Abstract
Rationale and Objectives
Multimodal imaging techniques for capturing normal and diseased human anatomy and physiology are being developed to benefit patient clinical care, research, and education. In the past, the incorporation of histopathology into these multimodal datasets has been complicated by the large differences in image quality, content, and spatial association.
Materials and Methods
We have developed a novel system, the large-scale image microtome array (LIMA), to bridge the gap between nonstructurally destructive and destructive imaging such that reliable registration between radiological data and histopathology can be achieved. Registration algorithms have been designed to align the multimodal datasets, which include computed tomography, computed micro-tomography, LIMA, and histopathology data to a common coordinate system.
Results
The resulting volumetric dataset provides an abundance of valuable information relating to the tissue sample including density, anatomical structure, color, texture, and cellular information in three dimensions. An image processing pipeline has been established to register all the multimodal data to a common coordinate system.
Conclusion
In this study, we have chosen to use human lung cancer nodules as an example; however, the flexibility of the image acquisition and subsequent processing algorithms makes it applicable to any soft organ tissue. A novel process model has been established to generate cross registered multimodal datasets for the investigation of human lung cancer nodule content and associated image-based representation.
Keywords: Computed tomography, lung adenocarcinoma, histopathology, image registration, multimodal image acquisition
The development of multimodal image acquisition and analysis continues to progress rapidly, driven by the potential to improve diagnostic and therapeutic aspects of medical care. Multimodal imaging, such as the combination of computed tomography (CT) and positron emission tomography, have been shown to improve the ability to identify potential disease states, such as lung nodules, and facilitate the planning of effective treatment approaches (1–5). However, the diagnostic “gold standard” in solid tumor oncology remains histological examination of diseased tissue, which requires invasive tissue sampling.
To understand and exploit all the information captured in noninvasive imaging techniques, accurate registration between the histological truth and the corresponding radiological appearance is required. This is complicated to achieve, because of the large differences in image quality, content, and loss of spatial association. Datasets that incorporate the corresponding histological truth are required to effectively evaluate the diagnostic ability of these systems.
For many years, it has been acceptable to correlate histopathological findings with external imaging observations by general comparison. Kennel et al presented histology sections of mouse lungs next to roughly corresponding computed micro-tomography (micro-CT) and used arrows to highlight suspected tumor foci in the micro-CT data and confirmed foci in the histology images (6). For this case, registration of the datasets may have made a more compelling statement, but it was not necessary to meet the objectives of the study. A prospective pilot study was conducted by Chan et al to correlate tumor size on CT with tumor sizes measured microscopically in non–small-cell lung cancers (7). In this basic study, the resected tumor was manually aligned to the approximate orientation of the in vivo CT imaging plane and transverse sections for histological preparation were made. The boundary from the CT data was then compared to that of the histological sections with the conclusion that CT over-estimated the size of the tumors to a degree greater than what could be accounted for by the significant tissue shrinkage through histological preparation. This study effectively avoids the possibility of creating false matching by using only rigid registration, but in doing so the investigators are unable to make any quantitative conclusions regarding boundary representation. Although the objectives of this study are highly important, it is clear that a methodology with greater precision, repeatability, and accuracy is required.
For more specific comparative studies, such as boundary representation or spatial distribution of tissues, the distortions caused by histological processing must be compensated for in some way. Some studies have investigated the non-rigid alignment of histology to nondestructive data, such as magnetic resonance imaging (MRI) and micro-CT. Clarke et al composed a multimodal mapping of carotid atherosclerotic plaque components (8). The approximate matching MRI and micro-CT slices were located by visual inspection and nonrigidly registered using the Delaunay triangle method. Registration approaches such as these, involve a high risk of introducing false information due to the nonrigid registration of the histology data to a noncorresponding image slice. A spatial reference system is required such that the correct corresponding base image can be located for each warped image.
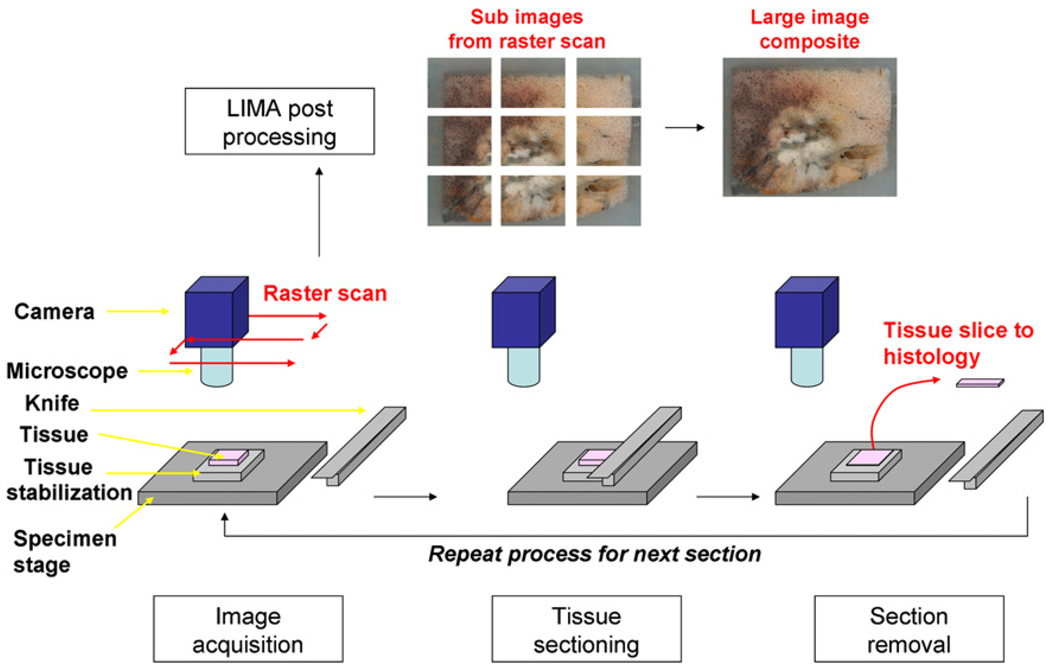
We previously developed a large image microscope array system (LIMA) for whole organ sectioning, which is used in this study as the spatial reference for registration of the datasets to a common coordinate system. Briefly, the LIMA is a purpose-built sectioning and imaging microscopy system consisting of a microscope coupled with a digital camera. The microscope and camera coupling raster scan over the tissue block surface acquiring many overlapping, high-resolution images. After the complete tissue surface is captured, a customized microtome sections the tissue, removing the top surface for histological processing. The process of sequentially imaging and then sectioning the tissue is automated via a computer-interfaced control system. A diagram summarizing this process is presented in Figure 1. Sectioning and imaging of the tissue via the LIMA system produces a volumetric image set in which no distortion of the tissue occurs between sequential images, and the corresponding histological section for each image is known. Hence the LIMA dataset can serve as a reliable basis for the accurate alignment of the histopathological data into the volumetric dataset.
