proteins went back to basal expression levels after cotransfection with Snai1 siRNA (Figure 7, A and B), suggesting a Snai1-independent action. Similarly, to analyze the role of Snai1 in Pit-1–induced cell motility, we also evaluated migration and invasion in MCF-7 cells. Knockdown of Snai1 in these cells significantly (P < 0.001) reduced migration and invasion (Figure 7C), but cotransfection with pRSV-hPit-1 did not bring the levels back to those observed in the control cells (Figure 7C).
Figure 7. Pit-1, independently and through Snai1, modifies expression of proteins involved in EMT and cell motility.
(A) Western blots of E-cadherin, β-catenin, vimentin, Snai1, and Pit-1 in control MCF-7 cells 48 hours after transfection of pRSV-hPit-1 and/or Snai1 siRNA. (B) Immunohistochemistry of E-cadherin, β-catenin, and vimentin, as indicated in A. Scale bar: 25 μm. (C) Migration and invasion assay in MCF-7 control cells and cells transfected as described in A. (D) Western blots of β-catenin, vimentin, Snai1, and Pit-1, and β-actin for control MDA-MB-231 cells, as well as for cells 48 hours after (i) Snai1 overexpression, (ii) Pit-1 knockdown, and (iii) both Snai1 overexpression and Pit-1 knockdown. (E) Immunohistochemistry of E-cadherin, β-catenin, and vimentin, as described in D. Scale bar: 25 μm. (F) Migration and invasion assay in MDA-MB-231 control cells and cells transfected as described in D. Values are expressed as mean ± SD. ***P < 0.001 versus controls.
Overall, our data strongly suggest that Pit-1 regulates β-catenin and vimentin protein expression independently of Snai1 but not that of E-cadherin. On the other hand, we found that increased cell motility seems to be mediated by Pit-1–induced Snai1, suggesting that Pit-1 by itself does not play a major role in cell motility.
To study the effects of Pit-1 on protein expression related to EMT and cell phenotype in more depth, we evaluated whether the effects of Pit-1 knockdown could be reversed by Snai1 overexpression in MDA-MB-231 cells. E-cadherin immunopositivity was found in neither control cells nor transfected cells after 48 hours (Figure 7E). Pit-1 knockdown induces cell death, as shown in MCF-7 cells (Figure 1C) and in tumors of mice induced by injection of MDA-MB-231 cells transfected with Pit-1 shRNA (see Figure 5E). In line with this, the decreased vimentin protein expression observed after transfection with Pit-1 siRNA could be related — though not necessarily exclusively — to Pit-1–induced apoptosis (Figure 7, D and E). Cotransfection of cells with both Pit-1 siRNA and the pcDNA3-Snai1 overexpression vector brought vimentin expression nearly back to levels observed in control cells (Figure 7, D and E). No significant changes in β-catenin expression were observed after cotransfection of the Snai1 overexpression vector and Pit-1 siRNA. Immunohistochemistry analysis indicated that β-catenin expression was weak in membranes and appeared mainly as dots or cytoplasmic aggresomes, suggesting a misfolding or a partially denatured protein (Figure 7, D and E). Migration and invasion values in MDA-MB-231 cells transfected with the Snai1–overexpressing vector significantly (P < 0.001) increased, with respect to those of control cells (Figure 7F). However, knockdown of Pit-1 in Snai1–overexpressed MDA-MB-231 cells did not reduce migration and invasion levels below those observed in control cells (Figure 7F), suggesting a Snai1-dependent Pit-1 role in cell motility, as described above for MCF-7 cells.
Pit-1 promotes breast cancer-associated lung metastasis.
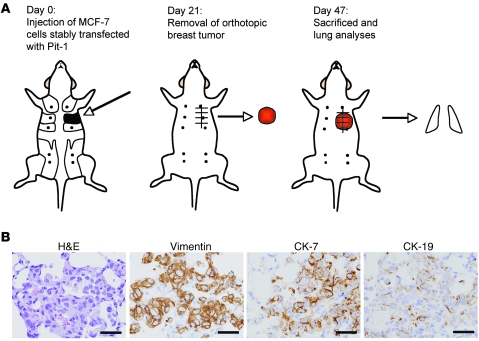
Given that Pit-1 transcriptionally raises Snai1 expression and Snai1 is a key EMT-inducing factor, we next tested the potential of MCF-7 cells, stably transfected with Pit-1, to metastasize in vivo. It has been shown that MCF-7 cells present very low levels of metastasis (31, 32). Therefore, SCID female mice were injected in the mammary fat pad with the MCF-7 cells stably transfected with the pTRE2-Pit-1–overexpressing vector (n = 12) or with the pTRE2 control vector (n = 11). Twenty-one days after injection, primary mammary tumors were excised under anesthesia in both groups of animals. Experiments were terminated 26 days after tumor excision (day 47 after injection of MCF-7 cells), and mice were sacrificed and lungs were removed, fixed, paraffin embedded, sectioned, and stained either with H&E or with specific antibodies for immunohistochemistry analysis (Figure 8A). Nine out of the twelve mice injected with Pit-1–overexpressing cells developed micrometastasis in lung, while 2 out of the 11 mice injected with control MCF-7 cells showed micrometastasis in lungs (P = 0.012). Micrometastases showed immunopositivity for vimentin, CK-7, and CK-19 (Figure 8B). These antibodies react only with human and not with mouse vimentin and cytokeratin.
