Abstract
The human E3 ubiquitin ligase murine double minute 2 (MDM2) targets the tumor suppressor p53 for ubiquitination and degradation but also promotes its own ubiquitination and subsequent degradation. As the balance between MDM2 and p53 levels plays a crucial role in regulating cell proliferation and apoptosis, we sought to identify factors selectively inhibiting MDM2 self-ubiquitination. Here we have shown that the LIM domain protein Enigma directly interacts with MDM2 to form a ternary complex with p53 in vitro and in human hepatoma and colon carcinoma cell lines and mouse embryonic fibroblasts. We found that Enigma elicited p53 degradation by inhibiting MDM2 self-ubiquitination and increasing its ubiquitin ligase activity toward p53 in cells. Moreover, mitogenic stimuli such as serum, FGF, and HGF increased Enigma transcription via induction of serum response factor (SRF), leading to MDM2 stabilization and subsequent p53 degradation. We observed similar results in the livers of mice treated with HGF. In humans, we found SRF and Enigma coexpressed with MDM2 but not p53 in several liver and stomach tumors. Finally, we showed that Enigma promoted cell survival and chemoresistance by suppressing p53-mediated apoptosis in both cell lines and a mouse xenograft model. Our findings suggest a role for Enigma in tumorigenesis and uncover a mechanism whereby mitogens attenuate p53 antiproliferative activity through an SRF/Enigma/MDM2 pathway.
Introduction
The p53 tumor suppressor is a transcription factor that activates many genes involved in cell growth arrest and apoptosis and thereby prevents incidence of abnormal cells and tumorigenesis (1). Tight regulation of the antiproliferative activities of p53 is essential for cell viability. This is mostly achieved by the mouse/human double minute 2 (Mdm2/Hdm2) E3 ubiquitin ligase, which targets p53 for degradation (2–4). MDM2 also targets itself for ubiquitination and degradation (5, 6). Since MDM2 is a transcriptional target of p53, these two molecules form an autoregulatory loop (7, 8). Recently, the p300/CBP-associated factor (PCAF) and/or unidentified E3 ligases have been shown to promote ubiquitin-mediated proteolysis of MDM2 (9, 10).
Factors that differentially regulate ubiquitination of MDM2 and p53 may affect cell fate profoundly, as they can change the ratio between MDM2 and p53 (11). Many proteins have been discovered that regulate MDM2 and p53 levels by multiple mechanisms (12–14). However, cellular factors that selectively inhibit self-ubiquitination of MDM2 and thereby promote ubiquitination and degradation of p53 have been elusive. These factors likely play a positive role in cell growth and tumorigenesis. Indeed, MDM2 is overexpressed in various human tumors and correlates with increased cell proliferation and tumor growth (15).
The LIM domain protein Enigma, PDLIM7, structurally contains one amino-terminal PDZ domain and three carboxyterminal LIM domains (16). The PDZ domain binds actin-binding proteins such as β-tropomyosin (17). The Enigma LIM domains interact with proteins involved in mitogenic or insulin signaling such as protein kinases (18–21). Enigma enhances bone morphogenetic responsiveness by interacting with Smad ubiquitin-regulatory factor 1 (22). Recently, pdlim7 in zebrafish has been shown to be involved in heart and skeletal muscle development (23). However, the exact cellular functions of Enigma remain enigmatic.
The Enigma gene is one of many candidates that have been identified from human liver and stomach tumors through the Functional Genome Project in Korea. We found that overexpression of Flag-tagged Enigma (F-Enigma) significantly reduced the expression of luciferase (luc) reporter gene under the control of an artificial promoter containing multiple p53 response elements (REs). We hypothesized, therefore, that Enigma might regulate the MDM2/p53 pathway.
Results
Enigma regulates p53 and p21 protein levels through MDM2.
To identify association of Enigma with the MDM2/p53 pathway, we examined MDM2 and p53 protein levels in HLK3 hepatoma cells with or without expression of F-Enigma in the presence or absence of MG132, a proteasome inhibitor (Figure 1A). We also examined protein levels of p21, a transcriptional target of p53, which induces cell growth arrest (24). The MDM2 level was increased without F-Enigma expression in the presence of MG132, confirming previous reports that MDM2 is susceptible to proteasomal degradation (5, 6). In the absence of MG132, F-Enigma expression resulted in a dose-dependent increase in MDM2 levels, with concomitant decrease in p53 and p21 levels (Figure 1A). To test whether alterations in p53 level are reflected in its functionality, we assessed luciferase activity of p53-responsive reporter constructs, such as p53RE-luc, p21-luc, and pBax-luc, in which the reporter gene expression is driven by p21 or Bax gene promoters (25). p53 was shown to induce expression of Bax that activates apoptosis (26). F-Enigma expression decreased reporter activity of all the p53-responsive constructs in a dose-dependent manner in the absence of MG132 (Figure 1B). These effects of Enigma on p53-responsive reporter activity were observed in p53+/+ cells but not p53–/– cells (Figure 1B), suggesting that the effects of Enigma are caused by p53.
Figure 1. Enigma regulates p53 and p21 protein levels through MDM2.
(A) Enigma stabilizes MDM2. HLK3 cells were transfected with F-Enigma vector (0, 2, 5 μg [designated –, +, ++]), incubated for 12 hours in the presence or absence of 10 μM MG132, and immunoblotted as indicated. (B) p53 levels are reflected in p53 activity. We transfected cells with the indicated reporter constructs (0.5 μg each) and with or without F-Enigma vector, and incubated them with or without MG132 for 12 hours. A luciferase assay was performed. Rel. luc. unit, relative luciferase units. (C) The effects of Enigma on mRNA levels of MDM2, p53, and p21. We transfected HLK3 cells with or without F-Enigma vector, prepared cell lysates or extracted total RNAs after 48 hours, and performed Northern blotting (NB) or IB as indicated. We stained 28S RNA with ethidium bromide as a control to indicate equivalent loading. (D) Enigma regulates p53 levels Mdm2-dependently. We transduced MEFs with Ad-p53 at an MOI of 100. After 16 hours, we transduced them with Ad–F-Enigma or Ad-LacZ at an MOI of 50, or Ad-siEnigma or Ad-siControl at an MOI of 100, and incubated them for 32 hours before IB as indicated. (E) Enigma regulates p53 and p21 levels MDM2-dependently. We transfected cells with F-Enigma (5 μg), F-EniΔLIM3 (5 μg), siEnigma (10 μg), or siControl (10 μg) vectors, and incubated them for 48 hours before IB as indicated.
We tested whether Enigma-mediated alterations in MDM2, p53, and p21 levels occurred at a transcriptional level (Figure 1C). The MDM2 gene has two promoters, one (P1) that is constitutively active and p53 independent and one (P2) that is activated by p53 under most conditions of stress (7, 8). Overexpression of F-Enigma did not greatly affect mRNA levels of p53 and P1-MDM2, but decreased those of p21 and P2-MDM2, most likely due to a decrease in p53 protein level caused by F-Enigma expression (Figure 1, A and C).
To test whether the effects of Enigma on p53 and p21 levels are Mdm2-dependent, we expressed p53 in mouse embryonic fibroblasts (MEFs) lacking either Mdm2 and p53 genes or p53 gene with adenoviral vector encoding p53 (Ad-p53) (Figure 1D). We validated the integrity and target-specificity of siRNA targeting Enigma (siEnigma) (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI42674DS1). p53 and p21 levels were decreased by F-Enigma overexpression and were increased by depletion of Enigma in Mdm2+/+ MEFs (Figure 1D). These effects of Enigma on p53 and p21 levels were not detected in Mdm2–/– MEFs (Figure 1D).
p53 induces MDM2 expression at the mRNA level (7, 8). p21 expression was shown to be regulated by a p53-independent mechanism (27). We therefore tested whether Enigma-mediated changes in MDM2 and p21 levels occurred p53-dependently (Figure 1E). We used an F-Enigma mutant without the LIM3 domain (F-EniΔLIM3) as a control, as the LIM3 domain was essential for binding of Enigma to MDM2 in vitro and in vivo (Figure 2, C, E, and F). The MDM2 level was increased by F-Enigma overexpression and was decreased by Enigma depletion in both p53+/+ and p53–/– cells (Figure 1E). The p21 level was decreased by F-Enigma overexpression and was increased by depletion of Enigma in p53+/+ but not p53–/– cells (Figure 1E).