Figure 1.
Summary of the large image microscope array (LIMA) system. This system serves to section the tissue while recording the spatial correspondence between sections by imaging the tissue block before the removal of a section. Multiple subimages are acquired as the camera and microscope coupling rastor scan over the tissue surface. The surface is then sectioned and removed from the tissue block, via a vibrating knife, for subsequent histological processing. This process results in the generation of a color dataset which can be used as the ground truth for linking nondestructive image sets, such as computed tomography to destructive histopathology image sets.
The aim of this study was to develop a process model for the acquisition and registration of multimodal data. The resulting datasets were to include in vivo multidetector CT (MDCT), MDCT scans of the resected lobe both fresh and fixed, and the resected nodule imaged via MDCT, micro–CT, and the LIMA system with associated histology. Through these datasets density, structure, color, and cellular information relating to the specimen can be gained.
The process model developed could be applied to nodules of any organ; however, for this study, the focus was directed to lung cancer nodules. The mortality from lung cancer is higher in both men and women than from any other form of cancer, despite the fact that the most commonly diagnosed cancers are prostate cancer in men and breast cancer in women. MDCT is playing an increasing role in the identification and monitoring of suspicious lung nodules. Some computer-aided detection and diagnosis algorithms based on MDCT data have been developed and achieved promising results. However, a considerable barrier to assisting with diagnosis is that the ground truth of lung nodule structure is not known, significantly inhibiting the development of more sophisticated algorithms with further informative outputs.
MATERIALS AND METHODS
Patients
Between October 2005 and May 2008, 17 patients were identified and consented for participation in the study (8 males and 9 females). Complete lobectomy specimens were required for inclusion into this study because a large airway was required for appropriate inflation and fixation of the tissue. Six of the 17 potential datasets were not collected because of changes in surgical plan or surgical pathology processing that required immediate dissection of the lung nodule. Changes in the surgical plan included a cancellation of surgery or removal of the nodule via wedge resection prior to lobectomy. Lobectomy specimens were obtained for 11 cases (7 adenocarcinomas, 3 squamous cell carcinomas, and 1 neuroendocrine carcinoma) as per the protocol approved by our Institutional Review Board. Table 1 lists the demographics of the 11 patients for which complete datasets were generated.
TABLE 1.
Patient Demographic Information
| Characteristic | ||
|---|---|---|
| Mean | Range | |
| Age (y) | 66 | 51–79 |
| Body mass index | 26 | 21–33 |
| Nodule size from radiology report | ||
| Maximum diameter | 27 | 14–50 |
| Minimum diameter | 19 | 10–15 |
| n | % | |