Figure 8. Orthotopic injection of Pit-1–overexpressing MCF-7 cells in SCID mice induces metastasis in lung.
(A) Schematic representation of experimental induction of metastasis. At day 0, eleven SCID mice were injected with MCF-7 cells (controls) and twelve SCID mice were injected with MCF-7 cells stably transfected with Pit-1–overexpressing vector into the second mammary fad pat. At day 21, animals were anesthetized and breast tumors were resected. On day 47, mice were sacrificed and lungs were removed for analysis. Nine out of twelve SCID mice injected with MCF-7 cells overexpressing Pit-1 showed micrometastasis in lung. Two micrometastasis were observed in mice (n = 11) injected with control MCF-7 cells. (B) Representative example of micrometastasis in lung. H&E staining and immunopositivity for vimentin, CK-7, and CK-19. Scale bar: 40 μm.
Pit-1 expression in human invasive ductal carcinomas of the breast is associated with distant metastasis.
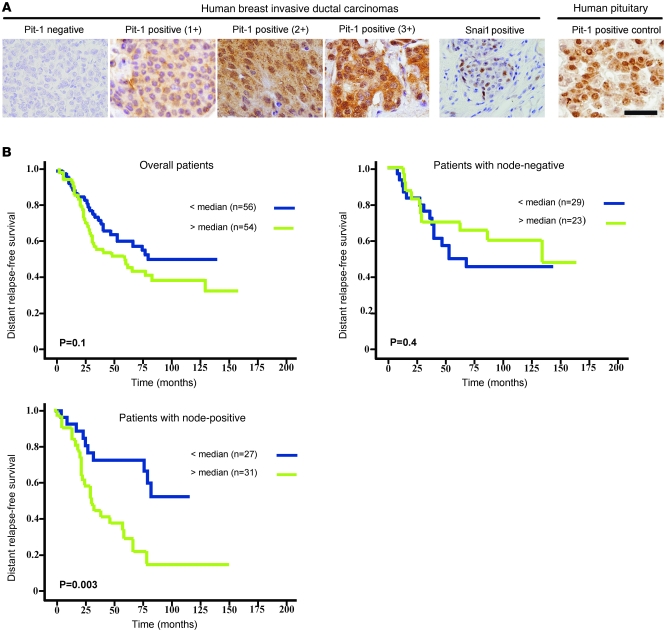
To further evaluate the clinical value of Pit-1 expression in human breast tissue, Pit-1 was analyzed by immunohistochemistry in 110 invasive ductal carcinomas of the breast. Representative examples of Pit-1 immunostaining in tumors are shown in Figure 9A. Pit-1 protein expression was detected mainly in the nuclei of tumor or control epithelial cells. A total of 89 tumors (80.9%) stained positively for Pit-1, with clear differences regarding intensity and the percentage of stained cells. The mean score value was 35.4 (range 0–194.5). Distribution of Pit-1 score values is shown in Supplemental Figure 2. In the group of 89 Pit-1–positive tumors, 49 tumors were weakly stained (score < 50), 26 were moderately stained (50 < score < 100), and the remaining 14 tumors were strongly stained (score > 100). In addition, to investigate the possible relationship between Pit-1 and Snai1 expression, we also analyzed the staining pattern for both proteins in 65 breast carcinomas using immunohistochemistry. A total of 14 cases (21.1%) showed positive immunostaining for Snai1 in the nucleus in at least 5% of cancerous cells. These Snai1-positive tumors showed higher scores for Pit-1 expression (score values: median, 77.9 [range, 13.06–143.19]) than the 51 Snai1-negative tumors (median, 31.6 [range, 0–156.7]) (P < 0.001). A representative example of Snai1 immunostaining in breast tumors is shown in Figure 9A.
Figure 9. High expression of Pit-1 in human ductal invasive carcinomas of the breast correlates with occurrence of distant metastasis.
(A) Representative set of negative and positive (1+, 2+, and 3+) Pit-1 and Snai1 immunodetections in human breast invasive ductal carcinomas. Pit-1 immunopositivity in human pituitary gland was used as positive control. Scale bar: 50 μm. (B) Kaplan-Meier survival curves as a function of the immunostaining expression of Pit-1 in the overall group of patients (n = 110), in the subgroup with node-negative breast cancer (n = 52), and in the subgroup with node-positive breast cancer (n = 58). The median of the immunostaining score values in the overall group of patients (score 35.4) was chosen as the cut-off point for both node-negative and node-positive patient subgroups.