Figure 2. Enigma interacts with p53 through MDM2 in vitro and in vivo.
(A) We immunoprecipitated and/or immunoblotted HCT116p53+/+ or p53–/– cells. (B) We transduced Mdm2–/–p53–/– (Mdm2–/–) and Mdm2+/+p53–/– (Mdm2+/+) MEFs with Ad-p53 at an MOI of 100 and incubated them for 24 hours before IP and/or IB. (C) We mixed His-MDM2 (1 μg), Enigma (0.5 μg), EniΔLIM3 (0.5 μg), GST-p53 (1 μg), and/or GST (1 μg) proteins for IP or GST pull-down and IB. (D and E) Various GST-MDM2 fusion (D) or F-Enigma (E) vectors are illustrated in the top panel. (D) We transfected 293–F-Enigma cells with the indicated vectors (5 μg each), and performed GST pull-down/IB and IB at 24 hours after transfection. MDM2 domains necessary for the interaction with Enigma and for binding to p53 are indicated at the top. The RING domain of MDM2 is shown in gray. (E) We transfected 293T cells with or without various F-Enigma (5 μg each) and with GST-MDM2 (5 μg each) vectors and incubated them for 24 hours before IB and IP/IB. The PDZ and 3 LIM domains of Enigma and the Enigma domain required for binding to MDM2 are shown at the top. The results from the binding assay are summarized on the right of the top panels in D and E. (F) We transfected HLK3 cells with the indicated vectors (5 μg each), which are illustrated in E, and incubated them for 48 hours before GST pull-down/IB and IB. (G) Schematic illustration of the Enigma-MDM2-p53 ternary complex.
Overexpression of Enigma increased the half-life of MDM2 and decreased that of p53, whereas depletion of Enigma induced the opposite effect (Supplemental Figure 2). Collectively, these results suggest that Enigma stabilizes MDM2 p53-independently and regulates the p53/p21 pathway negatively through MDM2 at a posttranscriptional level.
Enigma interacts with p53 through MDM2 in vitro and in vivo.
To identify how Enigma regulates the p53 pathway MDM2-dependently, we first tested whether Enigma interacted with MDM2 and/or p53. Endogenous Enigma was coimmunoprecipitated with endogenous MDM2 in the presence or absence of endogenous p53 and formed a ternary complex with endogenous MDM2 and p53 proteins (Figure 2A). To test whether Enigma directly interacts with p53, we expressed p53 in p53–/– MEFs with or without the Mdm2 gene and analyzed the interactions between them (Figure 2B). Enigma interacted with Mdm2 and formed a complex with p53 in Mdm2+/+ but not Mdm2–/– MEFs, suggesting that the interaction between Enigma and p53 is Mdm2-dependent. To substantiate these results in vitro, we analyzed the interactions among Enigma, MDM2, and/or p53 proteins purified from bacteria (Figure 2C). MDM2 interacted with p53, indicative of functionality of MDM2 and p53 proteins from bacteria. Enigma interacted with MDM2, but not with p53, and bound to p53 when MDM2 was present (Figure 2C). These results suggest that Enigma interacts with p53 through MDM2 in vitro and in vivo.
To identify the molecular basis for MDM2-mediated interaction of Enigma with p53, we delineated domains for binding between MDM2 and Enigma. The C terminus of MDM2 (amino acids 401–491) including the RING finger domain was sufficient for its binding to Enigma (Figure 2D). The LIM3 domain of Enigma, but not the PDZ, LIM1, or LIM2 domain, was found to bind to MDM2 (Figure 2, C, E, and F). The interaction between MDM2 and p53 was shown to occur through their N termini (28). Thus, these findings suggest that Enigma, MDM2, and p53 proteins form a ternary complex through the interaction between the C termini of Enigma and MDM2 and through the contact between the N termini of MDM2 and p53 (Figure 2G).
Enigma homolog (ENH) has a single PDZ domain and 3 LIM domains and is closely related to Enigma (16). Moreover, the LIM domains of these two proteins were shown to interact with identical protein kinases (20, 21). Therefore, we tested whether ENH could act like Enigma. ENH neither interacted with MDM2 nor stabilized it (Supplemental Figure 3), suggesting that Enigma specifically interacts with MDM2.
Enigma inhibits MDM2 self-ubiquitination and enhances MDM2-mediated ubiquitination of p53.
Enigma-mediated regulation of MDM2 and p53 levels and formation of the Enigma-MDM2-p53 ternary complex led us to test whether Enigma affected self- and p53 ubiquitination of MDM2. Overexpression of F-Enigma but not F-EniΔLIM3 reduced MDM2-Ub and increased p53-Ub conjugates in a dose-dependent manner in cells, while depletion of Enigma increased MDM2-Ub and decreased p53-Ub conjugates (Figure 3A). Since MDM2 levels were not greatly affected in the presence of MG132 (Figure 1A), the finding that Enigma dose-dependently increased p53-Ub conjugates suggests that Enigma may activate MDM2-dependent ubiquitination of p53 in cells.
Figure 3. Enigma inhibits MDM2 self-ubiquitination and enhances MDM2-mediated ubiquitination of p53.
(A) We transfected HLK3 cells with His-Ub (5 μg), F-Enigma (0, 5, 10 μg), F-EniΔLIM3 (10 μg), siEnigma (10, 15 μg), or siControl (15 μg) vectors and treated them with 10 μM MG132 for 12 hours before analysis as indicated. (B) We performed an in vitro ubiquitination assay with GST-MDM2 (2 μg), GST-MDM2(C464A) (2 μg), Enigma (0.5, 1 μg), His-PCAF (1 μg), GST (2 μg). After assay, we analyzed the reaction mixtures as indicated. F-Ub, Flag-tagged Ub. (C) We transfected MEFs with or without F-PCAF (5 μg), HA-Enigma (2, 5 μg), GST-MDM2 (10 μg), GST-MDM2(C464A) (10 μg), or GST (10 μg) vectors and prepared cell lysates at 48 hours after transfection before IB as indicated. (D) We performed an in vitro ubiquitination assay with His-MDM2 (2 μg), Enigma (0.5, 1 μg), EniΔLIM3 (1 μg), GST-p53 (1 μg), or GST (1 μg) and analyzed the reaction mixtures as indicated. Protein components in B and D were purified from bacteria. Molecular size markers are indicated at the left in A, B, and D.
Next, we examined the effects of Enigma on self-ubiquitination of MDM2 and on PCAF-mediated ubiquitination of MDM2 in vitro using protein components and in cells. Virtually all the MDM2 molecules appeared to bind to Enigma in MDM2 self-ubiquitination assay conditions (Supplemental Figure 4). In line with this, Enigma, but not EniΔLIM3, efficiently inhibited self-ubiquitination of MDM2 in vitro (Figure 3B and Supplemental Figure 5). Enigma bound to MDM2(C464A), a catalytically inactive mutant of Hdm2, but had no effect on it (Figure 3B). PCAF ubiquitinated MDM2(C464A) in vitro (Figure 3B), and PCAF expression decreased the protein level of MDM2(C464A) in Mdm2–/– MEFs (Figure 3C), as previously reported (10). However, Enigma did not greatly affect the PCAF-mediated ubiquitination and degradation of MDM2(C464A) in vitro and in cells.
