Abstract
The magnitude and durability of immunity to human cytomegalovirus (HCMV) following natural infection is compromised by the presence of immune modulation genes that appear to promote evasion of host clearance mechanisms. Since immunity to HCMV offers limited protection, rational design of effective vaccines has been challenging. In this issue of the JCI, Slavuljica and colleagues employ techniques to genetically modify the highly related mouse CMV (MCMV), in the process generating a virus that was rapidly cleared by NK cells. The virus functioned as a safe and highly effective vaccine. Demonstration of the ability to engineer a safe and highly effective live virus vaccine in a relevant rodent model of CMV infection may open the door to clinical trials of safer and more immunogenic HCMV vaccines.
The urgent need for a vaccine against human CMV
Human CMV (HCMV) is an important cause of disease in immunologically compromised individuals, including recipients of solid organ and hematopoietic stem cell transplants and patients with advanced HIV disease. At greatest risk for HCMV-associated injury, however, is the developing fetus. HCMV is the most common agent of congenital viral infection in the United States, and among all infectious diseases is the most common cause of childhood neurological disability, including deafness, in the developed world (1, 2). Since severe and symptomatic congenital HCMV infections can be associated with a lifetime of disability, the economic burden associated with this infection is striking. When the Institute of Medicine was commissioned to prioritize vaccine development for the new millennium based on, among other factors, quality-adjusted life years (a marker of economic benefit), a vaccine for HCMV was ranked “head-and-shoulders” above all other potential new vaccines with respect to overall cost-effectiveness (3).
Although the need for an HCMV vaccine is compelling, it is less clear to whom such a vaccine should be administered, and what the constituents of such a vaccine should be. The correlates of protective immunity remain undefined, both for the nonpregnant individual and for the developing fetus. Subunit vaccines, typically based on recombinant expression of key targets of humoral and cellular immune responses to HCMV infection, have been evaluated in clinical trials, as have live-attenuated vaccines (4). Until recently, clinical trials have yielded little information about the potential for protective efficacy, largely because most studies have focused on the end points of safety and immunogenicity and not the end point of protection against HCMV infection. A breakthrough in the HCMV vaccine field came in 2009, when, for the first time, a vaccine was shown to be effective against acquiring primary infection (5). This vaccine was based on a purified, recombinant formulation of the major envelope glycoprotein B, administered with a proprietary adjuvant known as MF59. In a phase II, placebo-controlled study, 18 of 225 (8%) women who received gB/MF59 vaccine acquired a primary HCMV infection in the one-year study window, compared with 31 of 216 in the placebo group (14%), for an overall vaccine efficacy of 50% (5). These data represent a major advance in the field, since they are the first evidence of efficacy for prevention against infection for any HCMV vaccine. However, questions remain about whether an immune response targeting a single HCMV protein can be sufficient for long-term disease control through vaccination and if a live-attenuated vaccine might represent a better solution, by conferring the broadest possible blanket of immunity against multiple HCMV gene products.
HCMV genes compromise immunity and complicate vaccine design
One of the most remarkable aspects of the biology of HCMV that has only recently begun to be elucidated is the role that multiple virus-encoded immune modulation genes play in the establishment (or lack thereof) of long-term immunity to this infection (6). Many of these immune modulation genes appear to be homologs of normal cellular immune effectors, presumably having been “hijacked” from the host genome during the course of coevolution of virus and host. Several of these gene products interfere with host adaptive immune responses, with the apparent goal of enabling immune evasion; others appear to play a proinflammatory role, seemingly to promote inflammation that presumably could facilitate widespread dissemination of CMV in the infected host. These gene products, which include cytokines, chemokines, GPCR homologs, and inhibitors of MHC class I and class II antigen presentation, are summarized in Table 1.
Table 1 .