| Gender | ||
| Male | 2 | 18 |
| Female | 9 | 82 |
| Race | ||
| Caucasian | 10 | 91 |
| African American | 1 | 9 |
| Site | ||
| Right upper lobe | 1 | 9 |
| Right lower lobe | 6 | 55 |
| Left upper lobe | 3 | 27% |
| Left lower lobe | 1 | 9 |
| Diagnosis | ||
| Adenocarcinoma | 7 | 64 |
| Squamous cell carcinoma | 3 | 27 |
| Neuroendocrine carcinoma | 1 | 9 |
| Stage | ||
| Stage IA | 5 | 46 |
| Stage IB | 3 | 27% |
| Stage IIB | 2 | 18% |
| Stage IIIA | 1 | 9 |
| Smoking history | ||
| Never | 1 | 9 |
| <1 pack per week | 2 | 18 |
| 1 pack per week | 6 | 55 |
| 2 pack per week | 1 | 9 |
| 3 pack per week | 1 | 9 |
Data Acquisition Process
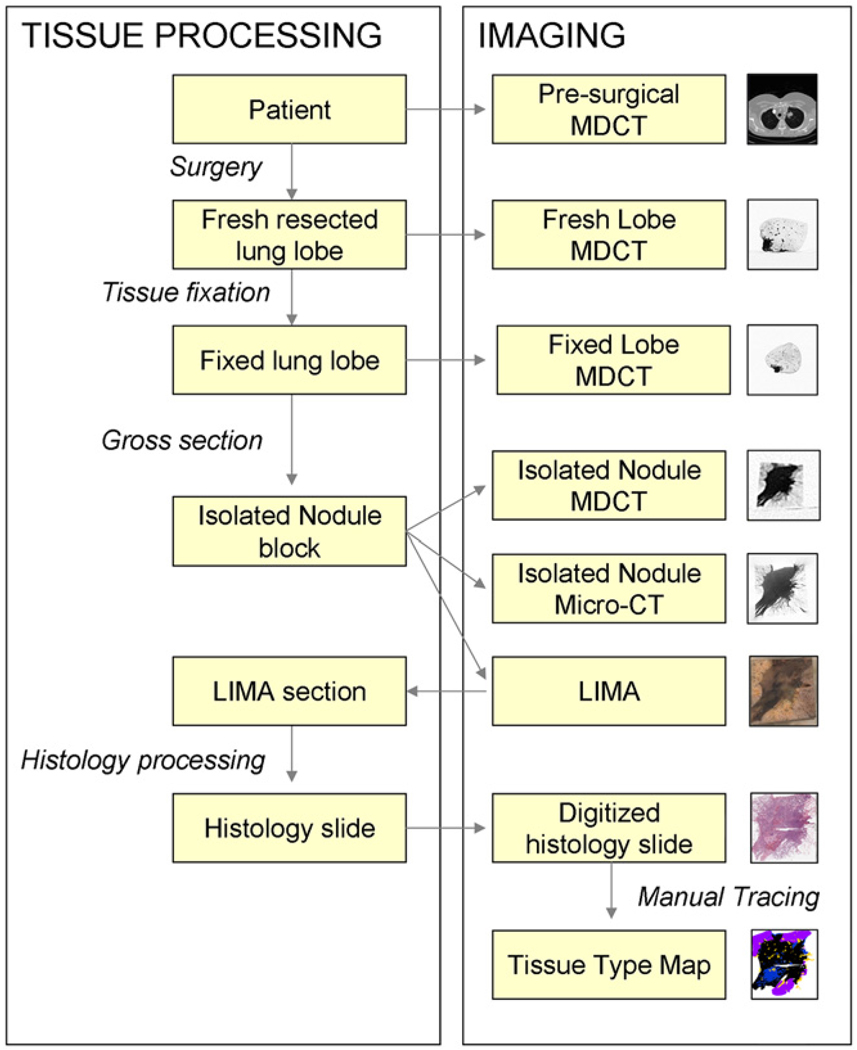
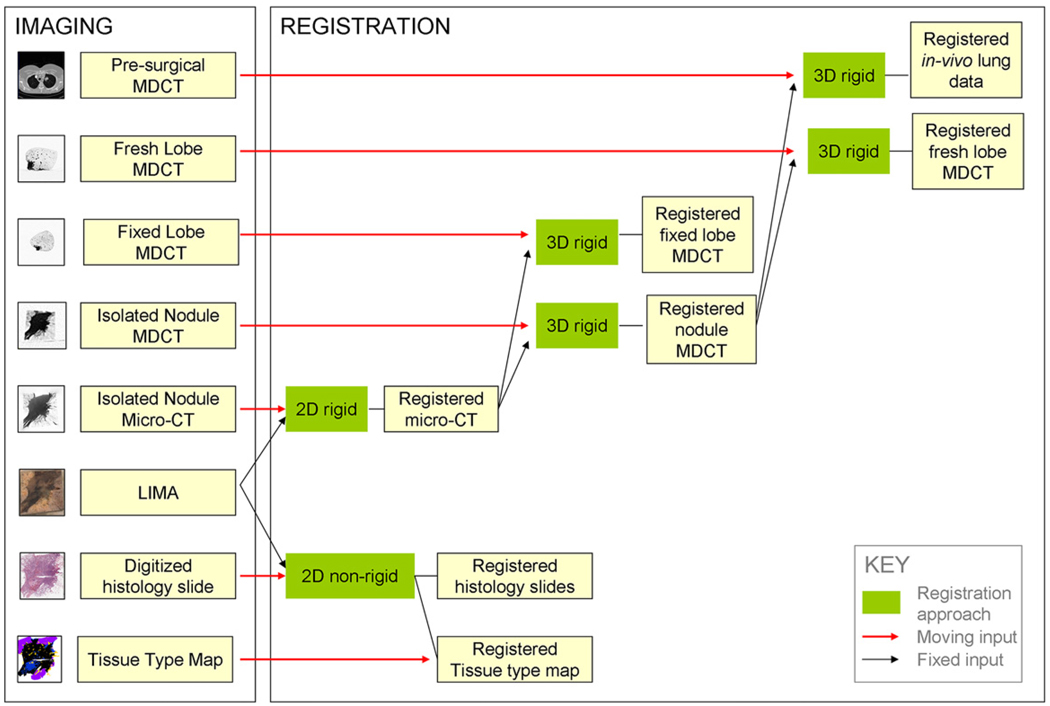
Figure 2 provides a summary of the data acquisition process developed, incorporating in vivo MDCT, fresh and fixed resected lobe MDCT and micro-CT, LIMA, and histopathological imaging for the isolated nodule.
Figure 2.
A summary diagram outlining the image acquisition and tissue processing procedure for the application of investigating human lung cancer nodules.
As a component of a lung cancer patient’s clinical care, a presurgical MDCT scan is required by the multidisciplinary cancer service at the University of Iowa. These presurgical scans were incorporated into the study to provide some in vivo data. Clinical MDCT protocols vary based on institution, producing slice thicknesses between 3 mm and 5 mm and a pixel size of 600 × 600 microns to 1 × 1 mm.
MDCT data was also obtained for the isolated lobe. To prevent degradation of the tissue, fixation was performed within an hour of resection. When no scheduling conflicts arose, the freshly resected lobe was inflated to 15 cmH2O through the main, cannulated airway and MDCT scanned before fixation. In some cases, the resection procedure extended beyond the planned completion time such that the MDCT facility was unavailable when the tissue was obtained and thus the tissue was fixed without the acquisition of a fresh MDCT scan. For all cases, a MDCT scan of the fixed isolated lobe was obtained. As radiation dose was not an issue after the tissue had been removed from the patient, a high-resolution thorax protocol with adjustments made to the voltage because of the removal of surrounding bone from the tissue was used. The isolated lobes were imaged using a Siemens Somatom Sensation 64 MDCT scanner (Siemens Medical Solutions, Forchheim, Germany) scanning 120 kV, 140 mAs to produce isotropic voxels of 300 microns.
The nodule was isolated from the surrounding lobe to allow for the subsequent high-resolution data acquisition. The high resolution MDCT protocol has a field of view (FOV) restriction of 5 cm × 5 cm, the micro-CT FOV was 4 cm × 4 cm, the LIMA was very accommodating with an FOV restriction of 30 cm × 30 cm and the maximum field of view for a histological section was 2 cm × 3 cm: the size of a standard histology slide. Hence the smallest of these FOV restrictions was used to determine the dimensions of the isolated nodule, with a length and width within 2 cm × 3 cm and flexibility in the height (z-axis).