We next tested the effect of Enigma on MDM2-mediated ubiquitination of p53 in vitro (Figure 3D). Enigma bound to MDM2 and p53, stimulated MDM2-mediated ubiquitination of p53 in a dose-dependent manner, and inhibited self-ubiquitination of MDM2. This effect of Enigma on p53 ubiquitination was not detected with MDM2(C464A) (Figure 3D). Collectively, these results suggest that Enigma specifically inhibits self-ubiquitination of MDM2 and increases the stability and activity of MDM2 E3 ligase, thereby enhancing MDM2-dependent ubiquitination and degradation of p53 in cells.
Proliferation signal attenuates p53 via the SRF/Enigma/MDM2 pathway.
Activation of cell proliferation or survival pathways has been shown to increase MDM2 levels by various mechanisms (29). We identified serum response element (SRE) in the upstream regulatory region of the Enigma gene (Supplemental Figure 6). Many immediate early genes responsive to growth factors contain SRE in their promoters to which serum response factor (SRF) binds (30). Therefore, we hypothesized that increased MDM2 levels in response to cell proliferation signals might be attributed to SRF-mediated induction of Enigma expression. Indeed, we found that serum, FGF, or HGF activates the Enigma promoter through the SRE to which SRF binds (Supplemental Figure 6). This SRF requirement for Enigma expression is consistent with the previous finding that Enigma is downregulated in the embryonic heart of Srf-knockout mice (31).
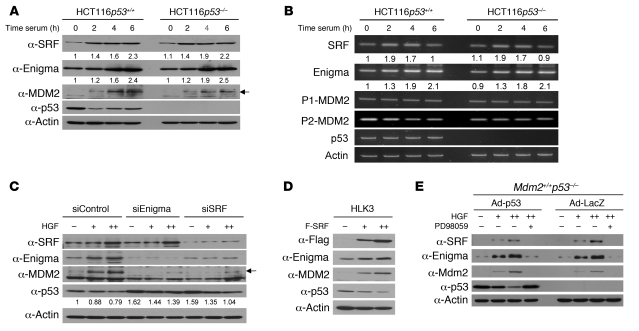
To test whether cell proliferation signals negatively regulate p53 through the SRF/Enigma/MDM2 pathway, we stimulated serum-starved cells with serum and examined time-dependent alterations in mRNA and protein levels of those molecules. Serum induced Enigma expression at the mRNA and protein levels following early induction of SRF expression in the presence or absence of p53 (Figure 4, A and B). Serum did not greatly affect the mRNA level of P1-MDM2, but decreased that of P2-MDM2 (Figure 4B), presumably due to the decreased protein level of p53 after serum stimulation (Figure 4A). The protein level of p53 was gradually and slightly increased after an initial decrease at 2 hours after serum stimulation, while the MDM2 level was steadily increased by 6 hours (Figure 4A). This may have been the result of molecular events caused by proliferation signaling, which negatively regulates MDM2-mediated degradation of p53 (32). However, serum stimulation increased protein levels of MDM2, which led to an overall decrease in p53 levels (Figure 4A). Serum withdrawal from proliferating cells resulted in decreasing mRNA and protein levels of SRF and Enigma in a p53-independent manner, and concomitantly resulted in a decreasing MDM2 protein level and increasing p53 protein level (Supplemental Figure 7). These alterations in MDM2 and p53 protein levels in serum-starved cells occurred at the posttranscriptional level, as serum removal increased the P2-MDM2 mRNA level in p53+/+ cells and did not significantly affect mRNA levels of P1-MDM2 and p53 (Supplemental Figure 7B).
Figure 4. Proliferation signal attenuates p53 by the SRF/Enigma/MDM2 pathway in cells.
(A and B) We incubated serum-starved cells in DMEM with 10% FBS (serum) for the indicated times, prepared protein lysates or total RNAs, and analyzed them by IB (A) or RT-PCR (B) as indicated. SRF and Enigma levels from 3 independent experiments were quantified by densitometry. Mean values are expressed relative to the band intensity of the first lane in Figure 4. A and B, respectively, which was arbitrarily defined as 1. (C) We transfected HLK3 cells with indicated siRNA vectors (10 μg each), incubated them in DMEM without serum for 48 hours, and incubated them in DMEM with HGF (0, 40, 60 ng/ml) for 4 hours before IB as indicated. p53 levels were quantified by densitometry, and mean values are expressed relative to the band intensity of the first lane in Figure 4C, which was arbitrarily defined as 1. Arrows in A and C indicate MDM2 band. (D) We transfected cells with F-SRF vector (0, 5, 10 μg) and incubated them for 48 hours before IB as indicated. (E) We transduced MEFs with Ad-p53 or Ad-LacZ at an MOI of 100, incubated them in DMEM without serum for 48 hours, and further incubated them in DMEM with HGF (0, 40, 60 ng/ml) in the presence or absence of 25 μM PD98059 for 4 hours before IB as indicated.
Treatment of serum-starved cells with HGF also increased protein levels of SRF, Enigma, and MDM2 in a dose-dependent manner, resulting in a decrease in p53 levels (Figure 4C). When SRF or Enigma was depleted, the effects of HGF on MDM2 and p53 levels were attenuated (Figure 4C). Forced expression of SRF increased protein levels of Enigma and MDM2 in a dose-dependent manner and decreased p53 protein level (Figure 4D), suggesting that SRF is directly involved in transcriptional activation of the Enigma gene. Growth factor–mediated activation of the MAP kinase cascade induces or activates expression of SRF (30, 33). We therefore used a specific MEK inhibitor, PD98059, to examine whether the MAP kinase pathway might regulate p53 through the SRF/Enigma pathway. When Mdm2+/+p53–/– MEFs expressing p53 were treated with PD98059, the effects of HGF on SRF, Enigma, Mdm2, and p53 levels were abrogated (Figure 4E). Collectively, these results suggest that cell proliferation signals induce SRF expression through activation of the MAP kinase cascade and subsequently increase Enigma expression, resulting in stabilization of MDM2 that enhances ubiquitin-mediated degradation of p53.
HGF infusion has been shown to induce liver growth in normal mice (34). We identified that the SRE is conserved in the Enigma promoters of human and mouse and then examined whether HGF might regulate p53 levels through the SRF/Enigma/Mdm2 pathway in vivo. To detect HGF-mediated alterations in Mdm2 and p53 levels more clearly, we initially injected mice with or without Ad-p53, treated them with HGF, and examined time-dependent alterations in SRF, Enigma, Mdm2, and p53 levels in mouse liver. HGF increased mRNA and protein levels of Enigma following induction of SRF expression with or without Ad-p53 injection (Figure 5, A and B). The protein level of Enigma was positively correlated with that of Mdm2 between 3 and 16 hours, at which time p53 protein levels were decreased (Figure 5A). HGF treatment decreased exogenous or endogenous p53 protein levels at 2 hours, at which time Enigma but not Mdm2 protein was detected (Figure 5A). This may result from the fact that the Mdm2 level at 2 hours might have been too low to be detected. Alternatively, there are unidentified mechanisms by which HGF may decrease p53 levels together with the Enigma/Mdm2 axis in mouse liver (35). To further verify that Enigma is directly involved in the HGF-mediated regulation of the Mdm2/p53 pathway in vivo, we pretreated mice with or without Ad-p53 and then injected one group of mice with Ad–F-Enigma and another group with Ad-siEnigma. Mice with Ad–F-Enigma were left untreated. Mice with Ad-siEnigma were treated with HGF. We then examined protein levels of Enigma, Mdm2, and p53 in mouse liver (Figure 5C). HGF increased endogenous Enigma and Mdm2 protein levels and decreased exogenous and endogenous p53 levels (Figure 5C). These effects of Enigma on Mdm2 and p53 levels were abolished when endogenous Enigma was depleted by Ad-siEnigma. Overexpression of F-Enigma increased endogenous Mdm2 protein levels and decreased exogenous and endogenous p53 protein levels (Figure 5C). Thus, these results suggest that Enigma is directly involved in the HGF-mediated decrease in p53 levels and support the in vivo presence of an SRF/Enigma/Mdm2 pathway by which mitogenic signals negatively regulate p53.