HCMV immune modulation genes
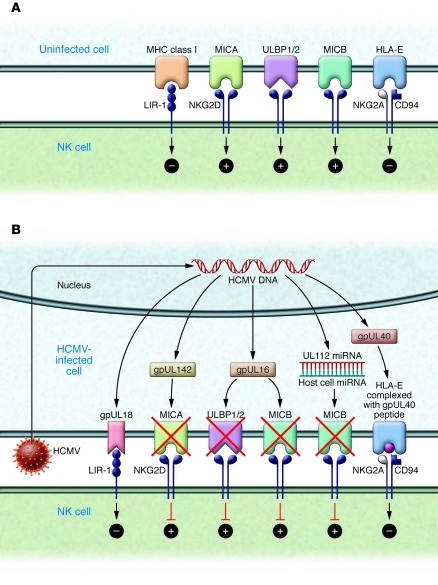
In addition to gene products that appear to modulate adaptive immune responses, HCMV also encodes proteins that impact innate responses, particularly the function of NK cells (Figure 1 and ref. 7). HCMV has evolved several genes that interact with NK cell–mediated pathways, modifying pathways of both inhibition and activation. One such protein, gpUL18, is encoded by the UL18 gene and appears to exert its evasive effect by binding to the NK cell inhibitory receptor leukocyte Ig-like receptor–1 (LIR-1) with a higher affinity than does the host MHC class I molecule (8, 9). Recently it has been shown that cells expressing another HCMV MHC class I homolog, the product of the UL142 gene, are protected from NK cell lysis (10). The NK cell evasion effect is related to the ability of gpUL142 to downregulate MHC class I polypeptide–related sequence A (MICA), which is a ligand for NK cell activation receptor NK group 2, member D (NKG2D) (11). A number of other CMV gene products, including those encoded by UL40, UL83, and UL141, also appear to function as NK cell evasins. The UL40 protein contains an epitope in its leader sequence that binds to HLA-E and is presented at the surface of infected cells. The gpUL40 peptide/HLA-E complex binds to CD94/NKG2A, resulting in NK cell inhibition (7). Remarkably, not all HCMV NK cell evasion genes function by encoding proteins: some gene products can promote evasion of innate immunity at the RNA level. Recently, a novel mechanism of NK cell evasion was attributed to an HCMV microRNA encoded by the HCMV UL112 gene (12). This effect appears to require interaction between the UL112 miRNA and a cellular miRNA (13).
Figure 1. Impact of HCMV gene products on NK cell–mediated pathways of inhibition and activation.
(A) Receptor-ligand interactions modulating NK cell function. MHC class I interacts with the inhibitory receptor LIR-1, and HLA-E interacts with the CD94/NKG2A receptor; both interactions result in inhibition of NK cell function. The ligands MICA, ULBP1/2, and MICB all interact with the NKG2D receptor, resulting in NK cell activation. (B) HCMV genes encode proteins that modify NK cell responses. NK cell evasion can be mediated by the HCMV UL18 gene product (gpUL18), a ligand for the inhibitory receptor LIR-1; the UL16 gene product (gpUL16), which downregulates the NKG2D activating ligands MICB and ULBP1/2; the UL142 gene product (gpUL142), which downregulates the NKG2D ligand MICA; an epitope from the leader peptide segment of the UL40-encoded protein, which loads HLA-E, driving an inhibitory signal to the NK cell via the CD94/NKG2A receptor; and a miRNA from the UL112 gene, which interacts with a cellular miRNA to inhibit MICB expression by preventing translation of MICB mRNA.
What is the clinical significance of the presence of this complex array of HCMV immune modulation genes? Key to the problem of HCMV immunity is the issue of reinfection. Mounting evidence indicates that people who have cleared primary viral infection and have latent, clinically quiescent infections can be reinfected with novel strains of HCMV. Such reinfections can occur in women of child-bearing age, even in the face of long-standing immunity, and can lead to symptomatic transmission to the developing fetus, with its attendant morbidity (14–16). The role that immune evasion genes play in the context of in vivo infection was recently authenticated in rhesus macaques (17). In this study, macaques that had been naturally infected with rhesus CMV (RhCMV, the rhesus macaque homolog of HCMV) were infected with a recombinant RhCMV virus expressing the SIV Gag protein. Despite their preexisting immunity, all inoculated macaques developed SIV Gag–specific T cell responses and shed the recombinant virus for up to three years after challenge. The ability of this virus to superinfect immune macaques, however, was abrogated by deletion of the RhCMV-encoded inhibitors of MHC class I antigen presentation. The authors pessimistically concluded that this ability to evade CTL-mediated immune memory “renders it unlikely” that an effective vaccine against HCMV is an “achievable goal” (17).