The wide variability in Pit-1 values obtained suggested a wide variability in biological and clinical behavior of breast carcinomas, thus the potential value of Pit-1 expression as a prognostic marker. Statistical analysis showed that Pit-1 score values were not significantly related to age, tumor size, nodal status, histological grade, estrogen or progesterone receptor status, desmoplastic reaction, peritumoral inflammation, tumoral advancing edge, or tumoral invasion (Supplemental Table 2). These facts prompted us to evaluate the potential role of Pit-1 as an independent prognostic factor in breast cancer.
The potential association between Pit-1 immunostaining and distant relapse-free survival was retrospectively evaluated in the 110 patients included in the present study. Using statistical analysis, we used the median of the Pit-1 scores as the cut-off point. Using this cut-off point, no significant association was found between Pit-1 expression and relapse in either the overall group of patients or the node-negative patient subgroup (Figure 9B). However, in the subgroup of patients with node-positive breast cancer, relapse was confirmed in 10 out of 27 (37%) patients with tumors showing low levels of Pit-1 expression and in 23 out of 31 (74.2%) patients with tumors showing high levels of Pit-1 expression. Statistical analysis showed that Pit-1 expression was significantly associated with the occurrence of distant metastasis in patients with node-positive breast cancer (hazard ratio, 3.02; 95% CI, 1.4–6.9; P = 0.004). In addition, differences between distant relapse-free curves for these 2 groups of patients were significant at level P = 0.003 (Figure 9B). On the other hand, multivariate analysis according to the Cox model demonstrated that tumor stage (tumor stage III, relative risk [RR], 2.20; CI, 1.05–4.60; P = 0.037), ER status (ER-positive status, RR, 0.37; CI, 0.17–0.79; P = 0.011), and PgR status (PgR-positive status, RR, 0.40; CI, 0.17–0.90; P = 0.028) were significantly and independently associated with distant relapse-free survival in the subgroup of patients with node-positive tumors. Nevertheless, this same analysis also demonstrated that Pit-1 expression was the most potent independent factor associated with distant relapse-free survival in patients with node-positive tumors (RR, 2.71; CI, 1.20–6.00; P = 0.015).
Discussion
In this study, we demonstrate that overexpression of Pit-1 in breast cancer cell lines and in SCID mice in vivo induces malignant features, tumor growth, and metastasis in lung. Our findings provide evidence that Pit-1 regulates the Snai1 transcription factor and induces EMT. In addition, elevated expression of Pit-1 in patients with invasive ductal carcinomas of the breast is significantly correlated with the occurrence of distant metastasis.
Several studies have suggested that Pit-1 is directly involved in pituitary cell proliferation (25), apoptosis (33), and possibly in the pathogenesis of pituitary adenomas (34, 35). However, other studies have not found any relation between Pit-1 expression and pituitary tumorigenesis (36). Given that the original studies describing Pit-1 expression and function were restricted to the pituitary gland and pituitary cell lines, the function of Pit-1 in extrapituitary tissues had not been well evaluated previously. In the mammary gland, Pit-1 also regulates GH and PRL, increases cell proliferation, and has higher expression in breast tumors than in normal mammary gland, suggesting that Pit-1 could be involved in mammary tumorigenesis (20, 23). In the present study, we demonstrate that Pit-1 was also involved in extrapituitary regulation of cell death. Our data show that Pit-1 overexpression in MCF-7 cells significantly reduced apoptosis and, conversely, Pit-1 knockdown increased cell death 14 fold. In addition, we show that Pit-1 upregulated the antiapoptotic protein Bcl-2. Regarding this point, it has been demonstrated that Bcl-2 overexpression occurs in 40% to 80% of human breast tumors (37).
Given the Pit-1 involvement in breast cell proliferation and cell death, two key processes in cancer (38), our next objective was to evaluate the function of Pit-1 in other steps of breast oncogenesis. Our experimental cell model was designed to overexpress Pit-1 in the noninvasive human breast adenocarcinoma cell line MCF-7 and evaluate the metastatic potential of Pit-1 and, on the other hand, to knockdown Pit-1 in the highly invasive human breast carcinoma cell line MDA-MB-231 and evaluate the possible regression of its invasive properties. Our results demonstrated that Pit-1 regulates cell migration, invasion, and anchorage-independent growth as well as the expression of MMP-1, Snai1, and E-cadherin. MMP-1, also known as collagenase-1, plays an important role in diverse physiologic processes, such as development, tissue morphogenesis, and wound repair. Likewise, it seems to be implicated in a variety of human diseases, including cancer (39). In cancer, it has recently been demonstrated that MMP-1 is part of a group of 4 genes that when expressed in human breast cancer cells facilitate the assembly of new tumor blood vessels, the release of tumor cells into the circulation, and the breaching of lung capillaries by circulating tumor cells to seed pulmonary metastasis (30).