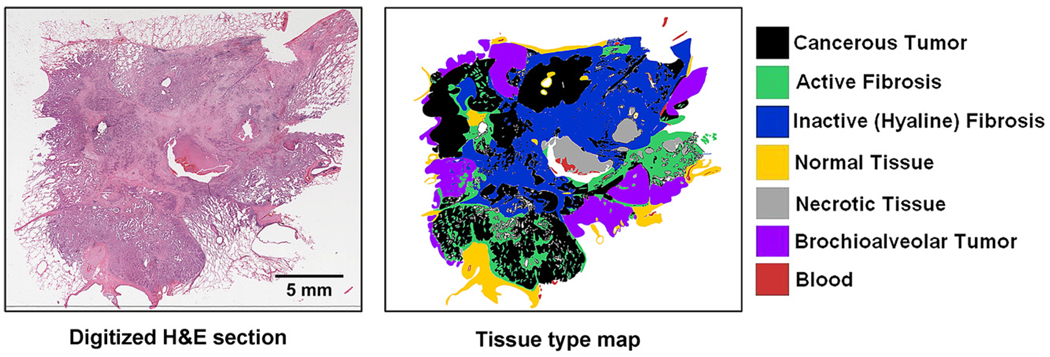
For the MDCT imaging of the isolated nodule, a high-resolution inner ear protocol was used. The protocol again used 120 kV, 140 mAs to produce a reconstructed dataset with 100 micron isotropic voxels. The nodules were then imaged using the Siemens MicroCAT II system (Siemens Pre-Clinical, Knoxville, TN) at 80 keV, 100 microA to produce isotropic 28 micron voxels. LIMA subsequently sectioned the tissue at 252 microns and imaged the cut tissue surface at a resolution of 8.5 × 8.5 microns (10 × magnification) (9–12). The LIMA system imaged the tissue surface before cutting and removing the section, hence retaining the spatial correspondence between tissue sections as described in the introduction (Fig 1). Each tissue slice generated by the LIMA system underwent standard histopathological processing to generate hematoxylin and eosin (H&E) slides. The whole field of view of each slide was digitized using ScanScope (Aperio Technology Inc, Vista, CA) at 20 × magnification to produce a pixel size of 0.5 × 0.5 microns. The large field of view, coupled with the ultra high resolution of the digitized histology slide results in an image file that was too large to be manipulated on a standard computer. To reduce the resolution of the dataset to a more manageable magnitude, bicubic resampling was used, resulting in a pixel size of 2.5 × 2.5 microns. A surgical pathologist with a subspecialty expertise in pulmonary pathology deemed this resampled resolution as adequate for the analysis the digitized H&E histopathological images. For the analysis, an Intuos graphics tablet and pen (Wacom, Vancouver, WA) were used to generate mask images corresponding to the different tissues identified. The cellular based tissue types identified and segmented include solid regions of cancerous tumor cells, cancerous tumor cells in a bronchoalveolar carcinoma configuration, necrotic cells, active fibroblastic stromal tissue, inactive (hyaline) fibrosis, red blood cells, and normal tissue (Fig 3).
Figure 3.
An example of a digitized hematoxylin and eosin histology section of an adenocarcinoma case and the corresponding tissue type map created via manual tracing by a surgical pathologist with a pulmonary subspecialty.
Some Specific Challenges in Multimodal acquisition
Specific challenges are presented when performing multimodality studies of organs using in vivo and ex vivo image data, including primarily tissue fixation, maintenance of orientation, and spatial correspondence.
In regard to fixation, a process was required which preserved the organ from degradation while also maintaining the structure and radiodensity of the organ for comparisons to the in vivo state. The lung in particular is difficult to study ex vivo because of the inflated in vivo state of the organ. Hence for the study of lung nodules, we used a modification of the Heitzman fixation technique in which glycol was substituted for the water in the tissue, allowing for comparable radiodensities between the in vivo organ and the fixed specimen (13). The fixative solution comprised 25% polyethylene glycol 400 (Fisher Chemicals, Fairlawn, NJ), 10% formaldehyde solution (Fisher Chemicals), 10% ethyl alcohol (190 Proof, 95%, Pharmco Products, Brookfield, CT), and 55% distilled water. Immediately after the MDCT imaging of the freshly resected lung lobe, the fixative solution was applied through the cannulated major airway at a pressure of 15 cmH2O and the lobe was concurrently submersed in fixative solution for 52 hours. After instillation of the fixative solution, the tissue was air dried at 15 cmH2O for at least 90 hours.
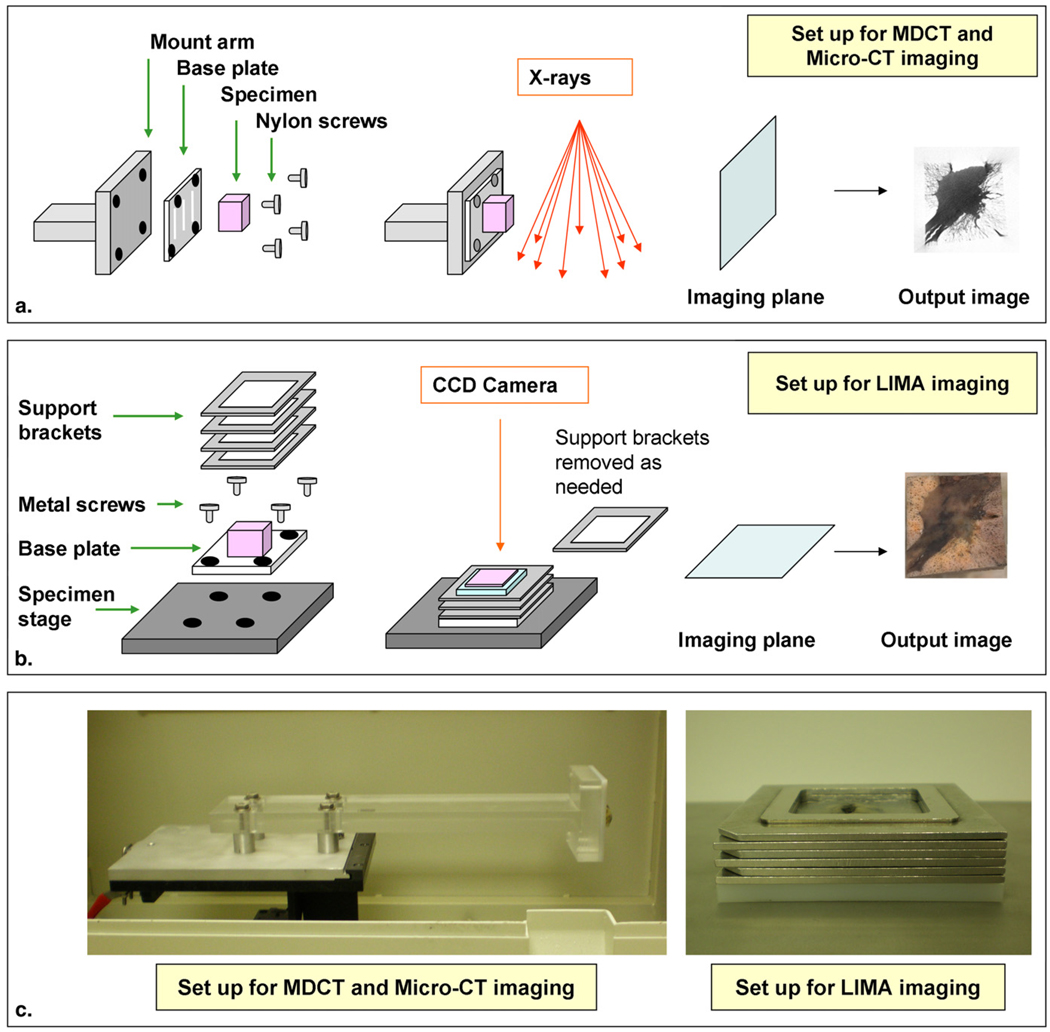
Retaining the exact orientation of tissues through in vivo and ex vivo imaging is impossible. However, it is advantageous to minimize the complexity of future registration requirements by physically maintaining tissue orientation where possible. This is of particular importance when the imaging datasets to be compared contain nonisotropic voxels. A nodule stabilization system was developed consisting of a base plate, mount arm, and specimen stage (Fig 4). The MDCT and micro-CT imaging systems acquire data in the transverse plane, whereas the LIMA system cuts and images along the coronal plane. To ensure there was a consistency between the direction and angle of the imaging planes, the specimen was mounted to a purposefully designed polyethylene base plate. The polyethylene material was selected due to its very low attenuation in radiological imaging systems, such that it would not generate artifact (14). Parallel grooves were etched into the base plate to facilitate tissue attachment. Nylon screws were used to vertically attach the base plate to a polymethyl methacrylate mount arm. The specimen was imaged in this configuration on both the MDCT and micro-CT systems.
