Abstract
Retinoblastoma (RB; encoded by RB1) is a tumor suppressor that is frequently disrupted in tumorigenesis and acts in multiple cell types to suppress cell cycle progression. The role of RB in tumor progression, however, is poorly defined. Here, we have identified a critical role for RB in protecting against tumor progression through regulation of targets distinct from cell cycle control. In analyses of human prostate cancer samples, RB loss was infrequently observed in primary disease and was predominantly associated with transition to the incurable, castration-resistant state. Further analyses revealed that loss of the RB1 locus may be a major mechanism of RB disruption and that loss of RB function was associated with poor clinical outcome. Modeling of RB dysfunction in vitro and in vivo revealed that RB controlled nuclear receptor networks critical for tumor progression and that it did so via E2F transcription factor 1–mediated regulation of androgen receptor (AR) expression and output. Through this pathway, RB depletion induced unchecked AR activity that underpinned therapeutic bypass and tumor progression. In agreement with these findings, disruption of the RB/E2F/nuclear receptor axis was frequently observed in the transition to therapy resistance in human disease. Together, these data reveal what we believe to be a new paradigm for RB function in controlling prostate tumor progression and lethal tumor phenotypes.
Introduction
Retinoblastoma (RB; encoded by RB1), a tumor suppressor protein, is a critical negative regulator of tumor development. RB prevents tumorigenesis by suppressing cell cycle progression (1). However, the role of RB in tumor progression is poorly understood, and the clinical importance of RB loss during this process has not been well considered. Here, we identified a clinically relevant function for RB in tumor progression, manifest through control of hormone signaling networks.
The function of RB in cell cycle control has been well described (1). Conditions favoring cell cycle arrest induce RB hypophosphorylation and activation. Active RB binds to promoters of genes required for S-phase entry (e.g., CCNA2 and MCM7) and, through association with the SWI/SNF complex and corepressor molecules (e.g., Sin3B), elicits transcriptional corepression. Many RB target genes are positively regulated by activator E2F transcription factors, supporting the current model that RB acts by suppressing E2F-mediated transcriptional activation. Indeed, the minimal transcriptional repression and tumor suppression domain of RB contains the E2F binding motif. E2F-independent functions of RB have been identified (2), but the contribution of these functions to tumor suppression is uncertain. Thus, contemporary views of RB suggest that the protein prevents cell cycle deregulation and tumor development through suppression of activator E2Fs.
Given the importance of RB in regulating cell cycle transitions, stringent control mechanisms are employed in untransformed cells to regulate proliferation (1). When intra- and extracellular conditions favor cell cycle progression, cascades of cyclin-dependent kinase/cyclin (CDK/cyclin) complexes sequentially phosphorylate and inactivate RB. Resultant RB phosphorylation events relieve the ability of RB to suppress E2F function at critical target genes, allowing for expression of downstream G1 cyclins. Once produced, active CDK2/cyclin E or cyclin A complexes complete the RB phosphorylation cascade, rendering the tumor suppressor ineffective at inhibiting E2F and facilitating S-phase entry. During M-phase, RB function is reset through phosphatase activity. Not surprisingly, the mechanisms that control RB regulation are frequently altered during the course of human tumor development (3). For example, RB is sequestered by viral oncoproteins during cervical cancer development, aberrantly hyperphosphorylated and inactivated in other tumor types by amplification or overexpression of cyclin D1 (e.g., mantle cell lymphoma), or inactivated indirectly via loss of the CDK4/cyclin D inhibitor p16ink4a, such as occurs in melanoma. Finally, loss of heterozygosity at the RB1 gene locus is causative for retinoblastoma development (4). Somatic loss of RB1 has also been reported in tumors that do not harbor p16ink4a loss or aberrant D-cyclin expression, which suggests that individual tumor types demonstrate distinct preferences for engaging mechanisms to perturb RB function (3, 5). The underlying basis for selectivity has not been defined. Nonetheless, while there is strong precedent for RB disruption in initiating tumorigenesis, the role of RB dysfunction in tumor progression is poorly understood.
Given the importance of RB in human disease, it is imperative to define the underlying basis for selective RB disruption, and to assess the impact of RB perturbation in the context of clinically relevant outcomes. Here, exploration of the RB pathway revealed an unexpected consequence of RB dysregulation in prostate cancer (PCa) and defined what we believe to be a novel role of RB in controlling tumor outcomes via nuclear receptor networks. Importantly, the RB/nuclear receptor axis revealed a critical role for RB in tumor progression rather than tumor development, demonstrating the clinical relevance of this paradigm.
Results
RB loss is overrepresented in PCa metastases and castration-resistant disease.
While previous studies demonstrated a significant role for RB loss in tumor development, the function of RB in protecting against tumor progression is not known. PCas undergo a discrete set of transitions, from carcinoma in situ to adenocarcinomas to metastatic disease that results in patient mortality (6). Intriguingly, PCa is refractory to standard chemotherapy and is treated based on the androgen dependence of this tumor type. First-line therapeutic intervention for non–organ-confined tumors is hormone deprivation therapy, which is designed to ablate androgen receptor (AR) activity. Although initially effective, hormone therapy–resistant tumors arise, representative of the transition to incurable, castration-resistant PCa (CRPC; ref. 7). Although RB plays an important role in the response to hormone therapy in vitro (8), the frequency and impact of RB dysregulation during PCa development and progression is largely unknown.
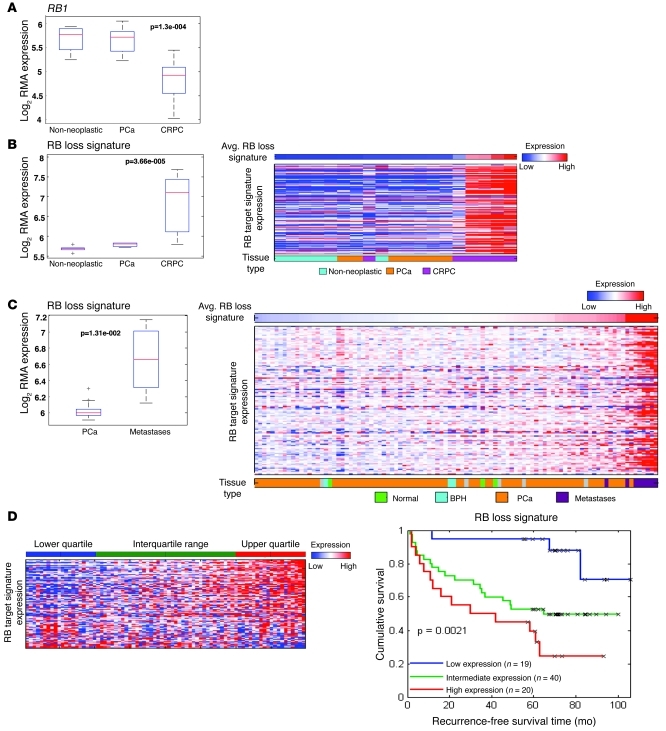
Initially, the expression profile of the RB1 gene was determined by comparing transcript abundance in non-neoplastic prostate tissue versus primary tumors and CRPCs. Notably, there was no significant alteration in RB1 transcript abundance upon comparison of benign tissue with primary PCa (Figure 1A). However, a marked reduction in RB1 mRNA was observed in CRPC. These data suggested that RB deficiency may be specifically associated with the transition to castration resistance, rather than with tumor initiation. However, since RB function can be compromised by mechanisms independent of altered RB1 gene expression (e.g., aberrant RB phosphorylation), it was imperative to assess overall RB function using additional, stringent metrics of RB activity. We previously developed a gene expression signature of RB loss, using genetically defined models of RB deletion. The gene signature has been validated across multiple platforms and is a reliable measure of RB transcriptional corepressor function (9–12). Analyses showed that loss of RB function was significantly overrepresented in CRPC but not primary disease (Figure 1B), consistent with Figure 1A. Unbiased cluster analyses of individual specimens are also shown in Figure 1B (samples were stratified based on the magnitude of the RB loss signature). Most CRPC specimens clustered with high representation of the RB loss signature. Analysis was expanded to consider a larger dataset, including both primary and metastatic disease. In agreement with the findings above, the RB loss signature was overrepresented in metastatic disease, and clustering analyses of individual samples further supported the contention that metastatic samples cluster with significantly elevated RB loss signatures (Figure 1C). Strikingly, quartile analyses for specimens wherein clinical outcome was available revealed that the RB loss signature was strongly associated with reduced recurrence-free survival (Figure 1D). Together, these data indicate that RB loss is markedly overrepresented in the transition to CRPC and is associated with poor outcome.
Figure 1. RB loss is overrepresented in CRPC and metastatic PCa and is associated with tumor recurrence.