Building it better: engineering enhanced effectiveness in a CMV vaccine
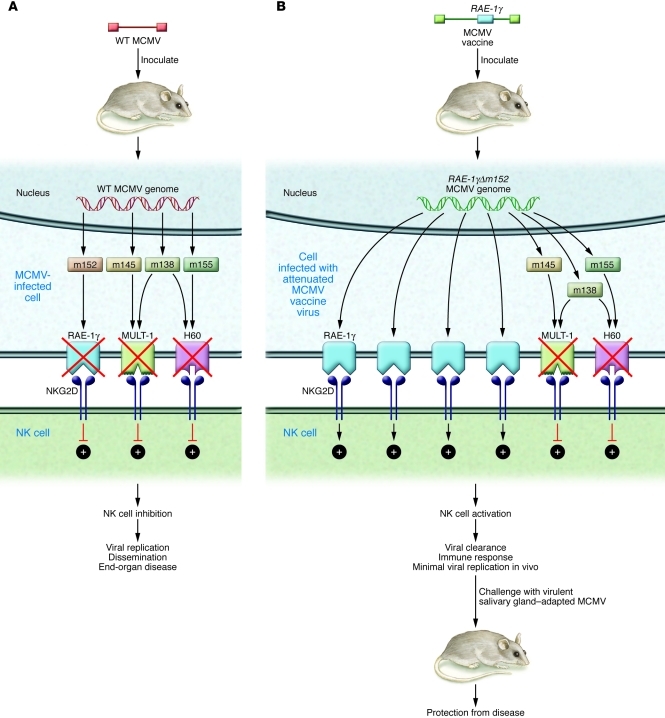
In light of HCMV-mediated immune evasion mechanisms, what strategies could enable development of an effective vaccine? Does the presence of immune evasion genes make the effort to develop a live-attenuated vaccine hopeless? Or, do we have the technology to bioengineer a vaccine against HCMV that is “better” than natural immunity? The work of Slavuljica and colleagues reported in this issue of the JCI suggests that the answer, at least for a related CMV (mouse CMV [MCMV]), may be “yes” (18). In this study, MCMV was studied in a vaccine/challenge model. Like HMCV, MCMV encodes a wide array of immune evasion genes: some are highly homologous to those encoded by HCMV, and others are unique and species-specific. In particular, MCMV encodes a variety of gene products that interfere with NK cell–mediated clearance following infection, either by downregulating ligands for NKG2D or by encoding homologs of MHC class I molecules capable of binding NK cell inhibitory receptors. Examples include the m145-encoded protein, which decreases expression of the NKG2D ligand MULT-1 (19); the m155-encoded glycoprotein, which downregulates expression of the NKG2D ligand H60 (20); and the m152-encoded glycoprotein, which downregulates expression of proteins of the RAE-1 family of NKG2D ligands (21). The m138 protein functions by downmodulating multiple NKG2D ligands, including MULT-1 and H60 (22). MCMV mutants lacking these genes are attenuated in vivo, but if mice are depleted of NK cells prior to viral challenge, replication of MCMV can be restored.
Using this knowledge, how did Slavuljica and colleagues design a vaccine with the potential to confer more robust immune responses than natural infection (18)? The key was to modify MCMV in such a fashion as to override inhibitory signals delivered by self-MHC class I proteins or potentiated by viral proteins that downregulate NKG2D ligands (Figure 2). Implicit in this strategy is the hypothesis that not only could such a vaccine have an outstanding safety profile, due to improved innate immune clearance following inoculation, but that it also could potentially be equally or more immunogenic than “natural” infection with wild-type MCMV, by virtue of increased NK cell activation leading to stronger adaptive immune responses. To engineer such a viral vaccine, Slavuljica and colleagues utilized the powerful mutagenesis techniques enabled by the availability of the MCMV genome cloned as an infectious BAC in Escherichia coli to insert a NKG2D ligand into the MCMV genome (18), an approach that has been exploited in designing other vaccine approaches in the MCMV model (23). The ligand employed in this study was a member of the RAE-1 family of NKG2D ligands, RAE-1γ (18). Insertion of the gene encoding RAE-1γ into the MCMV genome generated a recombinant virus with a profoundly attenuated phenotype in vivo, including in transgenic mice lacking type I interferon receptor and mice that were immune suppressed. Despite this attenuation, the recombinant vaccine efficiently primed and maintained adaptive immune responses, including CD8+ T cell responses, that were comparable to those induced by wild-type MCMV. Intriguingly, in a challenge infection model, mice immunized with the RAE-1γMCMV recombinant resisted challenge with virulent salivary gland–adapted virus, with improved survival compared with mice initially infected with wild-type MCMV, suggesting that expression of the NKG2D ligand RAE-1γ in the context of the recombinant MCMV provides an innate immune stimulus that results in enhanced effectiveness of the adaptive immune response compared with infection with wild-type MCMV. Adoptive transfer of CD8+ T cells from vaccinated mice to immunodepleted, infected mice resulted in protection against disease. In addition to the protective T cell effect, Slavuljica and colleagues identified an important role for antibody in a newborn pup challenge model mimicking neonatal HCMV disease (18). It was noted that pups born to vaccinated, but not control, mice were protected against MCMV infection and disseminated disease following challenge in the newborn period, indicating a role for transplacental antibody in prevention of disease.