Figure 5. Proliferation signal attenuates p53 by the SRF/Enigma/Mdm2 pathway in mouse liver.
(A and B) We injected mice with or without 5 × 108 PFU Ad-p53. After 24 hours, we injected mice with HGF (100 μg/kg), excised livers at the indicated times, and prepared protein extracts and total RNAs for IB (A) and RT-PCR (B), respectively, as indicated. Similar results were obtained from 3 independent experiments. (C) We injected mice with or without 5 × 108 PFU of Ad-p53. After 24 hours, we injected mice with or without 5 × 108 PFU Ad–F-Enigma or Ad-LacZ, or 109 PFU Ad-siEnigma or Ad-siControl. After 40 hours, we administered HGF (100 μg/kg) to mice received Ad-siEnigma or Ad-siControl. We excised livers from all the mice (n = 3 per group) at 8 hours after HGF administration and prepared protein extracts for IB as indicated. We injected mice with 0.1 ml PBS with or without adenoviruses or HGF via tail vein. Arrows in A and C indicate pertinent bands of indicated proteins.
SRF and Enigma are coexpressed with MDM2 in human tumors.
SRF is highly expressed in advanced human liver cancer and correlates with tumor progression (36) and increases expression of E2-EPF UCP, which targets the VHL tumor suppressor for degradation and promotes tumor growth (33, 37). It was shown that MDM2 is highly expressed in tumors with wild-type p53 (38). Thus, activation of the SRF/Enigma axis in proliferating cells (Figures 4 and 5) suggests that SRF and Enigma are likely coexpressed with MDM2 but not p53 in certain human tumors, presumably because MDM2 targets wild-type p53 for degradation. It was also shown that MDM2 levels are high in some human tumors where p53 levels are elevated (39–42). Moreover, the p53 gene is mutated in more than 50% of human tumors (38), and the mutated p53 proteins are unable to induce target genes through the p53RE and are often accumulated in tumor cells (43, 44). These previous findings suggest that the SRF/Enigma axis may be partly responsible for coexpression of MDM2 and p53 proteins in some tumors, where p53 proteins lose their ability to induce target genes or MDM2 interferes with transcriptional activation of p53 (45). We thus preselected SRF- or Enigma-positive human tumors from 27 liver and 18 stomach tumors of advanced stage by primary screening, and we examined SRF/Enigma/MDM2/p53 levels in those tumors and their normal counterparts (Figure 6A). Indeed, SRF and Enigma were coexpressed with MDM2 in 10 cases of human tumors, where p53 was undetected, and were also coexpressed with MDM2 and p53 proteins in 7 cases of human tumors. Thus, these results suggest that the SRF/Enigma axis may stabilize MDM2 in human tumors, thereby ablating wild-type p53.
Figure 6. SRF and Enigma are coexpressed with MDM2 in human tumors.
(A) Immunoblot analysis of human tumors. Protein extracts from tumor and nontumor (N) tissues adjacent to or apart from tumors (T) were immunoblotted as indicated. Arrows indicate pertinent bands of indicated proteins. T* indicates tumors expressing SRF, Enigma, and MDM2 but not p53. T** indicates tumors expressing SRF, Enigma, MDM2, and p53. (B) Immunofluorescence staining of tumor tissues. We stained human stomach and colon tumors on tissue arrays by H&E or analyzed them by immunofluorescence using relevant antibodies. The same tissue regions coexpressing Enigma and MDM2 proteins in tumor cells are shown. Original magnification, ×100. (C) HLK3 cells were incubated in DMEM without serum for 48 hours and then incubated with DMEM with 0% or 10% serum for 2 hours. Cells were stained as indicated. Original magnification, ×200.
Tissue array experiments revealed that Enigma was colocalized with MDM2 at apical cytoplasm of human stomach and colorectal tumor cells (Figure 6B). Immunostaining experiments also revealed that Enigma and MDM2 were colocalized mainly in the cytosol of the cells treated with 10% serum (Figure 6C). Coexistence of Enigma and MDM2 was also detected in the cytosol of the serum-starved cells (0% serum). Enigma appeared to exist in the nucleus in proliferating cells at low levels (10% serum). Since Enigma and MDM2 are colocalized in the cytosol in human tumor cells and a tumor cell line, we examined whether Enigma, MDM2, and p53 proteins were present as a ternary complex in the cytosol. To that end, we prepared cytosolic and nuclear fractions from cells incubated with or without 10% serum. Protein levels of Enigma, MDM2, and p53 and the interaction between them were examined in those fractions (Supplemental Figure 8). After serum stimulation, protein levels of Enigma, MDM2, and p53 were increased in the cytosol compared with those in the nucleus. Enigma was detected in the nuclear fraction from cells with 10% serum, albeit at a low level, which is consistent with the immunostaining data (Figure 6C). The Enigma-MDM2-p53 ternary complex was detected in both the cytosolic and nuclear fractions from serum-starved and serum-stimulated cells (Supplemental Figure 8). Thus, these results suggest that the Enigma/MDM2 axis may function to attenuate p53 activity mainly in the cytosol in proliferating cells and may act also in the nucleus to suppress p53 functionality.
Enigma promotes tumor cell viability in vitro and in vivo.
The presence of the Enigma/MDM2 axis in proliferating cells and human tumors led us to test whether Enigma might indeed affect tumor cell viability. To that end, we treated HLK3 tumor cells with or without adriamycin (ADR). In the absence of ADR, overexpression of F-Enigma appeared not to significantly promote cell growth, but depletion of Enigma resulted in decreased cell viability (Figure 7A). The results suggest that MDM2 may act as a survival factor in proliferating cells, as previously reported (46). The ADR-mediated decrease in cell viability was rescued by F-Enigma overexpression and became severe with Enigma depletion. F-Enigma overexpression increased the MDM2 level, decreased the p53 level, and promoted cell cycle progression in the presence or absence of ADR, while Enigma depletion decreased the MDM2 level and increased the p53 level and population of apoptotic cells (Figure 7, B and C). To substantiate this effect of Enigma on cell viability in vivo, we generated subcutaneous tumors in nude mice and examined the effect of Enigma on tumor growth after ADR injection (Figure 7, D and E). ADR significantly inhibited tumor growth in mice (Figure 7D, lines 1 and 2). This ADR-mediated inhibition of tumor growth was weakened when F-Enigma was overexpressed (line 3) and became stronger with Enigma depletion (line 5). Protein levels of MDM2 and p53 in tumors excised from mice (Figure 7F) suggest that the in vivo tumor growth rate may be positively correlated with MDM2 level and be inversely linked with p53 level.
Figure 7. Enigma promotes tumor cell viability in vitro and in vivo.