(A) RB expression was determined in non-neoplastic tissue, localized PCa, and CRPC from an established dataset (54) plotted as log of robust multichip average (RMA) expression. For all box plots, the red line is the median, the bottom and top of the box are the 25th and 75th percentiles, respectively, and the whiskers capture data points within 1.5 interquartile range (IQR). Outliers beyond 1.5 IQR are plotted individually with gray plus symbols. (B) The RB loss signature was determined from data in A, and plotted as a box and whisker plot and heatmap. In the heatmap, tissue samples were ordered from left to right based on relative representation of the RB loss signature (above). Tissue type is provided below. (C) RB loss signature in primary PCa and metastatic PCa, from data first described elsewhere (55). Also shown is RB loss signature across normal, benign prostate epithelia (BPH), localized PCa, and metastatic specimens. As in B, tissue samples were ordered from left to right based on relative level of the RB loss signature, and sample type is provided below. Indicated P values were calculated using the Student’s t test. (D) Quartile analyses were used to determine the impact of RB loss on recurrence-free survival using the dataset in C. See also Supplemental Figure 8.
RB1 copy number loss is frequently observed in CRPC.
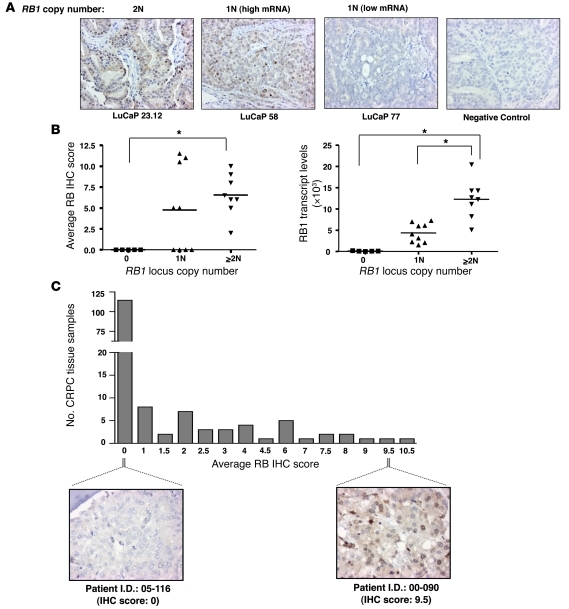
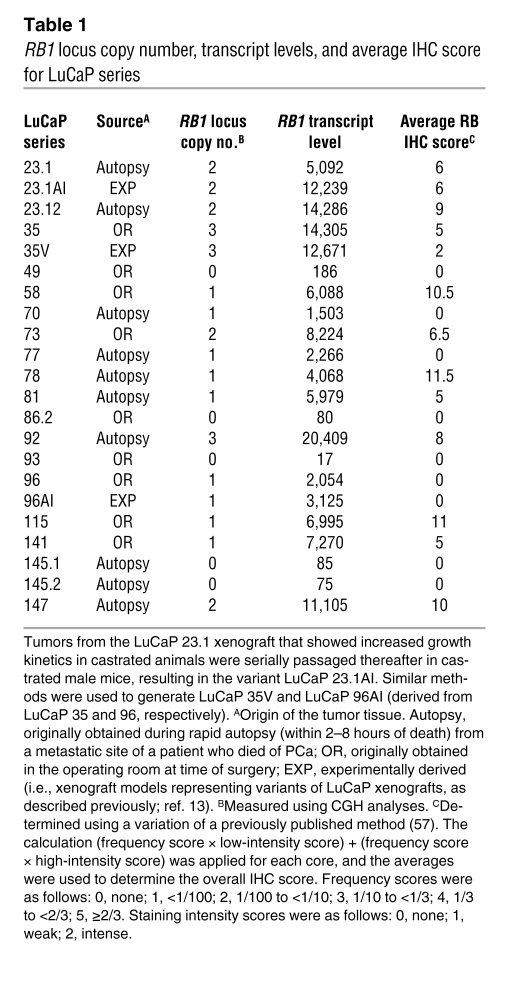
To complement these studies and address potential mechanisms of RB loss in CRPC, RB1 locus copy number and expression levels were initially measured in the LuCaP xenograft series, derived from rapid autopsy of patients who failed hormone therapy (13). As shown in Figure 2, A and B, and Table 1, tumors with no RB1 locus scored negative for nuclear RB immunodetection, whereas those tumors with at least 2 copies of the RB1 locus demonstrated the highest average levels of RB protein expression. As expected, tumors retaining only a single copy of the RB1 locus demonstrated a range of RB immunodetection (Figure 2B). Furthermore, tumors with 2 or more copies of RB1 locus displayed the highest levels of RB1 mRNA (Figure 2B). These data indicate that RB1 locus number significantly correlates with RB1 transcript levels and protein expression, validating the immunohistochemical (IHC) analyses. These analyses were therefore extended to a larger set of 156 individual metastatic CRPCs obtained from 44 patients (Figure 2C and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI44239DS1). As shown, 115 of 156 (73.7%) of the specimens scored negative for nuclear RB immunodetection, whereas 31 of 156 (19.8%) scored with levels similar to that observed in LuCaP tumors with 2 or more copies of the RB1 locus. These data further demonstrate that RB loss is highly represented in CRPC, as detected by both transcript and protein analyses, and loss of the RB1 locus itself may be a major mechanism of RB inactivation in CRPC.
Figure 2. Loss of RB expression in CRPC is frequently associated with RB1 deletion.
(A) RB1 mRNA expression, RB immunostaining, and RB1 copy number was concurrently measured using LuCaP xenografts derived from patients with CRPC. Representative images of RB immunostaining as a function of copy number are shown (original magnification, ×200). For samples retaining only a single RB1 locus, images for tumors found to harbor high and low RB1 mRNA expression are shown. LuCaP sample stained with control nonspecific anti–MOPC-21 antibody was used as a negative control. (B) Scatter plot for RB1 locus copy number versus average RB IHC score or RB1 transcript levels from the 22 LuCaP xenografts. *P < 0.05, ANOVA. (C) Average RB IHC immunostaining in 156 CRPC samples. Representative RB IHC images for tumor tissues with no RB staining (patient 05-116) and high RB staining (patient 00-090) are shown (original magnification, ×400).
Table 1 .
RB1 locus copy number, transcript levels, and average IHC score for LuCaP series
RB depletion is sufficient to induce castration-resistant tumor growth.
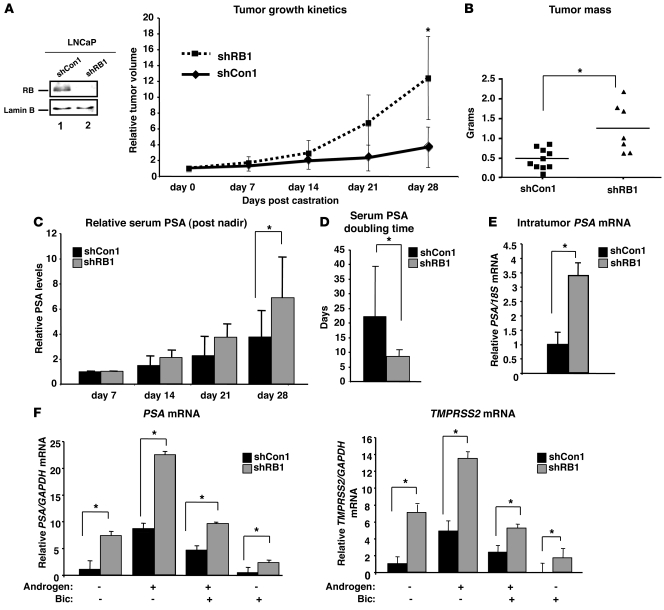
Given the frequency of RB perturbation during clinical tumor progression, the effect of this event was determined by modeling RB depletion in xenograft models of hormone therapy–sensitive PCa. Efficient RB knockdown in PCa cells was previously described (8), and validated herein (Figure 3A). RB deficiency alone did not confer a significant tumor growth advantage in vivo (Supplemental Figure 1A). However, castration of host animals with RB-depleted (shRB1) or control xenografts (shCon1) reaching 100–150 mm3 unmasked a growth advantage specific to RB-deficient tumors (Figure 3A). These data suggested that RB depletion is sufficient to induce castration-resistant tumor growth, as monitored by tumor growth kinetics. Consonantly, there was a significant increase in tumor mass among shRB1 compared with shCon1 tumors at sacrifice (Figure 3B). Additionally, serum prostate-specific antigen (PSA; also known as KLK3) was monitored. PSA is used clinically as a marker of PCa detection, burden, and progression (7), and is not expressed in mice; thus, serum PSA was monitored after castration as a measure of castration-resistant tumor growth. Serum PSA levels were significantly higher in animals carrying the shRB1 xenografts (Figure 3C). A weakened immediate response to castration was also observed, as manifest by a reduced drop in serum PSA at 1 week after castration (Supplemental Figure 1B). Finally, PSA doubling time after castration was determined, as this parameter is used clinically to detect disease recurrence, and rising PSA after hormone therapy (also known as biochemical failure) almost invariably precedes detectable recurrent tumor formation (14). RB depletion significantly shortened PSA doubling time (Figure 3D), and similar results were observed with a second xenograft model of hormone therapy–sensitive disease (Supplemental Figure 2, A and B, and data not shown). Combined, these results demonstrate that RB depletion reduces the time to biochemical failure and induces castration-resistant tumor growth. Both phenotypes are also frequently associated with alterations of the AR pathway, which suggests that RB loss may act in concert with or impinge upon the AR axis.