Figure 2. Recombinant MCMV overrides NK cell evasion genes, producing highly attenuated but immunogenic and protective vaccine.
(A) Like HCMV, MCMV encodes a variety of gene products that interfere with NK cell–mediated viral clearance by downregulating activating ligands for NKG2D. Examples include the m145 protein, which decreases expression of MULT-1; the m155 glycoprotein, which downregulates H60; and the m152 glycoprotein, which downregulates proteins of the RAE-1 family. The m138 protein functions by downmodulating multiple NKG2D ligands, including MULT-1 and H60. NK cell evasion genes impair activation of NKG2D as indicated, resulting in viremia, disseminated infection, and end-organ disease. (B) Recombinant MCMV attenuated vaccine described by Slavuljica and colleagues (18). Recombinant virus was engineered to express the NKG2D activating ligand RAE-1γ at high levels. This modification overcomes other NK cell evasion genes, resulting in minimal replication in vivo, but eliciting strong cellular and humoral immune responses that provide protection against MCMV disease following challenge with virulent, salivary gland–adapted virus.
Similar observations suggesting an enhanced immunogenicity and protective efficacy for a live-attenuated vaccine engineered to modify the impact of viral immune modulation genes have been made in vaccine studies in another rodent CMV model — the guinea pig CMV (GPCMV) (24). In this model, deletion of three MHC class I homologs, presumed NK cell evasins, was achieved using BAC technology. The resulting vaccine virus was highly attenuated in vivo, but vaccination was highly immunogenic and protective. Similar to the RAE 1-γ vaccine generated by Slavuljica and colleagues (18), these results suggested that the safety and efficacy of a live-attenuated HCMV vaccine could be enhanced by modification of the virus/NK cell interaction: in the case of the MCMV vaccine, by engineering a NKG2D ligand into the viral genome; and in the case of the GPCMV vaccine, by removing putative class I “decoy” genes that mediate evasion of NK cell–mediated clearance (24).
Time for human clinical trials? What’s the next step?
In light of these encouraging results in the MCMV and GPCMV models (18, 24), it seems reasonable to proceed with phase I studies of similarly constructed live-attenuated HCMV vaccines. Unfortunately, regulatory bodies have expressed some reluctance in moving forward with studies of live-attenuated HCMV vaccines, because of perceived long-term theoretical health risks associated with such vaccinations (25). In this context, the latent infection observed with the vaccine designed by Slavuljica and colleagues (18), which they hypothesized may help “boost” long-term immune responses during episodes of abortive reactivation, may not be advantageous for an HCMV vaccine. Additional studies are warranted in rodent models to try to optimize both attenuation and immunogenicity of live-attenuated vaccines. However, clinical trials of candidate live-attenuated HCMV vaccines should not be delayed until exhaustive evaluations are completed in animal models. The intriguing data reported by Slavuljica and colleagues (18) demonstrating feasibility and efficacy of a rationally engineered vaccine in a relevant animal model should help drive translational studies of analogous approaches in human clinical trials. The exciting advent of molecular technologies to “reengineer” live virus vaccines may be applicable to other live virus vaccines used in clinical practice. These improvements in vaccine technology — aimed at not only improving efficacy but also ensuring safety — should in turn help increase confidence in the safety and vital importance of all vaccines in clinical practice.
Acknowledgments
This work is supported by NIH grants HD038416 and HD044864.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(12):4192–4197. doi:10.1172/JCI45250
See the related article beginning on page 4532.