(A and B) We transfected HLK3 cells with pIRES2-eGFP (2 μg) and with or without F-Enigma (5 μg), F-EniΔLIM3 (5 μg), siEnigma (10 μg), or siControl (10 μg) vector. We assayed cell viability after transfection as described in Methods (A). At 36 hours after transfection, cells were incubated with or without 2 μg/ml ADR for 12 hours and immunoblotted as indicated (B). (C) We transfected HLK3 cells with the indicated vectors except pIRES2-eGFP as described in A and B. Cells were grown for 28 hours and then treated with or without ADR (20 μg/ml) for 20 hours. Cells were stained with both PI and FITC-conjugated Annexin V and immediately analyzed by flow cytometry. (D and E) We transduced HLK3 cells with adenoviral vectors and generated tumors in nude mice (n = 5 per group) by subcutaneous injection of the cells as described in Methods. After cell inoculation, we injected mice once with or without 4 mg/kg ADR through the tail vein, monitored tumor growth (D), and photographed tumors excised from mice (E). One mouse died in the group without ADR treatment during the experiment. (F) We generated tumors in mice (n = 2 per group) by subcutaneous inoculation of 5 × 106 HLK3 cells with or without adenoviral vectors as described in Methods, and treated mice with 4 mg/kg ADR at day 7 after cell inoculation and excised tumors from mice at day 3 after ADR injection for IB.
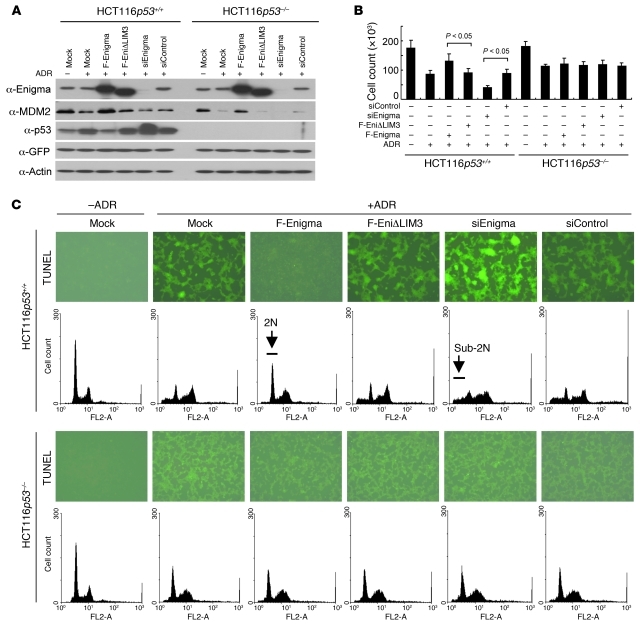
Next, we tested whether Enigma regulated tumor cell viability p53-dependently. To this end, we used HCT116 isogenic cell lines from human colon carcinoma, in which only p53 status is different. ADR decreased MDM2 level in both p53+/+ and p53–/– cells (Figure 8A), suggesting that damage-activated kinases may accelerate MDM2 autodegradation, as previously reported (47). This ADR-mediated decrease in MDM2 levels was rescued by F-Enigma overexpression and was further intensified by Enigma depletion in p53+/+ and p53–/– cells (Figure 8A). Viability of p53+/+ cells was increased by F-Enigma overexpression and was decreased by Enigma depletion (Figure 8B). This effect of Enigma on cell viability was not observed in p53–/– cells (Figure 8B). Depletion of Enigma increased the p53 level and population of apoptotic cells in p53+/+ but not p53–/– cells (Figure 8, A and C), suggesting that Enigma enhances tumor cell viability by suppressing p53-mediated apoptosis.
Figure 8. Enigma suppresses p53-mediated apoptosis.
(A) We transfected cells with pIRES2-eGFP (2 μg) and with or without F-Enigma (5 μg), F-EniΔLIM3 (5 μg), siEnigma (10 μg), or siControl (10 μg) vector. After 16 hours, culture media were changed, and cells were further grown for 20 hours and then treated with or without 2 μg/ml ADR for 12 hours. Cell lysates were immunoblotted as indicated. (B and C) We transfected cells with indicated vectors as described in A. We assayed cell viability as described in Methods (B). (C) At 16 hours after transfection, culture media were changed, and cells were further grown for 8 hours and then treated with or without 20 μg/ml ADR. After 24 hours, we performed TUNEL assay (original magnification, ×100) or analyzed cells by flow cytometry.
Discussion
Here we demonstrated that Enigma specifically inhibits self-ubiquitination of MDM2 and increases the stability and activity of MDM2 E3 ligase, thereby enhancing MDM2-mediated ubiquitination and degradation of p53 in cells. Moreover, Enigma promotes cell survival and chemoresistance through its effects on the MDM2/p53 pathway. In addition, our findings may provide a molecular basis for MDM2 overexpression in human tumors through the SRF/Enigma axis.
A number of factors have been shown to regulate MDM2 positively and p53 negatively (12–14). However, Enigma is unique and distinct among those factors in its abilities to form a complex with p53 through MDM2 and enhance the stability and activity of MDM2 E3 ligase. Mdm2-null mice are embryonic lethal but are rescued by p53 elimination (48, 49), indicating that Mdm2 is the central node in the p53 pathway. In this context, Enigma may act as an integral part of the MDM2/p53 pathway in proliferating cells through its ability to switch the E3 ligase specificity of MDM2 from itself to p53.
How does Enigma differentially regulate self- and p53 ubiquitination of MDM2? The MDM2 RING domain has been shown to be essential for oligomerization of MDM2 (50–52). MDM2 E3 ligase likely recognizes itself as a substrate via homo(di)merization to self-ubiquitinate (53). The binding of MDM2 to the N terminus of p53 has been proposed to cause a conformational change in MDM2 that enables binding of the central acidic region of MDM2 to a “ubiquitination signal” in the core DNA-binding domain of p53 (54). Thus, the interaction of Enigma with the RING domain of MDM2 may interfere with homo(di)merization of MDM2 and induce conformational changes in MDM2 that enhance p53 ubiquitination. This notion is supported by MDM2-dependent interaction of Enigma with p53 and by the previous findings that the self- and p53 ubiquitination activities of MDM2 reside in its RING domain and the self-ubiquitination activity of MDM2 is separate from its ability to drive the degradation of p53 (5, 6).
Data from knock-in mice of the RING mutant of Mdm2(C462A) suggest that Mdm2 is regulated by PCAF or another E3 ligases but does not regulate its own stability by self-ubiquitination (9). However, MDM2 exhibits many p53-independent functions (55) that may also be in part essential for survival of the knock-in mice with p53 elimination. Only the knock-in mice that can properly regulate Mdm2(C462A) levels are likely to survive, as uncontrolled accumulation of Mdm2(C462A) protein would be detrimental. Thus, MDM2 level is likely regulated by MDM2 itself, PCAF, and/or unidentified E3 ubiquitin ligases. It would depend on cellular status which E3 ubiquitin ligase acts dominantly for MDM2 and p53 levels to be properly balanced for cell viability. The balance between MDM2 and p53 levels in proliferating cells such as tumor cells is likely to be regulated by Enigma that inhibits self-ubiquitination of MDM2, as evidenced by this study.
Growth factor–mediated activation of the phosphatidylinositol 3-kinase/Akt or MAPK pathway has been shown to phosphorylate MDM2 and regulate nuclear import or export of MDM2, thereby negatively regulating p53 (56–59). Akt-mediated phosphorylation of MDM2 leads to inhibition of self-ubiquitination and stabilization of MDM2 (60). Phosphorylation of MDM2 and p53 during DNA damage inhibits the interaction of MDM2 with p53 and is associated with increased self-ubiquitination and degradation of MDM2 (47, 61). Activation of the p38 MAPK/MK2 pathway results in phosphorylation of Hdm2 Ser166, and this phosphorylation is a common response to mitogenic activation and UV-induced damage. However, these two treatments do not have the same effect on p53 stability (62). These findings therefore suggest that the MDM2/p53 pathway may be in part regulated in a phosphorylation-independent manner. We found here that serum or HGF can decrease p53 levels through the SRF/Enigma/MDM2 pathway (Figures 4 and 5). Moreover, a specific MEK inhibitor, PD98059, or depletion of SRF or Enigma diminished the HGF effects on MDM2 and p53 levels (Figure 4). Thus, the SRF/Enigma/MDM2 pathway we identified here may represent a novel mechanism by which the p53 level is regulated in a manner independent of phosphorylation of MDM2 and p53. This possibility is supported by the findings that MDM2 overexpression exhibits a more profound effect on cellular transformation (59) and phosphorylation of p53 plays only very subtle, fine-tuning roles in regulating its function in mouse models (14).