Disruption of the E-cadherin–mediated adhesion system is a major event in the transition from noninvasive tumors to invasive malignant carcinomas (40) and is a key biomarker for EMT (2). E-cadherin is directly repressed by Snai1 transcription factor, which in turn induces the acquisition of mesenchymal phenotype by epithelial tumor cells (27, 28). Snai1 is a key factor during embryogenesis, but its expression is also related to oncogenic processes. In fact, Snai1 expression is inversely correlated with the grade of differentiation of breast tumors, which seems to indicate that it is involved in the progression of breast carcinomas (41). It has also been demonstrated that Snai1 promotes mammary tumor recurrence, and high levels of Snai1 predict lower relapse-free survival in women with breast cancer (42). Our data demonstrate that Snai1 is transcriptionally regulated by Pit-1 and that Pit-1, independently and also through Snai1, could be involved in some important functions of EMT. Thus, overexpression of Pit-1 in Snai1 knocked down MCF-7 cells reduces β-catenin expression and increases vimentin expression but does not restore E-cadherin to basal levels. This suggests a Snai1-independent role of Pit-1 in the regulation of β-catenin and vimentin, while Snai1 induction is required for E-cadherin regulation. In addition, Snai1 also seems to mediate the role of Pit-1 in cell motility. However, we can not exclude Pit-1 modulation on other gene(s) involved in EMT.
Our in vivo results strongly support the in vitro data: after injection of Pit-1–overexpressing MCF-7 cells in SCID mice, a significant increase in tumor growth was observed with respect to controls. Immunohistochemistry analysis of tumors induced by Pit-1–overexpressing MCF-7 cells once again strongly indicated that Pit-1 is involved in the EMT, as shown by the decreased epithelial and increased mesenchymal markers. Transition between the epithelial and mesenchymal state is now known to be a central process that regulates cell plasticity in carcinomas, tumor recurrence, and metastatic progression (3). Our results showing the presence of micrometastasis in lungs of 75% of the SCID mice orthotopically injected with Pit-1–overexpressing MCF-7 cells seem to indicate that Pit-1 induces a phenotypic transformation in the originally nonmetastatic MCF-7 cells, leading them to acquire invasive and migratory properties and thus the capacity to develop metastasis in lung. The evaluation of breast metastasis using our experimental approach (i.e., orthotopic injection of MCF-7 cells, excision of tumor under anesthesia at day 21, and histological evaluation of lungs at day 47) seems to be a more physiological way to generate “primary tumors” that lead to spontaneous metastasis than injecting tumor cells directly into the bloodstream, which could bypass most of the steps in the invasion-metastasis cascade that depend on an EMT program.
In order to correlate our in vitro and in vivo results with Pit-1 expression in patients with breast cancer, we analyzed Pit-1 expression in invasive breast cancer using immunohistochemistry. In addition, we also evaluated Snai1 immunopositivity in these tumors and found significant correlation between Snai1 positivity and higher Pit-1 expression, supporting the regulation of Snai1 by Pit-1.
The wide variability obtained in Pit-1 values suggested that this transcription factor might influence the biological and clinical behavior of breast carcinomas. In addition, the fact that Pit-1 expression did not correlate with classical clinical-pathological parameters of breast cancer led us to consider Pit-1 as a potential independent prognostic factor in breast cancer. The potential association between Pit-1 immunostaining and distant relapse-free survival was retrospectively evaluated in the patients group. We found no significant association between Pit-1 expression and relapse, either in the overall patient group or in the node-negative patient subgroup. However, in the node-positive patient subgroup, high expression of Pit-1 was significantly and independently associated with the occurrence of distant metastasis. This finding could prove to be highly relevant in light of the fact that tumor involvement of axillary lymph nodes is considered to be the major prognostic factor of poor outcome in breast cancer (43, 44).
Human breast cancer has been related with deregulation of several embryonic transcription factors that play a crucial role in development (45, 46). For example, Oct3 and Brn3b proteins, which like Pit-1 belong to the POU family of transcription factors and have been implicated in embryonic stem cell and sensory nervous system development, are expressed at abnormally high levels in human breast cancer (47, 48). In addition, it has been recently demonstrated that EMT-inducing transcription factors may generate cells with stem-like cells characteristics (9). Thus, deregulation of EMT-regulating factors in the mammary gland could induce EMT in epithelial mammary cells and/or in stem-like cells, and thus, these cells could later acquire mesenchymal characteristics with invasive and self-renewal capabilities, enabling these cells to develop micrometastasis and macrometastasis in distant organs. These data also support the hypothesis that breast cancer cells may have a stem cell origin (49–51).