Figure 4.
Schematic diagram of the nodule stabalization system developed for maintaining the alignment between the imaging planes from multidetector computed tomography (MDCT), micro-CT, and the large image microscope array (LIMA) system. Shown are the components and imaging plane of the MDCT and micro-CT modalities (a), and the LIMA system (b). Photographs of the actual hardware is shown in (c), which show the detail of the mount arm and the support brackets.
For sectioning on the LIMA system, the base plate was removed from the mount arm and attached horizontally with metal screws to the specimen stage. Interlocking metal stacking support brackets were attached to the base plate and the interior space was filled with agarose. When set, the agarose formed a solid support media to prevent tissue motion during sectioning. The metal stacking support brackets and metal base plate become cold as the agarose solidifies in a refrigerated environment. During sectioning, the metal insulates the agarose, keeping it cool and hence at an ideal solid consistency. The 4-mm metal stacking brackets were removed one by one as the specimen is sectioned. As mentioned in the introduction, the LIMA system was specifically developed for the establishment of multimodal whole organ databases. The extremely valuable role this system serves is to provide a reliable spatial reference to which histopathological data can be nonrigidly registered.
Registration
In previous studies, a significant barrier to the registration of histological data to nondestructive data has been the lack of reliable spatial correspondence between the datasets. However, the development of the LIMA system served to bridge this information gap for this study. Figure 5 provides a summary of the registration pipeline developed to bring the multimodal lung nodule datasets to a common alignment. This pipeline incorporates three registration approaches; two-dimensional (2D) rigid registration, 2D nonrigid registration, and three-dimensional (3D) rigid registration.
Figure 5.
Summary diagram showing the registration approaches applied to map the multimodal image datasets to a common global image space.
2D Rigid Registration
The 2D rigid registration algorithm consisted of a similarity transform in which the registration landmark points were used to solve for the transform parameters by minimizing the distance between corresponding moving and fixed landmarks. The similarity transform fS consists of a translation vector, t, rotation matrix, A, and scale matrix λ (15).
| (1) |
The parameters of the similarity transform were found using the two corresponding registration landmark sets, Lrm and Lrf, so that the distance measure D(fs) was minimized, where;
| (2) |
Landmark points were used to calculate the transform as the large difference in image content in the micro-CT and LIMA datasets deemed them unsuitable for other intensity-based approaches. Landmark points were manually selected using identifiable anatomical features such as small blood vessels, distinctive airway wall features, or nodule spiculations. Four corresponding registration landmark points, Lr, and two corresponding evaluation landmark points, Le, were manually selected in the LIMA and micro-CT datasets for each nodule case. These points were saved in a registration landmark file and an evaluation landmark file and input to the registration algorithm along with the moving image set.
The output from the registration algorithm included the aligned moving dataset and the transform parameters applied. In addition, the fiducial registration error (FRE) and the target registration error (TRE) were output from the algorithm for validation purposes (16,17). The FRE directly equates to the distance measure D(fs) that was being minimized to solve for the transform using the registration landmark set.
| (3) |
The TRE was also calculated as a distance measure between fixed landmarks and transformed, moving landmarks. However, the TRE was based on the evaluation landmark set, Lem and Lef, which was not used to determine the registration parameters and hence reflects how well the determined transform parameters map other points in the image to their corresponding fixed target points.
| (4) |
In the ideal case where the determined transform exactly translated the provided moving image to the specified fixed image, the FRE and TRE values would be zero.
2D Nonrigid Registration
A landmark-based thin-plate spline algorithm was developed to nonrigidly register each histopathological moving image to its corresponding LIMA fixed image. The algorithm was based on the mathematics originally presented by Bookstein (18) in which a spline, fTPS, is found that interpolates the moving landmarks, Lrm to the fixed landmarks, Lrf (19,20). In thin plate spline registration, the smallest possible smooth deformation is found by minimizing the bending energy, which permits the landmarks to be exactly mapped to each other (18).
| (5) |
where U(r) = r2logr2, D is a matrix representing the affine transformation and wi is the warping coefficient matrix that represents the non-affine deformation. The bending energy function, E, is minimized to control the extent of warping to the smallest deformation possible:
| (6) |
The correspondence between the LIMA and histology images was inherently gained through the acquisition technique. For each LIMA and histology pair, registration landmarks, Lr, and evaluation landmarks, Le, were manually selected and saved to text files. The number of landmarks chosen was dependent on the size of the histology slide and the number of reliable landmark structures available, with the average number of landmarks being 36 ± 6. The landmark text files, the fixed base image, and the moving images were applied as inputs to the 2D thin plate spline registration algorithm. Multiple moving histological images could be input to the algorithm and have the same spline deformation applied. This was required in the case of registering the tissue type maps to the global coordinate system.