MDM2 was shown to shuttle between the nucleus and the cytoplasm constantly and p53-independently (63). It was shown that MDM2 mono- and poly-ubiquitinates p53-depending on MDM2 levels and monoubiquitinated p53 is MDM2-independently accumulated in the cytosol (64). Cocompartmentalization of MDM2 and p53 was shown to be a major determinant for MDM2-mediated degradation of p53 (65). Enigma is localized mainly in the cytosol and moderately in the nucleus and forms a trimeric complex with MDM2 and p53 in both compartments (Supplemental Figure 8). Thus, an Enigma-mediated increase in the level of MDM2 activity may promote polyubiquitination of p53 for degradation in both the cytosol and nucleus, where the 26S proteasomes are evenly distributed (66).
Enigma and MDM2 levels are increased in proliferating cells, and Enigma is colocalized with MDM2 mostly in the cytosol (Figure 6C and Supplemental Figure 8). MDM2 is mainly cytoplasmic in exponentially growing cells, whereas p53 is evenly distributed between the cytoplasm and nucleus (67). Enigma and MDM2 are colocalized at apical cytoplasm of human gastric and colorectal tumor cells (Figure 6B). Collectively, these findings suggest that Enigma and MDM2 are colocalized in the cytoplasm of actively growing cells to efficiently target p53 for degradation. Presumably, these tumor cells may represent rapidly proliferating cells in a tumor mass, where the antiproliferative activities of p53 have to be attenuated.
MDM2 is highly expressed in tumors that retain wild-type p53 (38). A slight reduction in Mdm2 levels leads to a significant reduction in intestinal adenoma formation in the mouse (68). A single nucleotide polymorphism identified in the MDM2 promoter that slightly increases MDM2 levels is associated with increased tumor incidence (69). These results suggest that even minor changes in MDM2 levels induce significant phenotypes (46). Enigma has been shown to be highly expressed in gastric tumor cells and bone metastasis (70, 71). Thus, Enigma may function as an oncoprotein by stabilizing and activating MDM2 E3 ligase and thereby ablating p53. Moreover, Enigma may be a new therapeutic target for selective activation of wild-type p53 in tumor cells without much affecting normal cells.
Methods
Plasmids.
Human Enigma and MDM2 cDNAs were supplied by 21C Frontier Human Gene Bank (KRIBB). We generated pEBG-Enigma, pHis-Enigma, pHA-Enigma, pGEX-53, pEBG-MDM2, pGEX-MDM2, pHis-MDM2, or pHis-PCAF by PCR-subcloning Enigma, Mdm2, p53, or PCAF cDNAs into pCMV-Taq2 (Stratagene), pET28a (Novagen), pCDNA (Invitrogen), pEBG, pGEX-4T-1 (GE Life Sciences), or pColdI (Takara). We constructed an MDM2(C464A) mutant by PCR-subcloning Hdm2 cDNA and inserted it into pEBG, pGEX-1, or pColdI. We generated deletion constructs of pF-Enigma, pEBG-Enigma, pHis-Enigma, or pEBG-MDM2 by PCR-subcloning each of them by a standard cloning procedure. His6-Ub, p53, and F-SRF vectors were previously described (33, 37, 72). F-Enigma, F-ENH, and F-PCAF constructs were supplied by W.K. Cho (KRIBB), A. Iavarone (Columbia University Medical Center, New York, New York, USA), and S.-B. Seo (Chung-Ang University, Seoul, South Korea), respectively. We synthesized an Enigma-specific siRNA (5′-AAAGACCTTCTACTCCAAGAA-3′) oligonucleotide duplex and cloned it into pSuper vector to generate siEnigma vector. The SRF-specific siRNA (siSRF) and a scrambled control siRNA (siControl) vectors were previously described (33, 37).
Recombinant proteins.
We induced expression of pColdI vector–based His-proteins with 1 mM IPTG for 16 hours at 18–20°C and expression of pET28a vector-based ones for 4 hours at 25°C. GST fusion and His-tagged proteins were expressed in Escherichia coli BL21DE3 and were affinity purified as previously reported (37). His-tag from His-proteins was removed by incubation with 100 U/ml thrombin for 16 hours at 16°C.
Adenoviral vectors.
We inserted the BamHI–XhoI fragment of F-Enigma vector or the XbaI–HindIII fragment of siEnigma vector into pCMV-Shuttle vector. Generation, amplification, purification, titer determination, and transduction of adenoviral vectors have been previously described (25). Ad-p53 and Ad-LacZ viruses were previously described (25, 72).
Antibodies and chemicals.
We purchased antibodies specific for ubiquitin (Cell Signaling Technology), Flag, actin, GST (Sigma-Aldrich), p53 (DO-1), SRF, His, HA (Santa Cruz Biotechnology Inc.), p21 (BD Biosciences — Pharmingen), and MDM2 (BD Biosciences — Pharmingen SMP14). We generated Enigma-specific antibody by immunizing mice with His-Enigma.
We purchased HGF (R&D), PD98059 (Cell Signaling Technology), Flag- or His-ubiquitin, rabbit E1, ADR, MG132, PMSF (Sigma-Aldrich), Lipofectamine (Invitrogen), thrombin (Merck), and proteinase inhibitor cocktail (Roche).
Cell culture.
Standard conditions and procedures were used for culturing cell lines. We obtained Mdm2+/+p53–/– and Mdm2–/–p53–/– MEFs (73) from G. Lozano (The University of Texas MD Anderson Cancer Center, Houston, Texas, USA). We constructed the 293–F-Enigma cell line by transfection of F-Enigma plasmid into 293 cells and selection of the cell using neomycin (1 mg/ml).
Immunoprecipitation and immunoblotting.
We transfected cells with calcium phosphate or Lipofectamine, treated them for the indicated times, lysed them with RIPA buffer for immunoblotting (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.5 mM EDTA, 1% NP40, 0.1% SDS, 0.1% sodium deoxycholate, 0.5 mM PMSF) or with IP buffer for immunoprecipitation (50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 0.5 mM EDTA, 1% NP40, 0.5 mM PMSF, one tablet proteinase inhibitor cocktail/100 ml). To remove IgG proteins in immunoprecipitates, we incubated IP samples with 50 μl of 0.1 M glycine buffer (pH 2) for 1–2 minutes, filtered them through a microspin column (QIAprep Spin Miniprep Kit, QIAGEN), immediately neutralized the filtrates with 5 μl of 1 M Tris (pH 10), and added them to 2× SDS-PAGE sample buffer. We lysed human tissues in a lysis buffer (iNtRON) containing 100 U Benzonase (Calbiochem) for 30 minutes at 4°C. Protein samples or cleared lysates were immunoblotted by a standard procedure.
Reporter plasmids and luciferase assay.
Luciferase reporter constructs p53RE-luc, p21-luc, and Bax-luc and reporter assay were previously described (25).
Northern blotting and RT-PCR.
We performed RNA preparation, Northern blotting, and RT-PCR using relevant primers as previously described (33, 37). In particular, pre-hybridization in Northern blot analysis was performed at 55°C for 2 hours in a pre-hybridization buffer (40% formamide, 5× SSC, 5× Denhart’s solution, 1% SDS, 100 mg/ml salmon sperm DNA). p53 and p21 transcripts were hybridized with the pre-hybridization buffer containing each labeled probe but not formamide at 55°C for 16 hours. P1- and P2-MDM2 transcripts were hybridized with the pre-hybridization buffer containing each labeled probe at 42°C. Primer sequences for Northern blotting probes or RT-PCR are shown in Supplemental Figure 9.