Figure 3. RB depletion enables bypass of hormonal therapy.
(A) Immunoblot for RB in shCon1 and shRB1 cells. Relative tumor volume of xenografts after castration is also shown. Hosts were castrated when tumors reached 100–150 mm3 (day 0). Data plotted are mean tumor size ± SD for each cohort. n = 10 (shCon1); 7 (shRB1). (B) Tumor mass at sacrifice (day 28). Individual data points represent the tumor mass of each xenograft at sacrifice, and the mean for each cohort is represented by a horizontal bar. (C) Relative serum PSA for 4 weeks after castration, beginning at the nadir (day 7 after castration, set to 1). (D) Serum PSA doubling time for each cohort was determined as follows: time (days) × loge (2)/[loge (PSA28) – loge (PSA7)], where PSA7 and PSA28 represent PSA levels at days 7 and 28 after castration. n = 9 (shCon1); 7 (shRB1). (E) Intratumor PSA mRNA levels in xenograft tissues at sacrifice were determined via qPCR. PSA relative to 18S is plotted; expression in shCon1 was set to 1. n = 5 (shCon1); 7 (shRB1). (F) PSA and TMPRSS2 mRNA levels were determined by qPCR in cells cultured for 48 hours in androgen-free (CDT) or androgen-containing (FBS) media and supplemented as indicated. Results are plotted for each treatment condition relative to that in shCon1 cells after androgen ablation (set to 1). Data reflect triplicate analyses of at least 2–3 independent biological replicates (mean ± SD). *P < 0.05, Student’s t test. See also Supplemental Figures 1 and 2.
Disruption of RB function results in ligand-independent and antagonist-resistant AR activation.
To address the impact of RB status on AR signaling, intratumor PSA mRNA levels were determined. Intratumor PSA mRNA levels were significantly elevated in shRB1 xenografts (Figure 3E), indicative of enhanced AR signaling. Subsequently, in vitro analyses of AR function were performed under conditions mimicking therapeutic intervention. Expression of 2 clinically relevant AR target genes, PSA and TMPRSS2, was examined under conditions of (a) androgen ablation (achieved by culture in steroid-deprived media); (b) complete serum; (c) introduction of the AR antagonist bicalutamide (Bic); or (d) maximum androgen blockade (androgen depletion and Bic). Under each condition, the RB-depleted cells showed significantly higher AR target gene expression (Figure 3F), which suggests that aberrations in RB function result in enhanced AR signaling. This effect was confirmed in a second model of hormone dependent PCa, LAPC4, in which RB depletion resulted in elevated AR target gene expression (Supplemental Figure 2D). Together, these data reveal an unanticipated consequence of RB depletion for AR function and provide the first evidence to our knowledge of a mechanism by which RB loss may promote a growth advantage that is specific to tumor progression.
AR occupancy is enriched at target gene loci as a consequence of RB downregulation.
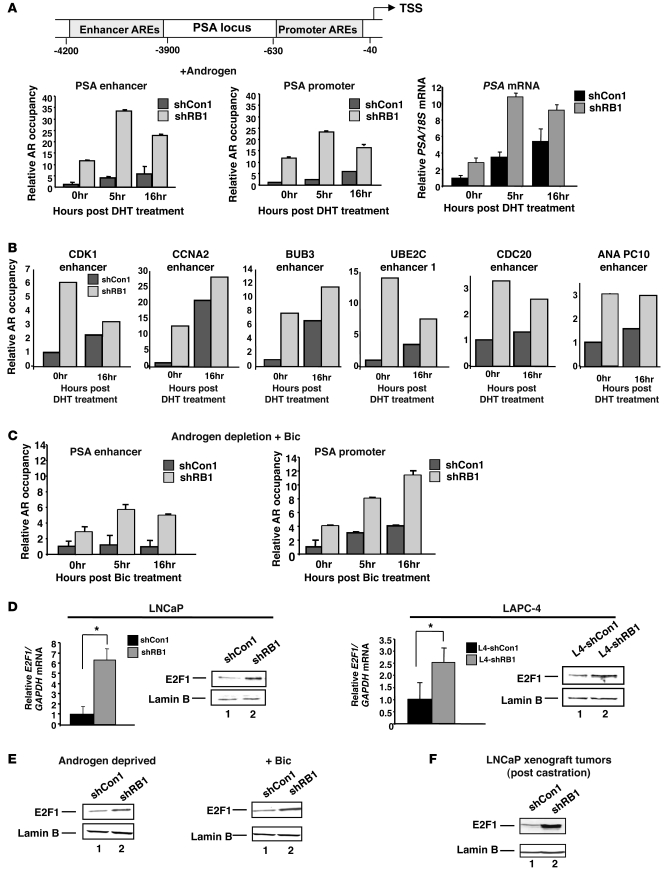
To address the mechanism by which RB dysfunction alters AR signaling, ChIP analyses were performed, wherein AR occupancy and output at the PSA locus was assessed after steroid depletion and no dihydrotestosterone (DHT) supplementation (0-hour time point) or at 5 and 16 hours after DHT addition. As expected, AR occupancy increased in control cells at both enhancer and promoter regions after DHT stimulation (Figure 4A). Concurrent analysis of PSA mRNA expression revealed a ligand-dependent induction of PSA transcript at the 5-hour time point and beyond (Figure 4A and data not shown). Notably, AR occupancy was enriched in shRB1 cells under both conditions, as shRB1 cells showed greater AR binding under ligand depletion and after DHT stimulation, and the change in AR occupancy mirrored that of PSA transcript levels. Moreover, a large subset of AR target genes associated with control of cell division were recently identified as enriched for receptor occupancy in CRPC cells (15), and ChIP analyses of these regions showed markedly enhanced AR occupancy after RB depletion (Figure 4B). Similar results were observed with CRPC-specific AR target genes not directly associated with cell cycle control (Supplemental Figure 3A). Importantly, a subset of these enhanced binding events was linked to enhanced expression of target genes (e.g., CDK1, CCNA2, BUB3, CDKN3, BCCIP, GNL3, and BTG3) after RB depletion, which indicates that RB loss–mediated induction of AR occupancy is sufficient to alter expression of CRPC-specific AR target gene expression (Supplemental Figure 3B). Based on these collective findings, it is evident that RB depletion promotes both ligand-independent and ligand-induced AR chromatin association and activity.
Figure 4. RB loss deregulates AR occupancy at target gene loci and E2F1 expression under therapeutic conditions.
(A) PSA locus depicting AREs relative to the TSS is illustrated. AR occupancy and output was determined at the indicated time points after 10 nM DHT stimulation; for comparison, AR occupancy in shCon1 at 0 hours was set to 1. In parallel, PSA mRNA was quantified relative to 18S control, and expression in shCon1 at the 0-hour time point was set to 1. (B) AR occupancy in both cell types was determined using loci specific to CRPC. (C) AR occupancy was determined as in A, but after stimulation with 10 μM Bic. (D) E2F1 expression was quantified by qPCR and immunoblot in LNCaP and LAPC4 cells after RB knockdown. qPCR data in A–D represent mean ± SD of 3 replicates; similar results were obtained in at least 2 independent experiments. (E) Immunoblots for E2F1 after 48 hours of androgen ablation or under parallel conditions supplemented with 1 μM Bic. (F) Immunoblot of E2F1 from representative tumors described in Figure 3 at sacrifice. See also Supplemental Figures 3 and 4.
Since AR antagonists used to treat PCa also promote AR recruitment to androgen response elements (AREs), the effect of RB status on the response to Bic was determined. Bic failed to promote AR occupancy at the PSA enhancer in shCon1 cells compared with the unstimulated (0-hour) condition (Figure 4C), consistent with previous reports showing that Bic primarily induces AR recruitment to the promoter (16). RB depletion altered this dynamic, as AR occupancy was significant at the enhancer region after Bic treatment in shRB1 cells and enriched above that in RB-proficient cells at the PSA promoter (Figure 4C). These data indicate that RB depletion affects AR recruitment in both unliganded and liganded states and alters the transcriptional response to both hormone depletion and hormone stimulation. Since RB is a known transcriptional corepressor, these data implicate downstream RB signaling as linked to AR-mediated transcriptional control.