The output from the registration approach included the transformed histopathology image. The deformation field, which represented the applied spline, was also output. For evaluation purposes, the TRE was also calculated and output. The nature of the spline calculation is to force the moving registration landmarks Lrm to the exact position of the fixed registration landmarks, Lrf. Hence the FRE is always equal to zero for this approach.
| (7) |
The TRE uses evaluation landmarks that are not used to calculate the spline warping and hence reflects how closely the deformation field matches target points in the translated moving image to the corresponding positions in the fixed image space.
| (8) |
3D Rigid Registration
Registration of the ex vivo MDCT data to the global coordinate space was achieved using the volume dataset. A rigid registration algorithm was deemed appropriate for the registration of the isolated lung nodule MDCT and the fixed lobe MDCT data to the micro-CT dataset as the tissue properties were not altered between imaging.
The in vivo MDCT datasets were acquired presurgically for clinical purposes and added to the study after a patient consented. Unfortunately, there was not consistency of the scanning protocol across the nodule cases, and the overall quality of the in vivo scans was less than ideal. Because different voltage, current, reconstruction kernels, and slice thicknesses were used, this dataset was deemed inappropriate for the comparison of Hounsfield unit variance to histopathological tissue type. Hence, the in vivo MDCT dataset was aligned to the global coordinate system using the 3D rigid registration approach. Although it could not be assumed that the tissue properties had not been altered between this data acquisition and the fixed tissue acquisition, the limited resolution of these datasets deemed more sophisticated registration approaches unnecessary.
Manual alignment between the in vivo MDCT and the fixed lobe MDCT was first achieved using the large airways and vessels in the lobe as structural landmarks. After a rough alignment was achieved, the 3D rigid registration approach was applied.
The 3D rigid registration transform, fR, involved translation, t, and rotation, A:
| (9) |
A Quasi-Newton optimizer was used to step the transformation toward a minimum solution. Because a significant difference in resolution was present between the micro-CT dataset and the MDCT dataset, a multiresolution optimization was included. This involved resampling the data to four levels; by a factor of 8 in the x and y plane and a factor of 3 in the z plane (8,8,3), then by (4,4,2), then by (2,2,1), and finally the original resolution (1,1,1). The registration transform was found first using the lower resolution data then that result was used as the initial setting for the next resolution level, and so on. Incorporating the multiresolution approach into the optimization decreased the computation time while increasing the robustness of the registration approach. Although scaling of the datasets was used to optimize the registration, in the resulting registered MDCT data, the original voxel matrices of the datasets were maintained (no scaling factor in the 3D rigid registration transform). In doing so, the introduction of false information through extensive resampling was avoided.
The normalized mutual information metric has been found to be more robust than the standard mutual information metric (21). This metric does not rely on the direct correspondence between gray-level values in the dataset but rather compares the overlap in information. The normalized mutual information of images A and B is calculated from the sum of the marginal entropies of A and B, divided by the joint entropy:
| (10) |
The entropy is based on the probability distribution of the gray values that are found from the histogram of the image.
| (11) |
Validation of this registration approach was performed qualitatively through checkerboard images and quantitatively by repeating the registration process three times and calculating the standard error between the resulting transformation parameters.
RESULTS
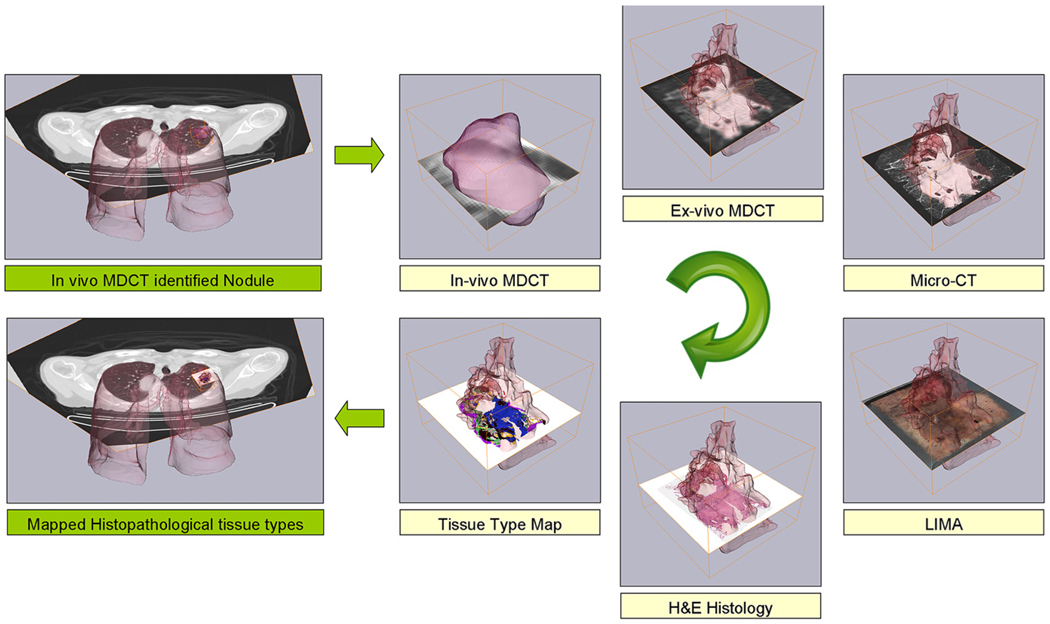
We have evaluated the 11 subjects enrolled in this study from which lobectomy specimens were obtained. The result of the data acquisition process was eight volumetric datasets per subject; 1: in vivo MDCT; 2: freshly resected lobe MDCT; 3: fixed lobe MDCT; 4: isolated nodule MDCT; 5: isolated nodule micro-CT; 6:– LIMA; 7: H&E histopathology; and 8: tissue type maps generated by the manual segmentation of the histopathology data. From these datasets, we obtain increasing resolution density information from the in vivo MDCT, isolated lobe MDCT, isolated nodule MDCT and nodule micro-CT. Color and structural information is contained in the LIMA dataset and cellular tissue content is established from the histopathology. The tissue type maps allowed the clear visualization of the cellular composition of the nodules. Figures 6 and 7 illustrate examples from the resulting cross-registered dataset for two adenocarcinoma cases. Figure 7 reveals the severity of the oversimplification of the nodule boundary as determined from the clinical, in vivo MDCT when compared to that from the higher resolution datasets.