The possibility of directly targeting Pit-1 has been opened by a recent in vitro study, showing a direct inhibition of the DNA-binding activity of Pit-1 and Brn3 by a small molecule (a phenyl-furan-benzimidazole dication, DB293) (52). In addition, our group has also demonstrated an inhibitory effect of 1,25-dihydroxyvitamin D3 (1,25[OH]2D3) on Pit-1 expression (53). 1,25(OH)2D3 and its analogs have antiproliferative and proapoptotic effects in a number of cell types, including breast cancer cells, through the vitamin D receptor (VDR). A link between VDR and Snai1 has also been demonstrated. Snai1 represses VDR gene expression in human colon cancer, and elevated Snai1 expression correlates with downregulation of VDR. In xenografted mice, Snai1 blocks the antitumor action of EB1089, a 1,25(OH)2D3 analog (54). Given that Pit-1 regulates Snai1 and 1,25(OH)2D3 inhibits Pit-1, treatment with 1,25(OH)2D3 or its analogs could be an interesting approach in breast tumors that overexpress Pit-1.
The in vitro data obtained from the experimental manipulation of breast cancer cell lines and the in vivo data from the SCID mouse studies indicate that Pit-1 induces EMT-associated changes and activates other cellular programs, such as cell survival and proliferation. The analysis of Pit-1 expression in human breast samples could contribute to improving the prognosis of patients with node-positive breast cancer. Taken together, our findings suggest that Pit-1 could represent a new therapeutic target in breast cancer.
Methods
Patients, breast cancer samples, and immunohistochemistry.
One hundred and ten patients with invasive breast cancer (without distant metastasis at the time of initial diagnoses), which were treated at Fundación Hospital de Jove between 1990 and 2003, were selected based on the availability of clinical details at presentation and a minimum of 5 years of follow-up for women without tumor recurrence. Patient sample characteristics are described in Supplemental Methods and Supplemental Table 1. Women were treated according to the guidelines used by Fundación Hospital de Jove. The study adhered to Spanish regulations and was approved by the Fundación Hospital de Jove Ethics and Investigation Committee. The end point was distant metastatic relapse. The median follow-up period was 88 months in patients without metastases and 45 months in patients with metastases. Breast carcinoma tissue samples were obtained at the time of surgery. Prior informed consent was obtained from patients. Routinely fixed (overnight in 10% buffered formalin), paraffin-embedded tumor samples stored in our pathology laboratories were used. Histopathologically representative tumor areas without necrosis were defined on H&E-stained sections. Serial 5-μm sections were consecutively cut with a microtome (Leica HM335S, Leica Microsystems) and transferred to adhesive-coated slides. Immunohistochemistry was performed on these sections using a TechMate TM50 autostainer (Dako), as previously described (23). Antibodies for Pit-1 and Snai1 were obtained from Santa Cruz Biotechnology Inc. and from Garcia de Herreros (Unidad de Biología Celular y Molecular, Universidad Pompeu Fabra, Barcelona, Spain), respectively. We used human pituitary gland (obtained from autopsy) as a positive control for Pit-1. Immunostaining levels were scored as detailed in Supplemental Methods.
Cell cultures, immunohistochemistry, constructs, and transfections.
MCF-7 Tet-Off cells were purchased from Clontech. MCF-7 and MDA-MB-231 (human breast adenocarcinoma cell lines) were obtained from the European Collection of Cell Cultures. Cells were maintained in DMEM supplemented with 10% FBS in 5% CO2. MCF-7 and MDA-MB-231 cells (2 × 104 cells) were transfected for 48 hours with the corresponding constructs, seeded in slides, and fixed for 10 minutes in 96% ethanol, before processing for immunohistochemistry, as described below for immunohistochemistry animal studies. Transient transfections were performed as previously described (52). Stable transfections were performed with the Cell Line Nucleofector Kit (Amaxa). Cell culture, plasmid construction, and transfection protocol are described in Supplemental Methods.
Western blot, real-time RT-PCR, ChIP, proliferation, and apoptosis assays.
Western blotting was performed as previously described (55). The primary antibodies and reagents are described in Supplemental Methods. cDNA synthesis and RT-PCR were performed as described previously (23). For quantitative RT-PCR, analyses were performed using iQ SYBR Green Supermix (Bio-Rad) on iCycler equipment (7500 PCR Systems, Applied Biosystems). The ChIP assay was performed as previously described (53). Primer sequences for PCR and ChIP assays are provided in Supplemental Methods. For proliferation assays, we treated cells with BrdU labeling reagent (BD Biosciences) for 1 hour as previously described (23). The number of apoptotic MCF-7 cells was measured 48 hours after transfection with pRc/RSV control vector, pRSV-hPit-1 expression vector, Pit-1 siRNA-1, or scrambled siRNA by labeling with PI and FITC-conjugated Annexin V (BD Biosciences) and performing TUNEL assay, followed by detection with flow cytometry (FACScan, BD Biosciences) (detailed in Supplemental Methods).
Soft agar, wound healing, and migration and invasion assays.