Elevated E2F1 is specifically associated with AR hyperactivity.
To address the putative mechanisms by which RB signaling affects AR output, the molecular consequence of RB depletion in PCa cells was considered. RB depletion results in differential, tissue-specific cellular responses (3); in PCa cells, RB depletion induces deregulation of selected E2F target genes (e.g., CCNE1 and E2F transcription factor 1 [E2F1]), whereas other E2F-regulated genes (e.g., PCNA and MCM7) remain unchanged (8). Here we observed that E2F1 was upregulated at the mRNA and protein levels in LNCaP and LAPC4 cells after RB depletion (Figure 4D). Importantly, E2F1 deregulation was also observed under conditions of androgen depletion and maximum androgen blockade (i.e., androgen depletion combined with Bic; Figure 4E), and E2F1 deregulation was maintained throughout tumor progression to castration resistance in vivo (Figure 4F). Thus, RB depletion results in aberrant E2F1 expression that is associated with the transition to lethal tumor phenotypes. Interestingly, it was previously reported in breast cancer cells that E2F1 induces expression of the SRC3 coactivator, providing a possible explanation for the observed increase in AR activity (17). However, SRC1 and SRC3 levels were not significantly increased in PCa cells after RB knockdown (Supplemental Figure 4). These collective observations demonstrate that whereas SRC cofactor expression is unaltered, gain of E2F1 expression is associated with RB depletion-mediated AR hyperactivation.
RB depletion induces AR mRNA deregulation and protein accumulation under conditions of clinical relevance.
Given the link between RB depletion and both AR hyperactivation and occupancy, the impact of RB deficiency on AR expression was examined. Strikingly, AR mRNA levels were significantly enhanced in RB-deficient compared with RB-proficient LNCaP-derived cells (Figure 5A). This was unexpected; to our knowledge, no previous link between endogenous AR gene regulation and the cell cycle machinery has been reported, and little is currently understood regarding the mechanisms that control AR gene transcription. RB knockdown resulted in markedly enhanced AR protein levels (Figure 5A), demonstrating the cellular consequence of the observed change in AR mRNA. Further validation in LAPC4 isogenic model systems of RB depletion confirmed the effect of RB suppression on mRNA and protein levels of AR (Figure 5B). The influence of RB depletion on AR was not limited to PCa, as shRNA directed against RB in MCF-7 and HUH-7 cells also resulted in elevated AR expression (Supplemental Figure 5, A and B). Conversely, depletion of RB family members p107 or p130 exerted no significant effect on AR transcript levels (Supplemental Figure 5C). Thus, RB depletion specifically resulted in upregulated mRNA and protein expression of AR in multiple in vitro model systems. To determine relevance in vivo and in castration resistance, AR expression levels were determined in tumor tissue using xenografts derived as in Figure 3. Transcript and protein levels of AR were significantly enhanced in RB-deficient tumor cells compared with controls after castration (Figure 5C), similar to that observed prior to castration in the LNCaP xenograft and with LAPC4 xenografts (Supplemental Figure 5, D and E). These data establish a link between AR regulation and the RB tumor suppressor and demonstrate that RB signaling controls AR gene expression.
Figure 5. AR expression is induced in RB-deficient cells.
(A) AR mRNA was monitored by qPCR, and AR protein levels by immunoblot, from cells cultured in the presence of androgen (FBS). For qPCR, expression in the shCon1 cohort was set to 1, and results represent triplicate analyses of at least 2–3 independent biological replicates (mean ± SD). *P < 0.05, Student’s t test. For immunoblots, Cdk4 served as a loading control. (B) Studies paralleling those of A were performed in L4-shCon1 and L4-shRB1 cells. (C) Intratumor AR mRNA levels monitored from xenograft studies described in Figure 3A. Immunoblots for intratumor AR protein levels are also shown. See also Supplemental Figure 5.
E2F1 is recruited to the AR regulatory locus and enriched upon RB depletion.
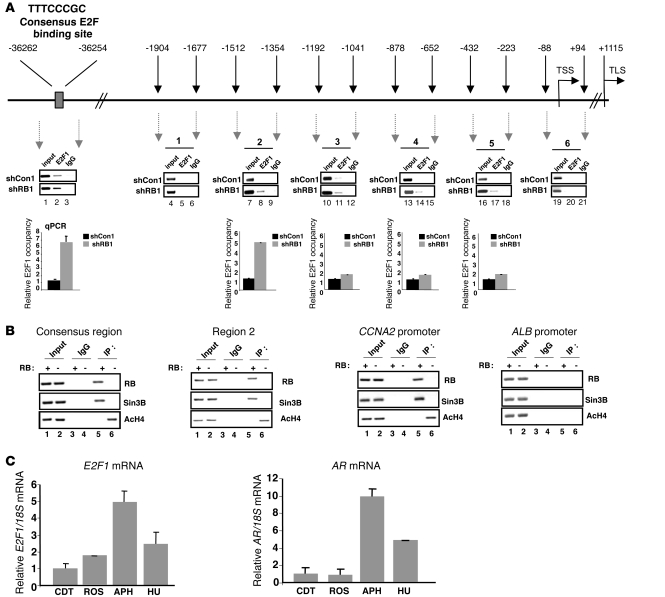
Since RB primarily elicited tumor suppressor function through negative regulation of activator E2Fs, and E2F1 was deregulated by RB suppression in PCa cells (Figure 4), these data put forward the provocative hypothesis that the AR gene is directly or indirectly regulated by activator E2F(s). Examination of the area 5′ to the AR transcriptional start site (TSS) revealed a putative E2F1 consensus binding site in a distal region (Figure 6A). ChIP analyses to detect E2F1 binding were performed, in which recruitment to the cyclin A2 (CCNA2) promoter served as a positive control (Supplemental Figure 6A). As expected, E2F1 occupancy at CCNA2 was enriched in RB-depleted cells. Concurrent analyses showed that E2F1 binds a consensus site in the AR locus, and E2F1 occupancy was significantly enriched in RB-deficient cells (Figure 6A). Subsequent analyses were performed in the 2-kb region 5′ to the TSS, as it was recently shown that E2F1 can use nonconsensus sites to modify transcription (18). Using chromatin sheared to less than 1 kb, E2F1 occupancy proved most significant after RB depletion in region 2 (Figure 6B and data not shown), which indicates that E2F1 also binds the proximal promoter region. Transient reporter assays revealed that activity governed by this region was sensitive to RB status (Supplemental Figure 6B). To assess functional regulation, both proximal and distal sites of E2F1 binding were analyzed for occupancy of a corepressor critical for RB function, Sin3B, and for histone acetylation (Figure 6B). Loss of RB at both regions resulted in commensurate dismissal of Sin3B and allowed for enhanced accumulation of histone H4 acetylation. Positive (CCNA2) and negative (ALB) control ChIPs for RB and Sin3B occupancy are also shown in Figure 6B. Together, these data identify the RB/E2F1 axis as a direct modulator of AR expression.
Figure 6. The RB/E2F1 axis regulates the AR locus, and the kinetics of E2F1 and AR expression are cell cycle dependent.
(A) Schematic of the AR regulatory locus, with the TSS and translational start site (TLS) shown. Results from ChIP analyses of E2F1 binding are shown, including semiquantitative and real-time PCR results, with occupancy in shCon1 cells set to 1. (B) ChIP analyses of RB binding, Sin3B, and histone H4 acetylation at sites enriched for E2F1 after RB depletion, and promoters of CCNA2 and ALB. (C) qPCR analyses of E2F1 and AR from LNCaP cells arrested in G0/G1 (CDT), G1 (ROS), early S-phase (APH), or mid–S-phase (HU). Transcript levels in CDT condition were set to 1. Data represent 3 replicates (mean ± SD); similar results were obtained in at least 3 independent analyses. See also Supplemental Figure 6.
AR expression is regulated as a function of the cell cycle.