Figure 6.
The result of the image acquisition and registration process for an adenocarcinoma case in which the tissue type map can be related to the other image datasets on a voxel to voxel basis.
Figure 7.
Because of the developed image acquisition sequence and image registration approach, the multimodal dataset is correlated back to the corresponding location in the in vivo multidetector computed tomography (MDCT) dataset. Note the boundary identified in the low-resolution, in vivo MDCT is an oversimplification of the nodule boundary as identified from the higher resolution datasets.
The registration of the multiple datasets to a common coordinate system was complicated by significant differences in image content as well as image resolution. Because the purpose of the study was to develop a methodology for examining the correlation between CT based HU and histopathological tissue type, an error less than the highest possible resolution MDCT dataset, 100 microns, was deemed acceptable.
The 2D rigid registration approach was employed for the registration of the LIMA and micro-CT data. The lung nodule tissue properties were not altered between the acquisition of the micro-CT data and the LIMA imaging and so a nonrigid registration was deemed unnecessary to correct the misalignment between the datasets. The average and standard deviation of the FRE was 1.59 ± 0.69 pixels and the TRE was 3.10 ± 1.40 pixels. In the global coordinate system, this equated to a physical FRE distance of 44.57 ± 19.22 microns and a TRE of 86.83 ± 39.23 microns.
The 2D nonrigid registration approach was used to align each digitized histopathology slide to the corresponding LIMA image. The average and standard deviation of the TRE for the landmark based thin plate spline warping of the histology image set to the LIMA base set was 5.04 ± 2.20 pixels, which translated to 43.08 ± 18.83 microns in global space.
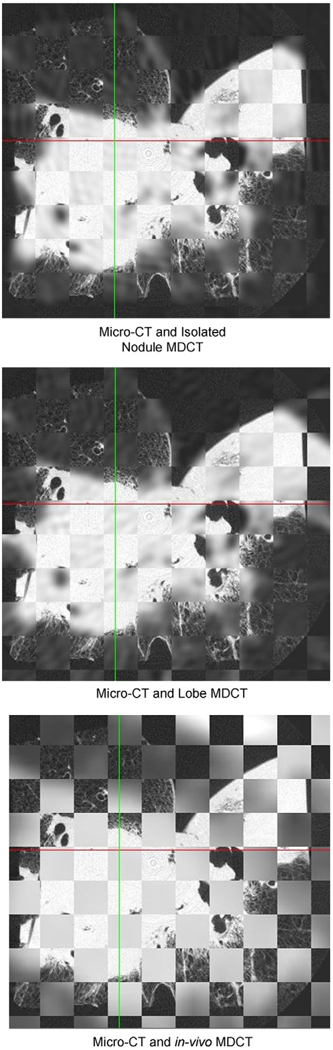
For the 3D rigid registration of the MDCT data to the micro-CT checkerboard images were used for qualitative validation and the standard error of repeated registrations was used for quantitative validation. Figure 8 illustrates an example of a set of checkerboard images for an adenocarcinoma case. These images show a good alignment between the identifiable structures in the micro-CT to isolated nodule MDCT and the micro-CT to lobe MDCT. The resolution of the in vivo MDCT data makes the evaluation of the registration difficult and no structural landmarks can be clearly visualized.
Figure 8.
The qualitative evaluation of the three-dimensional rigid registration approach was performed using checkerboard datasets in which alternating cubes of data were displayed from the two datasets being evaluated. Shown are three axial images through the same plane in the dataset comparing the alignment between the registration basis, the micro-computed tomography, and the aligned lower resolution datasets.
As a qualitative measure of registration, in the case of the isolated lung nodule MDCT to the micro-CT dataset, the standard errors for resulting translations in x, y, and z were 0.9 microns, 4.5 microns, and 5.4 microns, respectively. The standard errors for rotation around the x, y, and z axis were 6.3 microns, 7.3 microns, and 4.2 microns, respectively. For the registration of the fixed lung lobe to the micro-CT data, the standard errors in translation were 2.0 micron in x, 6.3 microns in y, and 2.0 microns in z. For rotation the standard errors in x, y, and z axis were 2.2 microns, 7.2 microns, and 4.7 microns, respectively, which are all subpixel registration errors. For the alignment of the in vivo MDCT to the global coordinate system the standard errors in the transform in x, y, and z were 1.0 mm, 0.6 mm, and 3 mm, respectively. The standard errors for rotation around the x, y, and z axis were 0.3 mm, 0.4 mm, and 3.8 mm, respectively. As one would expect, the standard errors reflect the difficulty in registering such sparse data. The overall tendency across the datasets, for the large slice separation to be in the global z-axis, is reflected by the large standard error in the z translation.
DISCUSSION
The creation of a process model for the acquisition and registration of histopathological data to corresponding ex vivo and in vivo MDCT data has high potential to impact clinical and research arenas. Because there is no complete database containing registered lung nodule pathology to corresponding MDCT data, it is impossible to evaluate the extent of information captured in an MDCT scan. This correlation may have a clinical significance in bridging radiological and pathological classifications of cancerous nodules. This is of particular interest in the case of lung cancer in classifying bronchoalveolar carcinoma subtypes that have been linked to differing prognostic outcomes (22).
Non–small-cell lung cancer nodules are considered by most clinicians to be topographically homogeneous (consisting only of cancer) with a well-defined boundary. In this study, delineating the different tissue types within the histopathological data revealed a highly complex intermixing of cancerous and noncancerous tissue types within the solid portion of the nodule (Fig 3). Because of the reliable structural basis of the LIMA dataset, the individual histopathological sections could be registered to a common coordinate system, generating 3D reconstructions of the histopathological content. To our knowledge, this is the first time this form of reconstruction has been established for lung adenocarcinoma nodules. We believe these histopathological reconstructions present a valuable opportunity to investigate relationships between histopathological content and patient diagnosis and prognosis with a greater accuracy than current preliminary studies (23–26).