References
- 1.Bale JF, Miner L, Petheram SJ. Congenital cytomegalovirus infection. Curr Treat Options Neurol. 2002;4(3):225–230. doi: 10.1007/s11940-002-0039-8. [DOI] [PubMed] [Google Scholar]
- 2.Cheeran MC, Lokensgard JR, Schleiss MR. Neuropathogenesis of congenital cytomegalovirus infection: disease mechanisms and prospects for intervention. Clin Microbiol Rev. 2009;22(1):99–126. doi: 10.1128/CMR.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stratton KR, Durch JS, Lawrence RS.Vaccines for the 21st Century: A Tool for Decision Making: Report of the Committee to Study Priorities for Vaccine Development . Washington, DC, USA: Institute of Medicine; 1999. [Google Scholar]
- 4.Schleiss MR. Cytomegalovirus vaccine development. Curr Top Microbiol Immunol. 2008;325:361–382. doi: 10.1007/978-3-540-77349-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pass RF, et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med. 2009;360(12):1191–1199. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller-Kittrell M, Sparer TE. Feeling manipulated: cytomegalovirus immune manipulation. Virol J. 2009;6:4. doi: 10.1186/1743-422X-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun JC, Lanier LL. The natural selection of herpesviruses and virus–specific NK cell receptors. Viruses. 2009;1(3):362–382. doi: 10.3390/v1030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Bjorkman PJ. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc Natl Acad Sci U S A. 2008;105(29):10095–10100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Occhino M, et al. Dissecting the structural determinants of the interaction between the human cytomegalovirus UL18 protein and the CD85j immune receptor. J Immunol. 2008;180(2):957–968. doi: 10.4049/jimmunol.180.2.957. [DOI] [PubMed] [Google Scholar]
- 10.Wills MR, et al. Human cytomegalovirus encodes an MHC class I–like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol. 2005;175(11):7457–7465. doi: 10.4049/jimmunol.175.11.7457. [DOI] [PubMed] [Google Scholar]
- 11.Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun. 2006;346(1):175–181. doi: 10.1016/j.bbrc.2006.05.092. [DOI] [PubMed] [Google Scholar]
- 12.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol. 2010;11(9):806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- 14.Ahlfors K, Ivarsson SA, Harris S. Report on a long-term study of maternal and congenital cytomegalovirus infection in Sweden. Review of prospective studies available in the literature. Scand J Infect Dis. 1999;31(5):443–457. doi: 10.1080/00365549950163969. [DOI] [PubMed] [Google Scholar]
- 15.Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med. 2001;344(18):1366–1371. doi: 10.1056/NEJM200105033441804. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto AY, et al. Human cytomegalovirus reinfection is associated with intrauterine transmission in a highly cytomegalovirus-immune maternal population. Am J Obstet Gynecol. 2010;202(3):297.e1–297.e8. doi: 10.1016/j.ajog.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen SG, et al. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science. 2010;328(5974):102–106. doi: 10.1126/science.1185350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slavuljica I, et al. Recombinant mouse cytomegalovirus expressing a ligand for the NKG2D receptor is attenuated and has improved vaccine properties. J Clin Invest. 2010;120(12):4532–4545. doi: 10.1172/JCI43961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krmpotic A, et al. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med. 2005;201(2):211–220. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. . J Exp Med. 2004;200(8):1075–1081. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto AK, Jamieson AM, Raulet DH, Hill AB. The role of NKG2D signaling in inhibition of cytotoxic T-lymphocyte lysis by the murine cytomegalovirus immunoevasin m152/gp40. J Virol. 2007;81(22):12564–12571. doi: 10.1128/JVI.01328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arapovic J, Lenac Rovis T, Reddy AB, Krmpotic A, Jonjic S. Promiscuity of MCMV immunoevasin of NKG2D: m138/fcr-1 down-modulates RAE-1epsilon in addition to MULT-1 and H60. Mol Immunol. 2009;47(1):114–122. doi: 10.1016/j.molimm.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Cicin-Sain L, et al. Targeted deletion of regions rich in immune-evasive genes from the cytomegalovirus genome as a novel vaccine strategy. J Virol. 2007;81(24):13825–13834. doi: 10.1128/JVI.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crumpler MM, Choi KY, McVoy MA, Schleiss MR. A live guinea pig cytomegalovirus vaccine deleted of three putative immune evasion genes is highly attenuated but remains immunogenic in a vaccine/challenge model of congenital cytomegalovirus infection. Vaccine. 2009;27(31):4209–4218. doi: 10.1016/j.vaccine.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Söderberg-Nauclér C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Intern Med. 2006;259(3):219–246. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]