E2F1 is regulated in a cell cycle–dependent manner; in G1, CDK-mediated RB inactivation causes E2F1 gene derepression, resulting in acute E2F1 accumulation and creation of a feed-forward loop. In S-phase, cellular mechanisms exist to attenuate this process (19). Since AR mRNA was enhanced by RB depletion, and E2F1 directly associated with AR regulatory locus, we examined the kinetics of AR mRNA expression as a function of the G1-S transition. Cells were synchronized in distinct cell cycle phases using established methods. Hormone depletion (resulting in G0/G1 arrest) was used as a baseline for AR expression; in the presence of androgen, G1-enriched pools of cells were obtained using the CDK inhibitor roscovitine, early S-phase populations using aphidicolin, and mid–S-phase populations using hydroxyurea. Flow cytometry confirmed enrichment of desired populations (Supplemental Figure 6C). As expected, E2F1 mRNA increased from G1 to early S-phase, and declined in later stages of S-phase (Figure 6C). A similar profile was observed with the AR transcript (Figure 6C), which indicates that the kinetics of E2F1 and AR mRNA levels are congruent. This was conserved at the protein level, as AR and E2F1 levels were enriched in early S-phase compared with G1 (Supplemental Figure 6D). Interestingly, in RB-competent CRPC cells resistant to androgen depletion (and therefore maintaining a high S-phase content in charcoal dextran treated serum [CDT]), cell cycle–dependent E2F1 expression was subverted, in that sustained high levels were observed in both G1 and S-phase: only a small increase was detected at the G1-S transition (APH arrested cells) compared with the G1-enriched (ROS) population (Supplemental Figure 6E). Remarkably, AR expression followed this same profile. These data reveal a critical relationship between E2F1 and AR and identify the AR gene as being regulated at the G1-S transition.
Selective activator E2F function controls AR expression.
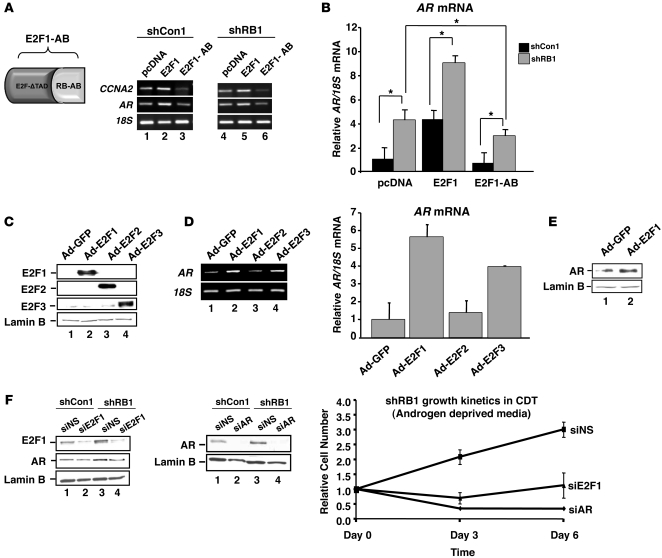
Since the findings herein linked E2F1 to AR regulation, the consequence of this network was determined through selective restoration of RB-mediated E2F suppression. To achieve this, we used a previously described chimeric protein, E2F1-AB (20), which fuses the E2F1 DNA binding and dimerization domains to the minimal transcriptional repression/tumor suppression domain of RB (Figure 7A); thus, E2F1-AB exclusively restores RB activity at sites of activator E2F1 function. In shCon1 cells, wild-type E2F1 induced deregulation of CCNA2, whereas E2F-AB suppressed CCNA2 expression below that of control (Figure 7A), as expected. A similar profile was observed for AR: ectopic E2F1 enhanced AR mRNA above control levels, and E2F1-AB suppressed AR expression. This signaling paradigm was also observed in shRB1 cells (Figure 7A), demonstrating that although these cells harbored elevated AR mRNA, elevation of E2F1 further heightened AR expression. Conversely, E2F1-AB significantly suppressed RB depletion–induced AR mRNA expression (Figure 7B). Therefore, activator E2F positively regulates AR expression, and RB depletion is sufficient to induce E2F-mediated AR deregulation.
Figure 7. Selective restoration of RB activity at sites of E2F1 action suppresses AR expression; AR regulation is E2F selective; and E2F1-induced AR deregulation is required for RB loss–mediated castration resistance.
(A) Schematic of the E2F1-AB chimera. Also shown is RT-PCR for CCNA2 and AR mRNA from cells transfected with plasmids encoding E2F1, E2F1-AB, or control (pcDNA). (B) qPCR analyses of data in A, with AR levels in shCon1/control–transfected (pcDNA) cells set to 1. Data represent triplicate analyses of at least 2–3 independent biological replicates. (C) Immunoblot analyses for transduced E2F1, E2F2, E2F3, and lamin B (loading control) in LNCaP. (D) Parallel semiquantitative RT-PCR and qPCR analysis of AR mRNA in cells from C. Results are plotted relative to control (Ad-GFP–transduced LNCaP) from triplicate replicates (mean ± SD); similar results were obtained in at least 3 independent analyses. (E) AR immunoblot in control and E2F1-transduced cells from C and D. (F) Immunoblot for E2F1 and AR after siRNA-mediated knockdown of E2F1; verification of AR knockdown is also shown. Impact of E2F1 and AR knockdown on castration-resistant proliferation in RB-depleted cells was determined by trypan blue exclusion and cell counting. Data reflect duplicate experiments, each with 3 independent biologic replicates. *P < 0.05, Student’s t test.
Given these findings, the ability of individual activator E2Fs to induce aberrant AR expression was examined under RB-proficient conditions. Virally transduced E2F1, E2F2, and E2F3 was validated by immunoblot (Figure 7C), and the consequence for AR mRNA expression was determined (Figure 7D). E2F1 and E2F3 (albeit to a lesser extent) induced the AR transcript above control levels. No increase was observed with E2F2, which indicates that only a subset of activator E2Fs can modulate AR expression. Interestingly, E2F1 alone was sufficient to elevate AR protein in RB-proficient cells (Figure 7E and data not shown). Conversely, siRNA-mediated knockdown of E2F1 suppressed both AR expression and castration-resistant cell proliferation in RB-depleted cells (Figure 7F). Similar results were observed upon AR knockdown (Figure 7F). These findings not only identify AR as an E2F1-responsive gene and demonstrate that RB suppresses E2F1-mediated AR expression, but establish E2F1-mediated AR deregulation as a critical facet of RB loss–mediated, castration-resistant proliferation.
Clinical evidence for AR regulation by the RB/E2F1 axis in CRPC.
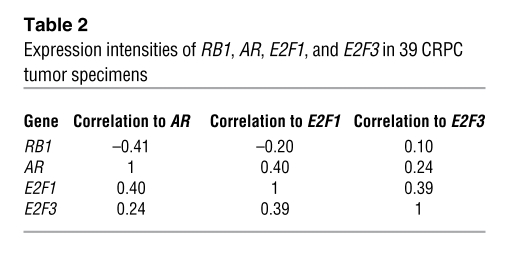
The above findings suggest a paradigm for AR regulation and the development of incurable CRPC, wherein loss of RB function induced E2F1-mediated deregulation of AR and resultant disease progression. To challenge this hypothesis, the relevance of RB and activator E2F status for AR expression was analyzed in mRNA from CRPC specimens. Access to such samples is rare, but gene expression profiling of 39 available specimens showed that loss of RB expression significantly correlated with increased AR mRNA (Figure 8A). Furthermore, a significant relationship was observed between E2F1 and AR transcripts (Figure 8A). These data were even more striking when assessed through clustering based on RB status: a significant inverse relationship was demonstrated between RB and both E2F1 and AR, and a direct correlation between AR and E2F1 was observed (Figure 8B and Table 2). The relationship of AR or RB to E2F3 showed a trend that did not reach significance (Supplemental Figure 7), which suggests that, consistent with the in vitro data (Figure 7), E2F1 is the primary activator E2F controlling AR expression. Combined, these analyses of clinical CRPC specimens strongly support a model wherein AR is under stringent E2F1 control and suggest that perturbations of this process are associated with the transition to lethal, advanced PCa.
Figure 8. AR levels correlate inversely with RB levels and directly with E2F1 levels in CRPC specimens.
(A) Log2 scaled expression ratios of AR versus RB1 and AR versus E2F1 from 39 human CRPC soft tissue metastases. (B) Heatmap depicting AR, RB1, E2F1, and E2F3 expression levels in 39 CRPC tumors. Pearson correlations are provided. See also Table 2 and Supplemental Figure 7.
Table 2 .
Expression intensities of RB1, AR, E2F1, and E2F3 in 39 CRPC tumor specimens
Discussion
The RB/E2F pathway regulates expression of genes that play crucial roles during cellular proliferation and growth and has a well-established role in controlling tumorigenesis. However, the contribution of RB to tumor progression remains largely unexplored. The current study is the first to our knowledge to provide a clinically relevant function for RB in mediating tumor progression by impinging on nuclear receptor expression and output. Our data strongly support a model wherein RB perturbation occurs during PCa progression and promotes lethal tumor phenotypes. This conclusion is supported by 5 key observations. (a) RB loss of function was overrepresented in CRPC and metastatic PCa and was associated with poor outcome. (b) Loss of the RB1 gene locus contributed significantly to RB inactivation in CRPC. (c) RB depletion was sufficient to induce hormone therapy resistance via AR deregulation, exemplified by resistance to hormone therapy in vitro and reduced time to restored AR activity, tumor progression, and castration resistance in vivo. (d) AR gene expression was under stringent E2F1-mediated regulation, and this process was deregulated by RB dysfunction. (e) Perturbation of the RB/E2F/AR axis was frequently observed in CRPC, resulting in enhanced AR expression. Based on these findings, we propose what we believe to be a new paradigm for RB function in protecting against the progression to lethal tumor phenotypes (Figure 9), wherein RB loss promotes PCa progression through a pathway linked to nuclear receptor expression and output. These data underscore the relevance of E2F deregulation for controlling progression to lethal phenotypes in PCa and reveal critical facets of RB function in protecting against tumor progression.