Soft agar assay was carried out with 5 × 104 MCF-7 Tet-Off cells and with MDA-MB-231 cells stably transfected with, on one hand, either the overexpression vector pTRE2-Pit-1 or the control vector pTRE2, and, on other hand, with the pSingle-tTS-shRNA-hPit-1 vector (detailed in Supplemental Methods). Colonies that grew beyond 50 μm in diameter were scored as positive. The wound healing assay is detailed in Supplemental Methods. The migration and invasion assays were performed in BD BioCoat Matrigel Invasion Chambers according to the manufacturer’s instructions (BD Biosciences), with uncoated porous filters (8-μm pore size) for estimation of cell migration and Matrigel-precoated filters to examine cell invasion (detailed in Supplemental Methods).
Animal studies.
All animal studies were approved by the University of Santiago de Compostela Ethics Committee for Animal Experiments. Female mice (age matched, between 6 to 8 weeks) homozygous for the severe combined immune deficiency spontaneous mutation (CB17-Prkdcscid, referred to as SCID, The Jackson Laboratory) were used for xenografting studies. Seven SCID mice were injected subcutaneously with 1 × 107 MCF-7 cells stably transfected either with the pTRE2 (control) vector in the left flank or with the pTRE-Pit-1–overexpressing vector in the right flank. Six mice were subcutaneously injected in the left and right flanks with 5 × 106 MDA-MB-231 cells stably transfected with the doxycycline-inducible pSingle-tTS-Pit-1 shRNA vector. Of these mice, 3 were fed with doxycycline in food pellets 15 days before injection and thereafter until sacrifice. Tumor growth was monitored externally using a vernier calliper every week, until 20 days after injecting MCF-7 cells or 23 days after injecting MDA-MB-231 cells, and tumor volume was calculated using the following formula: π/6 × length × width2. For experimental metastasis assays, we generated orthotopic primary tumors in 8-week-old SCID female mice by inoculation into the second mammary fat pad of 1 × 106 MCF-7 cells stably transfected with either the pTRE2 control vector (n = 11, controls) or the pTRE2-Pit-1–overexpressing vector (n = 12) in 0.15 ml of PBS and Matrigel (50:50, BD Biosciences). At day 21 after cell injection, orthotopic primary mammary tumors were removed under anesthesia (100 mg kg–1 ketamine, 10 mg kg–1 xylazine, intraperitoneally), and 26 days later (day 47 after cell injection), mice were sacrificed, and lungs were removed and examined for metastasis. Lung micrometastasis was explored in paraffin sections by H&E staining. For immunohistochemistry studies, mouse tumors and lungs were immersion fixed in 10% neutral buffered formalin for 24 hours and embedded in paraffin routinely. Sections (4-μm thick) were mounted on silanized slides (Dako). The immunohistochemical technique was automatically performed in an Autostainer Link 48 (Dako). FLEX Ready-to-Use Primary Antibodies to CKAE1/AE3, CK7, CK19, E-cadherin, vimentin, and Ki-67 (all from Dako) were used. A polyclonal antibody to Pit-1 (1:200; Santa Cruz Biotechnology Inc.), active caspase-3 (1:50; Abcam), and a monoclonal antibody to β-catenin (1:50; Dako) were also used. For detection we used EnVision FLEX/HRP (Dako). Quantitation of immunopositive cells for active caspase-3 expression was performed as previously described (56).
Statistics.
Each experiment was performed at least 3 times. Values are expressed as mean ± SD. Means were compared using 2-tailed Student’s t test or 1-way ANOVA, with the Tukey-Kramer multiple comparison test for post-hoc comparisons. For lung metastasis studies we used the Fisher’s exact test. Immunostaining score values for each protein were expressed as a median along with a range. Comparison of immunostaining values between groups was made with the Mann-Whitney or Kruskall-Wallis tests. For metastasis-free survival analysis, we used the Cox’s univariate method. Cox’s regression model was used to examine interactions of different prognostic factors in a multivariate analysis. The multivariate analysis only included parameters that achieved statistical significance for distant relapse-free survival in the univariate analysis as well as the type of systemic therapy. The SPSS 17.0 program (SPSS Inc.) was used for all calculations. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful to Anxo Vidal for helping us with flow cytometry analysis, Efi Arias for her invaluable help in surgical procedures, and Alberto Muñoz for critical reading of the manuscript. We also wish to thank Richard Bold and Veronique Bourgarel-Rey for their cooperation. This study was supported by a grant from the Ministerio de Ciencia e Innovación (SAF2009-07631).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(12):4289–4302. doi:10.1172/JCI42015.