In vitro binding assay.
We mixed relevant proteins in 500 μl IP buffer, adjusted the reaction volume up to 1 ml by the addition of IP buffer, incubated the mixtures at 4°C for 1 hour, and performed the binding assay.
Ubiquitination assay.
To detect Ub conjugates in cells, we lysed cells in a buffer (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl [pH 8.0], 5 mM imidazole, 10 mM β-mercaptoethanol) and incubated cell lysates with Ni2+-NTA agarose beads for 4 hours at room temperature. The beads were washed with buffer A (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl [pH 8.0], 10 mM β-mercaptoethanol) and buffer B (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl [pH 6.3], 10 mM β-mercaptoethanol). The bead-bound proteins were eluted with a buffer (200 mM imidazole, 0.15 M Tris-Cl [pH 6.7], 30% glycerol, 0.72 M β-mercaptoethanol, 5% SDS) and were analyzed with relevant antibodies.
We performed the in vitro ubiquitination assay with E1 (0.2 μg), GST-UbcH5C (E2, 0.2 μg), and His-Ub or Flag-Ub (1 μg) as previously reported (37). For detection of in vitro ubiquitinated proteins by GST pull-down/IB assay, we added 10% SDS to the reaction mixtures for a final concentration of 1% SDS. One-tenth of reaction mixtures were used for IB to identify input proteins. The rest was diluted up to 1 ml with IP buffer and incubated with GST-Sepharose beads (Amersham) at 4°C for 16 hours. Bead-bound proteins were analyzed by IB. For detection of in vitro ubiquitinated proteins by IP/IB assay, we boiled 1% SDS of reaction mixtures for 5 minutes, diluted them up to 1 ml with IP buffer, and incubated them with relevant antibodies at 4°C for 16 hours and then with protein A–Sepharose beads for another 2 hours. Bead-bound proteins were eluted by the addition of 0.1 M glycine buffer (pH 2), and the eluates were neutralized with 1 M Tris (pH 10) and then analyzed by IB.
Immunofluorescence staining.
Cells were grown in glass coverslips in a 6-well dish, fixed with ice-cold methanol for 15 minutes at room temperature, blocked with PBS containing 5% normal bovine serum (NBS) for 20 minutes, and then incubated with a mixture of rabbit anti-MDM2 (1:100 dilution) and mouse anti-Enigma (1:100) in PBS containing 0.5 % NBS for 90 minutes at room temperature. Cells were washed with PBST, incubated with FITC-conjugated goat anti-rabbit IgG plus rhodamine-conjugated goat anti-mouse IgG plus DAPI (0.1 μg/ml) in PBST containing 1% NBS for 1 hour at room temperature, washed with PBST, and mounted with VECTASHIELD HardSet. Images were taken with a microscope (Olympus).
Cell viability.
We transfected cells with relevant expression vectors. At 16 hours after transfection, 5 × 104 cells per 60-mm dish were seeded and grown for 12 hours and treated with or without 20 μg/ml ADR for 48 hours. We counted viable cells with a hemocytometer. We repeated the experiments 3 times, each in triplicate. Cell viability data are mean ± SD.
Cell cycle analysis.
Cells were washed with PBS, fixed in 70% ethanol at –20°C for 16 hours, incubated in PBST with 100 μg/ml propidium iodide (PI, Sigma-Aldrich) plus 100 μg/ml RNase A at 37°C for 30 minutes or 50 μg/ml PI plus 100 μg/ml RNase A plus FITC-conjugated Annexin V (1:100, Santa Cruz Biotechnology Inc.) in a binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). The DNA content of the cells was then assessed by flow cytometry with a FACScalibur (BD).
TUNEL assay.
Cells were fixed in 4% paraformaldehyde at 4°C for 20 minutes, washed 3 times in PBS for more than 30 minutes, and treated with permeation solution (0.1% sodium citrate and 0.1% Triton X-100) for 5 minutes. After being washed with PBS, cells were analyzed using the In Situ Cell Death Detection Kit (Roche) according to the manufacturer’s instructions.
Human samples.
All samples from humans were obtained with informed consent. The use of stomach and liver tumors and their normal counterparts was approved by the Research Ethics Committee of Chonbuk National University Medical School and Hospital, Jeonju, Korea; Keimyung University School of Medicine, Daegu, Korea; and Chungnam National University Hospital, Daejeon, Korea.
Mouse experiments.
We maintained 5- to 6-week-old BABL/c mice (Taconic) and nude mice (Japan SLC Inc.) in accordance with the guidelines and under approval of the Institutional Review Committee for Animal Care and Use, KRIBB.
Liver extracts.
We injected mice with PBS with or without HGF or relevant adenoviral vectors through the tail vein. Adenoviral vectors expressed transgenes in more than 90% hepatocytes. We prepared protein extracts and total RNAs from mouse liver as previously reported (33, 37).
Tissue array.
We performed tissue arrays as previously described (25). Tissue arrays were supplied by W.H. Kim (Seoul National University College of Medicine, Seoul, Korea).
Tumor growth.
We transduced HLK3 cells with or without Ad–F-Enigma or Ad-LacZ at an MOI of 100, or Ad-siEnigma or Ad-siControl at an MOI of 200, incubated them for 24 hours, and generated tumors in nude mice by subcutaneous inoculation of 106 cells with or without adenoviral transduction in 0.1 ml PBS. At day 14 after cell inoculation, we injected mice with or without 4 mg/kg ADR intravenously, monitored tumor growth for 28 days after ADR injection, and photographed excised tumors from mice. We calculated tumor size as previously described (37).
Statistics.
We performed statistical analysis using a paired, 1-tailed Student’s t test. We considered data statistically significant when P was less than 0.05.