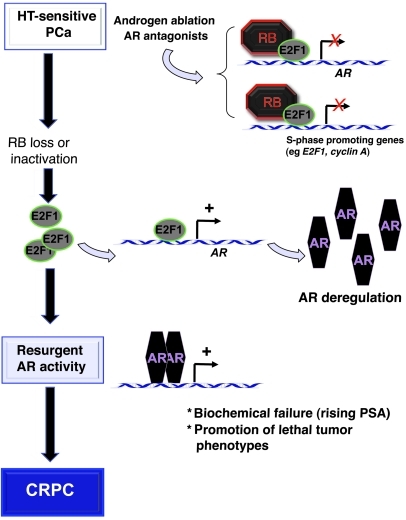
Figure 9. Model for RB loss in controlling tumor progression through nuclear receptor expression and output.
The present data suggest what we believe to be a new model for RB tumor suppressor function in tumor progression. While RB is actively engaged in response to hormone therapy, RB loss induces E2F1-mediated deregulation of the AR locus. This event is sufficient to induce unchecked AR activity and progression to CRPC. This model strongly suggests that RB plays a significant role in tumor progression that is independent of genes directly associated with cell cycle, manifest by controlling nuclear receptor expression, function, and output.
Identification of the RB/E2F/AR network is significant, as AR-directed therapeutics are the first line of intervention for patients with non–organ-confined PCa, and restored AR activity is the major mechanism of transition to the incurable CRPC stage (7). While incompletely understood, resurgent AR activity after hormone therapy can be induced by multiple mechanisms, which include somatic AR mutation, ligand-independent AR activation, alternative splicing of the AR transcript, deregulation of AR cofactors, aberrant AR posttranslational modification, and intratumor androgen production (7, 21). Each has disease relevance, but the most common AR pathway alteration in CRPC is via heightened accumulation of AR itself, as is observed in approximately 30% of CRPCs. Only a fraction can be accounted for by amplification of the AR locus (22), indicating that amplification-independent mechanisms of AR deregulation must exist. The data herein provide a mechanism by which AR overexpression is achieved in CRPC, as mediated by loss of RB function and/or deregulation of E2F1. This is of significance, as forced AR deregulation alone is sufficient to induce CRPC transition in tumor models (23), and high AR levels are strongly correlated with increased risk of death from PCa (24). Thus, the present findings suggest that aberrations in RB promote PCa progression and is associated with lethal tumor phenotypes. Supporting this view, the RB loss signature was significantly overrepresented in CRPC and PCa metastases. These data provide the impetus for future studies linking RB pathway status to PCa morbidity and suggest that RB could be developed as a metric for predicting the response to hormone therapy. Combined, these data establish a paradigm for RB in controlling tumor progression through regulation of nuclear receptor expression and signaling.
Copy number analyses revealed that the RB1 gene locus is lost in significant percent of CRPC tissues, suggesting that RB1 deletion may significantly contribute to the CRPC transition. While the present manuscript was under review, a genome-wide CGH analyses study by Taylor et al. reported that loss of RB1 is enriched in advanced PCa (25). In their study, deletion of the 13q14.2 locus was observed in only 5% of primary tumors but in 37% of the metastatic cases, consistent with loss of RB1 as highly overrepresented in CRPC. Moreover, these data are highly concordant with copy number analyses from the LuCaP series reported here, wherein significant number of the cases demonstrated loss of at least 1 RB1 copy. Notably, loss of RB expression occurred with much higher frequency at the transcript and protein levels, and significant variability in expression was observed in tumors retaining only a single RB1 allele (Figure 2 and Table 1). These data indicate that multiple mechanisms are employed in CRPC to ablate RB expression and function, and ongoing studies are in progress to discern the underlying mechanisms.
Identification of the RB/E2F pathway as a critical regulator of AR gene expression is clinically important, as there is scant knowledge of the factors that control increased AR expression during transition to CRPC. Negative regulation of AR gene expression can be mediated by ErbB kinases (26) or the Purα transcriptional repressor (27). Among the remaining known factors, androgen itself plays a complex role in regulating AR levels. Androgen increases overall AR protein levels and stabilizes the AR transcript, whereas androgen-activated AR can attenuate or enhance AR mRNA levels, dependent on cell context (28–30). Potential positive regulation of AR mRNA expression was reported as a consequence of Sp1, cAMP, NF-κB, and Twist1 (31–34). It is intriguing to speculate that these mechanisms may crosstalk with the RB/E2F1-mediated AR regulation identified here. For example, RB can suppress NF-κB signaling (35), and E2F1 binds to and promotes activity of the Rel-A subunit (36). Twist1 is a candidate RB/E2F1 target gene (37) and has been implicated in PCa progression (38). Sp1 can associate with and enhance E2F1 activity (39), which suggests that Sp1 may assist in E2F1-mediated deregulation of nuclear receptor signaling. Finally, the action of RB in controlling AR appears to be specific, as depletion of RB family members p107 and p130 yielded no effect on AR transcript levels. While the underlying basis of specificity should be discerned, there is little evidence for p107 or p130 alteration in PCa, and to date, no studies have rigorously addressed the impact of RB family member alteration in CRPC. Based on the present data, it will be imperative to determine what factor(s) cooperate with RB loss and/or E2F1 deregulation in promoting aberrant AR expression in CRPC.
The observation that not all activator E2Fs can support AR deregulation was unexpected, and is a basis of current investigation. While E2F1 and E2F3 were each sufficient to enhance AR in vitro, E2F2 proved deficient in this function, and only E2F1 significantly associated with AR mRNA in clinical CRPC. Accordingly, these data suggest that E2F1 is the primary effector of AR gene deregulation mediated by RB loss and is essential for castration-resistant cell proliferation after RB depletion. Previous studies supported the contention that the activator E2Fs harbor overlapping but distinct functions (40). The present study identified a selective function for E2F1 in mediating crosstalk between the RB axis and nuclear receptor signaling in tumor progression. Interestingly, a previous report showed that forced E2F1 overexpression in vitro using a single PCa cell line or 293T cells resulted in reduced AR mRNA expression (41). It is not clear how these results relate to the effects of E2F deregulation as mediated by RB loss, and downstream AR target gene expression was not considered. The same study suggested that E2F1 was associated in tissue samples with a subset of tumors that show low AR immunoreactivity, suggesting that in a subgroup of tumors, the RB/E2F-AR axis may not apply (41); however, abundant evidence demonstrates that the majority of CRPCs are AR positive and express high levels of the receptor (42). Herein, data obtained using multiple in vitro model systems, analysis of E2F1 binding at the endogenous AR locus, in vivo xenograft models of tumor progression, and analysis of human CRPC showed that gain of E2F1 function (as mediated by either E2F1 deregulation or RB depletion) strongly induced AR induction and resultant CRPC progression. Moreover, our findings are consistent with previous studies wherein E2F1 was associated with AR-positive, androgen-independent proliferation in vitro (43). Interestingly, although the RB loss signature only partially overlapped with E2F1-responsive gene expression profiles identified in other cell types, the 2 signatures were highly coregulated in tumor specimens (Supplemental Figure 8). It will be important to discern whether E2F1-independent functions of RB may also contribute to CRPC phenotypes. Nonetheless, the study herein clearly identified RB-loss mediated E2F1 deregulation as sufficient to drive AR-dependent castration resistance.
Finally, it is critical to consider that impairment of RB function and/or E2F1 activation resulted in AR deregulation sufficient to alter AR output and confer castration resistance. Enhanced recruitment of AR to target gene loci was observed in both the presence and the absence of ligand. While elevated AR may be sufficient to explain the gain of AR function, several findings suggest the existence of a possible feed-forward loop between E2F1 and AR. First, androgens induce G1 progression and E2F1 expression in vitro (44, 45). Second, we demonstrated that AR function was altered in CRPC, as the receptor was found to occupy what we believe to be new sites in CRPC cells, many of which are associated with mitosis (15). As reported herein, RB depletion also promoted enhanced occupancy at CPRC-specific sites of AR action. Strikingly, several of these (e.g., UBE2C, CDC20, and CDK1) have previously been identified as regulated by E2F1 (46–48), which suggests the provocative hypothesis that E2F1 may both control AR expression and act in concert with AR to alter downstream gene regulation. Consistent with this hypothesis, AR is known to bind the second intron of the AR gene through an ARE half-site (15). Preliminary studies indicate that E2F1 may co-occupy this site under selected conditions (our unpublished observations), providing some indication that E2F1 may not only regulate AR levels, but could modify AR function on chromatin. Ongoing studies will address the impact of cell cycle position on AR function, the relevance of E2F1 cooperation for this event, and the interplay between AR and E2F1 on regulatory loci of cancer relevance.