References
- 1.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 6.Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc Natl Acad Sci U S A. 2006;103(50):18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCoy EL, et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest. 2009;119(9):2663–2677. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen B, Rosenfeld MG. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev. 2001;22(1):2–35. doi: 10.1210/er.22.1.2. [DOI] [PubMed] [Google Scholar]
- 11.Dolle P, Castrillo JL, Theill LE, Deerinck T, Ellisman M, Karin M. Expression of GHF-1 protein in mouse pituitaries correlates both temporally and spatially with the onset of growth hormone gene activity. Cell. 1990;60(5):809–820. doi: 10.1016/0092-8674(90)90095-V. [DOI] [PubMed] [Google Scholar]
- 12.Nelson C, Albert VR, Elsholtz HP, Lu LI, Rosenfeld MG. Activation of cell-specific expression of rat growth hormone and prolactin genes by a common transcription factor. Science. 1988;239(4846):1400–1405. doi: 10.1126/science.2831625. [DOI] [PubMed] [Google Scholar]
- 13.Mangalam HJ, et al. A pituitary POU domain protein, Pit-1, activates both growth hormone and prolactin promoters transcriptionally. Genes Dev. 1989;3(7):946–958. doi: 10.1101/gad.3.7.946. [DOI] [PubMed] [Google Scholar]
- 14.Chen RP, Ingraham HA, Treacy MN, Albert VR, Wilson L, Rosenfeld MG. Autoregulation of pit-1 gene expression mediated by two cis-active promoter elements. Nature. 1990;346(6284):583–586. doi: 10.1038/346583a0. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Crenshaw EB3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347(6293):528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 16.Bamberger AM, Bamberger CM, Pu LP, Puy LA, Loh YP, Asa SL. Expression of pit-1 messenger ribonucleic acid and protein in the human placenta. J Clin Endocrinol Metab. 1995;80(7):2021–2026. doi: 10.1210/jcem.80.7.7608249. [DOI] [PubMed] [Google Scholar]
- 17.Schanke JT, Conwell CM, Durning M, Fisher JM, Golos TG. Pit-1/growth hormone factor 1 splice variant expression in the rhesus monkey pituitary gland and the rhesus and human placenta. J Clin Endocrinol Metab. 1997;82(3):800–807. doi: 10.1210/jc.82.3.800. [DOI] [PubMed] [Google Scholar]
- 18.Delhase M, Vergani P, Malur A, Hooghe-Peters EL, Hooghe RJ. The transcription factor Pit-1/GHF-1 is expressed in hemopoietic and lymphoid tissues. Eur J Immunol. 1993;23(4):951–955. doi: 10.1002/eji.1830230428. [DOI] [PubMed] [Google Scholar]
- 19.Gil-Puig C, Blanco M, Garcia-Caballero T, Segura C, Perez-Fernandez R. Pit-1/GHF-1 and GH expression in the MCF-7 human breast adenocarcinoma cell line. J Endocrinol. 2002;173(1):161–167. doi: 10.1677/joe.0.1730161. [DOI] [PubMed] [Google Scholar]
- 20.Gil-Puig C, et al. Pit-1 is expressed in normal and tumorous human breast and regulates GH secretion and cell proliferation. Eur J Endocrinol. 2005;153(2):335–344. doi: 10.1530/eje.1.01962. [DOI] [PubMed] [Google Scholar]
- 21.Costoya JA, Garcia-Barros M, Gallego R, Senaris R, Arce VM, Devesa J. Correlation of Pit-1 gene expression and Pit-1 content with proliferation and differentiation in human myeloid leukemic cells. Exp Cell Res. 1998;245(1):132–136. doi: 10.1006/excr.1998.4232. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Fernandez R, Seoane S, Garcia-Caballero T, Segura C, Macia M. Vitamin D, Pit-1, GH, and PRL: possible roles in breast cancer development. Curr Med Chem. 2007;14(29):3051–3058. doi: 10.2174/092986707782793943. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Batalla I, et al. The Pit-1/Pou1f1 transcription factor regulates and correlates with prolactin expression in human breast cell lines and tumors. Endocr Relat Cancer. 2010;17(1):73–85. doi: 10.1677/ERC-09-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wennbo H, Tornell J. The role of prolactin and growth hormone in breast cancer. Oncogene. 2000;19(8):1072–1076. doi: 10.1038/sj.onc.1203349. [DOI] [PubMed] [Google Scholar]
- 25.Castrillo JL, Theill LE, Karin M. Function of the homeodomain protein GHF1 in pituitary cell proliferation. Science. 1991;253(5016):197–199. doi: 10.1126/science.1677216. [DOI] [PubMed] [Google Scholar]
- 26.Gaiddon C, de Tapia M, Loeffler JP. The tissue-specific transcription factor Pit-1/GHF-1 binds to the c-fos serum response element and activates c-fos transcription. Mol Endocrinol. 1999;13(5):742–751. doi: 10.1210/me.13.5.742. [DOI] [PubMed] [Google Scholar]
- 27.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 28.Batlle E, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 29.Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21(12):2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta GP, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446(7137):765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 31.Price JE. Metastasis from human breast cancer cell lines. Breast Cancer Res Treat. 1996;39(1):93–102. doi: 10.1007/BF01806081. [DOI] [PubMed] [Google Scholar]