Supplementary Material
Acknowledgments
We are grateful to S.H. Leem, I.S. Chu, Y.S. Kim, and K.S. Song for their help in preparing human samples. This work was supported by a grant (FG08-32-01) of the 21C Frontier Functional Genome Project from the Ministry of Education, Science and Technology, South Korea; and a grant (no. 0920320) from the National R&D Program for Cancer Control, Ministry for Health and Welfare, South Korea.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(12):4493–4506. doi:10.1172/JCI42674.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 3.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase for tumor suppressor p53. FEBS Lett. 1997;420(1):25–27. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 4.Kubbutat KHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–302. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 5.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275(12):8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 6.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19(11):1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 7.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type-p53 activity. EMBO J. 1993;12(2):461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Bayle JH, Olson D, Levine AJ. The p53 mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 9.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12(4):355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Linares LK, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol. 2007;9(3):331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- 11.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13(1):49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 12.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26(9):1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 13.Kruse J-P, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 15.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2(1):1–8. [PubMed] [Google Scholar]
- 16.Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91(1–2):5–17. doi: 10.1016/S0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 17.Guy PM, Kenny DA, Gill GN. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol Biol Cell. 1999;10(6):1973–1984. doi: 10.1091/mbc.10.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrès R, et al. Enigma interacts with adaptor protein with PH and SH2 domains to control insulin-induced actin cytoskeleton remodeling and glucose transporter 4 translocation. Mol Endocrinol. 2006;20(11):2864–2875. doi: 10.1210/me.2005-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durick K, Gill GN, Taylor SS. Shc and Enigma are both required for mitogenic signaling by Ret/ptc2. Mol Cell Biol. 1998;18(4):2298–2308. doi: 10.1128/mcb.18.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda S, et al. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271(49):31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- 21.Wu R, Durick K, Songyang Z, Cantley LC, Taylor SS, Gill GN. Specificity of LIM domain interaction with receptor tyrosine kinases. J Biol Chem. 1996;271(27):15934–15941. doi: 10.1074/jbc.271.27.15934. [DOI] [PubMed] [Google Scholar]
- 22.Sangadala S, Boden SD, Viggeswarapu M, Liu Y, Titus L. LIM mineralization protein-1 potentiates bone morphogenetic protein responsiveness via a novel interaction with Smurf 1 resulting in decreased ubiquitination of Smads. J Biol Chem. 2006;281(25):17212–17219. doi: 10.1074/jbc.M511013200. [DOI] [PubMed] [Google Scholar]
- 23.Krcmery J, Camarata T, Kulisz A, Simon HG. Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. BioEssays. 2010;32(2):100–108. doi: 10.1002/bies.200900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 25.Jung CR, et al. Adenovirus-mediated transfer of siRNA against PTTG1 inhibits liver cancer cell growth in vitro and in vivo. Hepatology. 2006;43(5):1042–1052. doi: 10.1002/hep.21137. [DOI] [PubMed] [Google Scholar]
- 26.Miyashita T, et al. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9(6):1799–1805. [PubMed] [Google Scholar]
- 27.Moustakas A, Kardassism D.Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci. U. S. A. 199895158733–8738. 10.1073/pnas.95.15.8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13(7):4107–4014. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levav-Cohen Y, Haupt S, Haupt Y. Mdm2 in growth signaling and cancer. Growth Factors. 2005;23(3):183–192. doi: 10.1080/08977190500196218. [DOI] [PubMed] [Google Scholar]
- 30.Chai J, Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol. 2002;53(2):147–157. [PubMed] [Google Scholar]
- 31.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19(6):618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkovich E, Lamed Y, Ginsberg D. E2F and Ras synergize in transcriptionally activating p14ARF expression. Cell Cycle. 2003;2(2):127–133. doi: 10.4161/cc.2.2.293. [DOI] [PubMed] [Google Scholar]
- 33.Lim JH, Jung CR, Lee CH, Im DS. Egr-1 and serum response factor are involved in growth factors- and serum-mediated induction of E2-EPF UCP expression that regulates the VHL-HIF pathway. J Cell Biochem. 2008;105(4):1117–1127. doi: 10.1002/jcb.21914. [DOI] [PubMed] [Google Scholar]
- 34.Roos F, Ryan AM, Chamow SM, Bennett GL, Schwall RH. Induction of liver growth in normal mice by infusion of hepatocyte growth factor/scatter factor. Am J Physiol. 1995;268(2 pt 1):G380–G386. doi: 10.1152/ajpgi.1995.268.2.G380. [DOI] [PubMed] [Google Scholar]
- 35.Le MT, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park MY, et al. Expression of the serum response factor in hepatocellular carcinoma: implications for epithelial-mesenchymal transition. Int J Oncol. 2007;31(6):1309–1315. [PubMed] [Google Scholar]
- 37.Jung CR, et al. E2–EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12(7):809–816. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- 38.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–3458. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endo K, Ueda T, Ohta T, Terada T. Protein expression of MDM2 and its clinicopathological relationships in human hepatocellular carcinoma. Liver. 2000;20(3):209–215. doi: 10.1034/j.1600-0676.2000.020003209.x. [DOI] [PubMed] [Google Scholar]
- 40.Gorgoulis VG, et al. A molecular and immunohistochemical study of the MDM2 protein isoforms and p53 gene product in bronchogenic carcinoma. J Pathol. 1996;180(2):129–137. doi: 10.1002/(SICI)1096-9896(199610)180:2<129::AID-PATH646>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Kasper HU, Schneider-Stock R, Mellin W, Günther T, Roessner A. P53-protein accumulation and MDM2-protein overexpression in gastric carcinomas. No apparent correlation with survival. Pathol Res Pract. 1999;195(12):815–820. doi: 10.1016/S0344-0338(99)80103-4. [DOI] [PubMed] [Google Scholar]
- 42.Lianes P, et al. Altered patterns of MDM2 and TP53 expression in human bladder cancer. J Natl Cancer Inst. 1994;86(17):1325–1330. doi: 10.1093/jnci/86.17.1325. [DOI] [PubMed] [Google Scholar]
- 43.Cadwell C, Zambetti GP. The effects of wild–type p53 tumor suppressor activity and mutant p53 gain–of–function. Gene. 2001;277(1–2):15–30. doi: 10.1016/S0378-1119(01)00696-5. [DOI] [PubMed] [Google Scholar]
- 44.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the quardian of the genome. Cancer Res. 2000;60(24):6788–6793. [PubMed] [Google Scholar]
- 45.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-R. [DOI] [PubMed] [Google Scholar]
- 46.Marine JC, Francoz S, Maetens M, Wahl G, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13(6):927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 47.Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damaged kinases is required for p53 activation. EMBO J. 2004;23(7):1547–1558. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 49.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 50.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26(1):90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26(1):102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28(24):3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100(21):12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23(2):251–263. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 55.Ganguli G, Wasylyk B. p53-independent functions of MDM2. Mol Cancer Res. 2003;1(14):1027–1035. [PubMed] [Google Scholar]
- 56.Mayo LD, Donner DB.A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U S A. 2001982011598–11603. . 10.1073/pnas.181181198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3(11):973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 58.Ashcroft M, et al. Phosphorylation of HDM2 by Akt. Oncogene. 2002;21(13):1955–1962. doi: 10.1038/sj.onc.1205276. [DOI] [PubMed] [Google Scholar]
- 59.Jackson MW, Patt LE, LaRusch GA, Donner DB, Stark GR, Mayo LD. Hdm2 nuclear export, regulated by insulin-like growth factor-I/MAPK/p90Rsk signaling, mediates the transformation of human cells. J Biol Chem. 2006;281(24):16814–16820. doi: 10.1074/jbc.M511617200. [DOI] [PubMed] [Google Scholar]
- 60.Feng J, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem. 2004;279(34):35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- 61.Maya R, et al. ATM–dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15(9):1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber HO, Ludwig RL, Morrison D, Kotlyarov A, Gaestel M, Vousden KH. HDM2 phosphorylation by MAPKAP kinase 2. Oncogene. 2005;24(12):1965–1972. doi: 10.1038/sj.onc.1208389. [DOI] [PubMed] [Google Scholar]
- 63.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17(2):554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 65.Xirodimas DP, Stephen CW, Lane DP. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res. 2001;270(1):66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- 66.Shirangi TR, Zaika A, Moll UM. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 2002;16(3):420–422. doi: 10.1096/fj.01-0617fje. [DOI] [PubMed] [Google Scholar]
- 67.Wang YV, Wade M, Wong E, Rodewald LW, Wahl GM. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci U S A. 2007;104(30):12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mendrysa SM, et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20(1):16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bond GL, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 70.Klein A, et al. Identification of brain- and bone-specific breast cancer metastasis genes. Cancer Lett. 2009;276(2):212–220. doi: 10.1016/j.canlet.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 71.Yanglin P, et al. KCNE2, a down–regulated gene identified by in silico analysis, suppressed proliferation of gastric cancer cells. Cancer Lett. 2007;246(1–2):129–138. doi: 10.1016/j.canlet.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 72.Choi JY, et al. Adenovirus-mediated p53 tumor suppressor gene therapy against subcutaneous HuH7 hepatoma cell line nodule of nude mice. J Korean Med Sci. 1999;14(3):271–276. doi: 10.3346/jkms.1999.14.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barboza JA, Iwakuma T, Terzian T, El-Naggar K, Lozano G. Mdm2 and Mdm4 loss regulates distinct p53 activities. Mol Cancer Res. 2008;6(6):947–954. doi: 10.1158/1541-7786.MCR-07-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.