In summary, the findings herein present a paradigm for RB function in protecting against tumor progression and suggest that disruption of the RB/E2F1-AR network identified herein is sufficient to promote lethal tumor phenotypes of clinical relevance. These findings identify unexpected actions of E2F1 as a regulator of therapeutic resistance and unmask a mechanism to explain AR deregulation in human disease. Together, the present results provide understanding of RB/E2F1 function, demonstrate the importance of discerning RB function outside canonical cell cycle control, and reveal potential new avenues of therapeutic intervention.
Methods
Reagents and plasmids.
Reagents were obtained as follows: DHT from Sigma-Aldrich; Bic from AstraZeneca; roscovitine, aphidicolin, and hydroxyurea from Calbiochem. E2F viral constructs were a gift of J. Nevins (Duke University, Durham, North Carolina, USA). Plasmids encoding E2F1 and E2F1-AB were previously described (20). Cells were transfected as previously described (44). E2F1 and AR knockdown strategies are described in Supplemental Methods.
Cell lines and xenografts.
C4-2, LNCaP, shRB1, shCon1, LAPC4, L4-shRB1, and L4-shCon1 cells were previously described (8, 49). In vitro growth assays were performed as previously described (8), using trypan blue exclusion and a hemacytometer. For xenograft studies, 4 × 106 cells were combined with Matrigel (BD) and inoculated s.c. into the flanks of NCR/nu/nu (athymic) male mice. Tumor volumes were measured weekly with calipers, serum PSA levels were determined, and PSA doubling times were calculated as previously described (50). In shRB1 and L4-shRB1 xenografts, maintenance of RB knockdown in vivo was verified by quantitative PCR (qPCR) analysis of the human RB1 transcript in tumors at sacrifice, using mouse cDNA as a negative control. Tumors in which RB1 silencing was maintained were selected for study. Animal studies were conducted in accordance with the principles and procedures outlined by the NIH guidelines and the IACUC of Thomas Jefferson University.
Human tumor tissue acquisition.
The tissue samples used in this study were obtained from 44 patients who had died from advanced PCa and who underwent a rapid autopsy performed under the aegis of the Prostate Cancer Donor Program at the University of Washington. This program has been previously described and has received local IRB approval (51). Metastatic liver, lymph nodes, and bone cores were fixed in 10% buffered formalin, processed, and stored in paraffin as described previously (51). Briefly, grossly evident soft tissue metastases were removed, and bone cores were obtained using a cordless drill with attached trephine (11-mm-diameter drill bit) from 20 predetermined sites. The IRB of the University of Washington Medical Center approved all procedures, and all subjects signed written informed consent.
RB1 transcript and copy number analyses.
Freshly frozen samples of 22 LuCaP PCa xenografts were processed as described previously (13) to extract total RNA, which was amplified 1 round and hybridized to Agilent 44K whole human genome expression oligonucleotide microarrays (Agilent Technologies). Probe labeling and hybridization was performed following the Agilent-suggested protocols, and fluorescent array images were collected using the Agilent DNA microarray scanner G2565BA. Agilent Feature Extraction software was used to grid, extract, and normalize data. Normalized cy3 channel values for the RB1 probe on the array are reported. To determine genome copy number variation, samples were verified to contain greater than 80% tumor cells by histology, and DNA was extracted using a DNAeasy Blood and Tissue Kit (Qiagen). DNA was amplified and hybridized to Infinium HD Human 660W-Quad_v1_A BeadChips (Illumina) in the Fred Hutchinson Cancer Center Genomics Resource according to the manufacturer’s instructions. DNA copy number was determined using Genome Studio (Illumina) with the Genotyping and cnvPartition plug-ins.
RB IHC analyses.
Tissue microarrays of fixed paraffin-embedded tissues from 44 rapid autopsy patients (consisting of 4 tissue microarray slides with 2 replicate cores per site) and 22 LuCaP PCa xenograft lines (consisting of 1 tissue microarray slide with 2 replicate cores per line) were used for IHC analyses. 5-μm sections of the tissue microarrays were deparaffinized, antigen retrieval was performed in 10 mM EDTA (pH 9) for 10 minutes in a pressure cooker, and slides were incubated with 3% H2O2 for 10 minutes, then blocked with avidin/biotin blocking solution (Vector Laboratories) for 30 minutes and incubated in a 5% chicken/goat/horse serum solution for 2 hours. Sections were incubated with anti-RB antibody (MS-107-P0, Thermo Scientific) at a dilution of 4 μg/ml overnight at 4°C. Negative control slides were incubated with mouse anti-MOPC21 (generated from a hybridoma obtained from ATCC) at the same concentration as the primary antibody. Slides were then incubated with horse anti-mouse biotinylated secondary antibody (1:150, Vector Laboratories) for 30 minutes, developed using Vectastain ABC (Vector Laboratories) and stable DAB (Invitrogen) counterstained with hematoxylin, dehydrated, and mounted with Cytoseal XYL (Richard Allan Scientific). In tumors with loss of RB immunostaining, nuclear staining in endothelial and stromal cells served as a positive internal control. Immunostaining was assessed as described in Table 1.
Antibodies and immunoblots.
Antibodies were used against RB (for immunoblot, 554136, BD Biosciences; for ChIP, Ab-1 [1F8], Thermo Scientific), Lamin B (sc-6217, Santa Cruz Biotechnology Inc.), AR (N-20, Santa Cruz Biotechnology Inc.), E2F1 (sc-193, Santa Cruz Biotechnology Inc.), E2F2 (sc-9967, Santa Cruz Biotechnology Inc.), E2F3 (sc-878, Santa Cruz Biotechnology Inc.), cdk-4 (sc-601, Santa Cruz Biotechnology Inc.), AcH4 (06-866, Upstate), and Sin3B (sc-768, Santa Cruz Biotechnology Inc.). Immunoblot analyses and growth assays were performed as described previously (8).
RNA and PCR analyses.
RNA was isolated and cDNA generated as previously described (52). Q-PCR was performed with an ABI Step-One apparatus using Power SYBR Green Master Mix and primers described in Supplemental Table 2.
ChIP assay.
Cells were cultured in steroid-free (CDT) media for 72 hours and stimulated with 0.1% EtOH, 10 nM DHT, or 10 μM Bic as indicated. ChIP analyses and PCR were performed as previously described (52, 53). Quantification of the recovered DNA products was also determined by qPCR. Input and recovered products after ChIP were normalized to the respective negative control (IgG) using the formula ΔCt = Cttarget product or input – CtIgG. Comparative ΔΔCt values (difference between target product and input ΔCts) were used to calculate fold enrichment (2–ΔΔCt) over input. Primers are either previously described (15, 16) or shown in Supplemental Table 3.
Flow cytometry.
LNCaP and C4-2 cells were cultured in FBS-containing media or CDT media, respectively, and treated with roscovitine (5 μg/ml), aphidicolin (2 μg/ml), or hydroxyurea (1 mM) for 24 hours. Cells were then harvested and subjected to flow cytometry using a BD FACSCalibur and FlowJo analyses (Tree Star) as described previously (44). See Supplemental Methods for details.
Analyses of RB loss signature and recurrence-free survival.
The RB loss signature consists of 159 previously identified genes indicative of RB loss in other model systems, as previously described (9, 12). The RB loss signature has been was analyzed with previously described microarray datasets (54, 55), for which survival data was available for 1 set. See Supplemental Methods for details.
AR, RB, E2F1, and E2F3 expression in human CRPC prostate tumors.
Whole human genome expression oligonucleotide microarrays (Agilent) were used to profile 39 human castration-resistant soft tissue metastases of prostate adenocarcinomas (56) for depicting expression of AR, RB, E2F1, and E2F3. See Supplemental Methods for details.
Statistics.
Statistical analyses for comparison of RB IHC score and RB1 transcript levels to RB1 locus copy number was performed using ANOVA with Bonferroni multiple-comparison tests. All other results were analyzed using the 2-tailed Student’s t test or Mann-Whitney test. For all analyses, a P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
K.E. Knudsen was supported by NIH grants CA099996 and CA116777 and by the Prostate Cancer Foundation. Additional NIH grant support was provided to A. Sharma (PC073287), E.S. Knudsen (CA129341), and P.S. Nelson and C. Morrissey (Pacific Northwest SPORE P50CA097186 and RC1CA146849). We thank R. Vessella, D. Johnson, A. Biswas, L. True, E. Gelmann, S. Shah, S. Balasubramaniam, R. Dumpit, R. Den, the K.E. Knudsen lab, and the patients, families, and clinicians in the Rapid Autopsy Program at University of Washington.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(12):4478–4492. doi:10.1172/JCI44239.