- 32.Shafie SM, Liotta LA. Formation of metastasis by human breast carcinoma cells (MCF-7) in nude mice. Cancer Lett. 1980;11(2):81–87. doi: 10.1016/0304-3835(80)90097-X. [DOI] [PubMed] [Google Scholar]
- 33.Pellegrini I, et al. Involvement of the pituitary-specific transcription factor pit-1 in somatolactotrope cell growth and death: an approach using dominant-negative pit-1 mutants. Mol Endocrinol. 2006;20(12):3212–3227. doi: 10.1210/me.2006-0122. [DOI] [PubMed] [Google Scholar]
- 34.Asa SL, Puy LA, Lew AM, Sundmark VC, Elsholtz HP. Cell type-specific expression of the pituitary transcription activator pit-1 in the human pituitary and pituitary adenomas. J Clin Endocrinol Metab. 1993;77(5):1275–1280. doi: 10.1210/jc.77.5.1275. [DOI] [PubMed] [Google Scholar]
- 35.Sanno N, Teramoto A, Matsuno A, Osamura RY. Expression of human Pit-1 product in the human pituitary and pituitary adenomas. Immunohistochemical studies using an antibody against synthetic human Pit-1 product. Arch Pathol Lab Med. 1996;120(1):73–77. [PubMed] [Google Scholar]
- 36.Pellegrini I, et al. Pit-1 gene expression in the human pituitary and pituitary adenomas. J Clin Endocrinol Metab. 1994;79(1):189–196. doi: 10.1210/jc.79.1.189. [DOI] [PubMed] [Google Scholar]
- 37.Krajewski S, et al. Prognostic significance of apoptosis regulators in breast cancer. Endocr Relat Cancer. 1999;6(1):29–40. doi: 10.1677/erc.0.0060029. [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 39.Pardo A, Selman M. MMP-1: the elder of the family. Int J Biochem Cell Biol. 2005;37(2):283–288. doi: 10.1016/j.biocel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392(6672):190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 41.Blanco MJ, et al. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21(20):3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- 42.Moody SE, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Veronesi U, et al. A randomized comparison of sentinel-node biopsy with routine axillary dissection in breast cancer. N Engl J Med. 2003;349(6):546–553. doi: 10.1056/NEJMoa012782. [DOI] [PubMed] [Google Scholar]
- 44.van Deurzen CH, et al. Non-sentinel lymph node metastases associated with isolated breast cancer cells in the sentinel node. J Natl Cancer Inst. 2008;100(22):1574–1580. doi: 10.1093/jnci/djn343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abate-Shen C.Deregulated homeobox gene expression in cancer: cause or consequence ? Nat Rev Cancer. 2002210777–785. 10.1038/nrc907 [DOI] [PubMed] [Google Scholar]
- 46.Briegel KJ. Embryonic transcription factors in human breast cancer. IUBMB Life. 2006;58(3):123–132. doi: 10.1080/15216540600686870. [DOI] [PubMed] [Google Scholar]
- 47.Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE. Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer. 1999;81(1):104–112. doi: 10.1002/(SICI)1097-0215(19990331)81:1<104::AID-IJC18>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 48.Budhram-Mahadeo V, Ndisang D, Ward T, Weber BL, Latchman DS. The Brn-3b POU family transcription factor represses expression of the BRCA-1 anti-oncogene in breast cancer cells. Oncogene. 1999;18(48):6684–6691. doi: 10.1038/sj.onc.1203072. [DOI] [PubMed] [Google Scholar]
- 49.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7(3):86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawson JC, Blatch GL, Edkins AL. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res Treat. 2009;118(2):241–254. doi: 10.1007/s10549-009-0524-9. [DOI] [PubMed] [Google Scholar]
- 52.Peixoto P, et al. Direct inhibition of the DNA-binding activity of POU transcription factors Pit-1 and Brn-3 by selective binding of a phenyl-furan-benzimidazole dication. Nucleic Acids Res. 2008;36(10):3341–3353. doi: 10.1093/nar/gkn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seoane S, Perez-Fernandez R. The vitamin D receptor represses transcription of the pituitary transcription factor Pit-1 gene without involvement of the retinoid X receptor. Mol Endocrinol. 2006;20(4):735–748. doi: 10.1210/me.2005-0253. [DOI] [PubMed] [Google Scholar]
- 54.Palmer HG, et al. The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat Med. 2004;10(9):917–919. doi: 10.1038/nm1095. [DOI] [PubMed] [Google Scholar]
- 55.Seoane S, Ben I, Centeno V, Perez-Fernandez R. Cellular expression levels of the vitamin D receptor are critical to its transcriptional regulation by the pituitary transcription factor Pit-1. Mol Endocrinol. 2007;21(7):1513–1525. doi: 10.1210/me.2006-0554. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez B, Relova JL, Gallego R, Ben-Batalla I, Perez-Fernandez R. 1, 25-dihydroxyvitamin D3 administration to 6-hydroxy dopamine-lesioned rats increases GDNF and partially restores tyrosine hydroxylase expression in substantia nigra and striatum. J Neurosci Res. 2009;87(3):723–732. doi: 10.1002/jnr.21878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.