See the related Commentary beginning on page 4179.
References
- 1.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8(9):671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitaker LL, Su H, Baskaran R, Knudsen ES, Wang JY. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol Cell Biol. 1998;18(7):4032–4042. doi: 10.1128/mcb.18.7.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8(9):714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavenee WK, et al. Genetic origin of mutations predisposing to retinoblastoma. Science. 1985;228(4698):501–503. doi: 10.1126/science.3983638. [DOI] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1(3):222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 6.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. . J Clin Oncol. 2005;23(32):8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 7.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15(15):4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma A, et al. Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells. Cancer Res. 2007;67(13):6192–6203. doi: 10.1158/0008-5472.CAN-06-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markey MP, et al. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62(22):6587–6597. [PubMed] [Google Scholar]
- 10.Bosco EE, et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117(1):218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayhew CN, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133(3):976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Markey MP, et al. Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function. Oncogene. 2007;26(43):6307–6318. doi: 10.1038/sj.onc.1210450. [DOI] [PubMed] [Google Scholar]
- 13.Sun S, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120(8):2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan CJ, et al. Persistent prostate-specific antigen expression after neoadjuvant androgen depletion: an early predictor of relapse or incomplete androgen suppression. Urology. 2006;68(4):834–839. doi: 10.1016/j.urology.2006.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9(3):601–610. doi: 10.1016/S1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 17.Mussi P, Yu C, O’Malley BW, Xu J. Stimulation of steroid receptor coactivator-3 (SRC-3) gene overexpression by a positive regulatory loop of E2F1 and SRC-3. Mol Endocrinol. 2006;20(12):3105–3119. doi: 10.1210/me.2005-0522. [DOI] [PubMed] [Google Scholar]
- 18.Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18(11):1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sellers WR, Rodgers JW, Kaelin WG., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci U S A. 1995;92(25):11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery RB, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68(11):4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visakorpi T, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9(4):401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 23.Chen CD, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 24.Donovan MJ, et al. Androgen receptor expression is associated with prostate cancer-specific survival in castrate patients with metastatic disease. BJU Int. 2010;105(4):462–467. doi: 10.1111/j.1464-410X.2009.08747.x. [DOI] [PubMed] [Google Scholar]
- 25.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai C, Portnoy DC, Wang H, Jiang X, Chen S, Balk SP. Androgen receptor expression in prostate cancer cells is suppressed by activation of epidermal growth factor receptor and ErbB2. Cancer Res. 2009;69(12):5202–5209. doi: 10.1158/0008-5472.CAN-09-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang LG, et al. Androgen receptor overexpression in prostate cancer linked to Pur alpha loss from a novel repressor complex. Cancer Res. 2008;68(8):2678–2688. doi: 10.1158/0008-5472.CAN-07-6017. [DOI] [PubMed] [Google Scholar]
- 28.Yeap BB, Krueger RG, Leedman PJ. Differential posttranscriptional regulation of androgen receptor gene expression by androgen in prostate and breast cancer cells. Endocrinology. 1999;140(7):3282–3291. doi: 10.1210/en.140.7.3282. [DOI] [PubMed] [Google Scholar]
- 29.Krongrad A, Wilson CM, Wilson JD, Allman DR, McPhaul MJ. Androgen increases androgen receptor protein while decreasing receptor mRNA in LNCaP cells. Mol Cell Endocrinol. 1991;76(1–3):79–88. doi: 10.1016/0303-7207(91)90262-Q. [DOI] [PubMed] [Google Scholar]
- 30.Wiren KM, Zhang X, Chang C, Keenan E, Orwoll ES. Transcriptional up-regulation of the human androgen receptor by androgen in bone cells. Endocrinology. 1997;138(6):2291–2300. doi: 10.1210/en.138.6.2291. [DOI] [PubMed] [Google Scholar]
- 31.Mizokami A, Yeh SY, Chang C. Identification of 3’,5’-cyclic adenosine monophosphate response element and other cis-acting elements in the human androgen receptor gene promoter. Mol Endocrinol. 1994;8(1):77–88. doi: 10.1210/me.8.1.77. [DOI] [PubMed] [Google Scholar]
- 32.Shiota M, et al. Castration resistance of prostate cancer cells caused by castration-induced oxidative stress through Twist1 and androgen receptor overexpression. Oncogene. 2010;29(2):237–250. doi: 10.1038/onc.2009.322. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, et al. NF-kappaB regulates androgen receptor expression and prostate cancer growth. Am J Pathol. 2009;175(2):489–499. doi: 10.2353/ajpath.2009.080727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faber PW, van Rooij HC, Schipper HJ, Brinkmann AO, Trapman J. Two different, overlapping pathways of transcription initiation are active on the TATA-less human androgen receptor promoter. The role of Sp1. J Biol Chem. 1993;268(13):9296–9301. [PubMed] [Google Scholar]
- 35.Tamami M, Lindholm PF, Brady JN. The retinoblastoma gene product (Rb) induces binding of a conformationally inactive nuclear factor-kappaB. J Biol Chem. 1996;271(40):24551–24556. doi: 10.1074/jbc.271.40.24551. [DOI] [PubMed] [Google Scholar]
- 36.Zheng C, et al. E2F1 Induces tumor cell survival via nuclear factor-kappaB-dependent induction of EGR1 transcription in prostate cancer cells. Cancer Res. 2009;69(6):2324–2331. doi: 10.1158/0008-5472.CAN-08-4113. [DOI] [PubMed] [Google Scholar]
- 37.McClellan KA, et al. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol Cell Biol. 2007;27(13):4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwok WK, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65(12):5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 39.Lin SY, Black AR, Kostic D, Pajovic S, Hoover CN, Azizkhan JC. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16(4):1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Black EP, Hallstrom T, Dressman HK, West M, Nevins JR. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc Natl Acad Sci U S A. 2005;102(44):15948–15953. doi: 10.1073/pnas.0504300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis JN, et al. Elevated E2F1 inhibits transcription of the androgen receptor in metastatic hormone-resistant prostate cancer. Cancer Res. 2006;66(24):11897–11906. doi: 10.1158/0008-5472.CAN-06-2497. [DOI] [PubMed] [Google Scholar]
- 42.Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61(9):3550–3555. [PubMed] [Google Scholar]
- 43.Libertini SJ, Tepper CG, Guadalupe M, Lu Y, Asmuth DM, Mudryj M. E2F1 expression in LNCaP prostate cancer cells deregulates androgen dependent growth, suppresses differentiation, and enhances apoptosis. Prostate. 2006;66(1):70–81. doi: 10.1002/pros.20314. [DOI] [PubMed] [Google Scholar]
- 44.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. . J Biol Chem. 1998;273(32):20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 45.Schayek H, et al. Transcription factor E2F1 is a potent transactivator of the insulin-like growth factor-I receptor (IGF-IR) gene. Growth Horm IGF Res. 2010;20(1):68–72. doi: 10.1016/j.ghir.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Hardy K, et al. Transcriptional networks and cellular senescence in human mammary fibroblasts. Mol Biol Cell. 2005;16(2):943–953. doi: 10.1091/mbc.E04-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21(9):2112–2123. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- 48.Yu F, Megyesi J, Safirstein RL, Price PM. Involvement of the CDK2-E2F1 pathway in cisplatin cytotoxicity in vitro and in vivo. Am J Physiol Renal Physiol. 2007;293(1):F52–59. doi: 10.1152/ajprenal.00119.2007. [DOI] [PubMed] [Google Scholar]
- 49.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer. 1994;57(3):406–412. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- 50.Wetherill YB, et al. Bisphenol A facilitates bypass of androgen ablation therapy in prostate cancer. Mol Cancer Ther. 2006;5(12):3181–3190. doi: 10.1158/1535-7163.MCT-06-0272. [DOI] [PubMed] [Google Scholar]
- 51.Roudier MP, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34(7):646–653. doi: 10.1016/S0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 52.Link KA, et al. Targeting the BAF57 SWI/SNF subunit in prostate cancer: a novel platform to control androgen receptor activity. Cancer Res. 2008;68(12):4551–4558. doi: 10.1158/0008-5472.CAN-07-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103(6):843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 54.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8(5):393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holcomb IN, et al. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer Res. 2009;69(19):7793–7802. doi: 10.1158/0008-5472.CAN-08-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17(5):1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.