It is valuable to note that from this study it was found that the boundary complexity of the nodules was grossly underestimated in the clinical data. Some aspect of this was unavoidable due to the larger field of view required for clinical in vivo imaging, resulting in a lower in-plane resolution. However, the main contributing factor to this boundary oversimplification was the large slice thickness of the clinical MDCT scans. With a slice thickness of 3 mm or greater, much of the valuable information regarding the nodule shape is lost, even in these cases in which the nodules were approximately 2 cm in diameter. Boundary shape is commonly used by both radiologists and computer-aided diagnosis (CAD) systems as a feature for distinguishing cancerous from noncancerous lesions (27,28). In our study, however, the complexity of the nodule boundaries was not appropriately captured in the in vivo, clinical CT data.
Registration of multimodal, multiresolution datasets is a challenging task. In this study, both rigid and nonrigid registration approaches were used to align the datasets to a common coordinate system. These registration approaches were evaluated using TRE and FRE measures and standard errors of repeated registrations. The minimum registration accuracy for this study was determined to be the highest resolution of the MDCT dataset. This was the resolution of the isolated nodule MDCT that had a 100-micron pixel size. The greatest interest in this study was correlating histological tissue type to MDCT representations, and subpixel accuracy in the registration of these datasets was considered desirable. This was successfully achieved for all image sets, except for the in vivo MDCT data, in which the data quality was poor. In the future, with higher resolution presurgical MDCT imaging, we are confident a great improvement can be achieved in the alignment of this dataset. It is believed the major contributor to the TRE and FREs in the micro-CT to LIMA registration and the histopathology to LIMA registration was the manual selection of landmark points. In the different modality datasets, there exists a challenge in the precise location of corresponding landmark points. This is due to modality specific influences such as partial volume effect in the micro-CT datasets and physical distortion of the tissue in histopathology.
Considering the challenges in landmark selection, the 3D mutual information registration as opposed to 2D landmark based registration may seem a better approach for the registration of the micro-CT dataset to the LIMA. However, each slice from the LIMA dataset was acquired from the cut tissue block so that a large depth of field was incorporated into the image. This acquisition method was selected to allow the structural correspondence between LIMA sections to be preserved. Unfortunately, this large depth of field in the LIMA datasets complicated the multimodal registration problem because of the significant difference in airway representations between the LIMA and the micro-CT dataset. Hence, a landmark based approach was required to achieve reliable alignment results. Landmarks were not necessary to register the lower resolution MDCT datasets to the aligned micro-CT dataset because of the similarity in information content.
In the future, the quality and speed of the 2D rigid and nonrigid registration approaches may be improved by using semiautomated boundary detection approaches to delineate the nodule perimeter and then use the contour, rather than landmark points, to drive the registration. Some previous work in this form of registration was conducted by Jaccobs et al in the alignment between histology sections and MRI of rat brains (29). In this study, the Eigenimage filter technique was used to segment the MRI data, followed by an automated multiresolution contour extraction method. A head and hat surface matching approach mapped the lower resolution “hat” dataset (MRI) to the high resolution “head” (histology) (30). However, an extensive investigation into the applicability of a similar approach to our lung nodule study is required to determine feasibility and cost-to-benefit ratio.
After the registration of the histopathological tissue type maps and the radiological datasets to the common coordinate system, a direct correspondence can be established between the voxels in the radiological data and the tissue type labels. We plan to use this labeling to extract out the HU values corresponding to each tissue type and statistically evaluate the resulting HU histograms to determine if separability between the pathological tissue type classifications exists, a crucial piece of information that is unknown.
For future cases, we intend to acquire a presurgical contrast MDCT, for blood flow, as well as a positron emission tomography/CT scan, for glucose metabolism. Both blood flow and glucose metabolism are forms of functional data used to aid in distinguishing cancerous from noncancerous nodules. As enrollment continues, we aim to link patient outcomes to the histopathological content to determine if the proportion of each tissue type has prognostic significance. Additionally, all the datasets will be made publicly available to researchers and clinicians through inclusion in the Large Image Database Consortium database (31).
CAD systems are becoming increasingly important in the clinical setting, serving as a second reader in image interpretation, effectively improving detection accuracy and consistency (32,33). To develop a CAD system, a complete dataset is required containing numerous cases for which the final diagnoses are known, this is known as a training dataset. From the training set, a group of features is established to describe each of the diagnoses, or classes. The training dataset is used to guide the development of the CAD algorithm by comparing the features of an unknown case to the features of the training set and seeing which class (diagnosis) it most closely matches. Therefore, if the final diagnosis in the training set has only two classes—malignant and benign—then they are the two possible outputs when the CAD algorithm is applied to a new case. It is hoped that the datasets produced by the methodology described in this paper can be used in the future for training more sophisticated CAD systems. Through the reliable registration of the histopathology ground truth to other modalities, the possible classes for classification can extend beyond the currently used whole nodule descriptors, malignant and benign, to include specific subnodule pathological tissue types such as cancerous tumor, fibrosis, and necrosis.
In conclusion, a novel process model has been established to generate cross-registered multimodal datasets for the investigation of human lung cancer nodule content and representation. We believe this methodology produces datasets of high potential to increase the understanding of 3D histopathological tissue content and its relationship to patient prognosis, and also to provide a ground truth for the evaluation and further development of CAD systems using MDCT. In this process new image-based biomarkers relevant to lung cancer are likely (34).
ACKNOWLEDGMENT
Much appreciation is given to Mark Iannettoni, Timothy Van Natta, William Lynch, Kalpaj Parekh, Joan Rick-McGillin, and Kelley McLaughlin for facilitating patient identification and recruitment. The authors also thank Melissa Hudson, John Morgan, and Jan Rodgers for technical assistance.
Supported by NIH R01 CA129022-01 (7/1/2007-6/30/2012), Precise Correspondence of 3D Pathology with Radiological Features in Lung Nodules.
REFERENCES
- 1.Oehr P, Biersack J, Coleman RE. PET and PET-CT in oncology. Verlag Berlin Heidelberg, NY: Springer; 2004. [Google Scholar]
- 2.van Der Wel A, Nijsten S, Hochstenbag M, et al. Increased therapeutic ratio by 18FDG-PET CT planning in patients with clinical CT stage N2-N3M0 non-small-cell lung cancer: a modeling study. Int J Radiat Oncol Biol Phys. 2005;61:649–655. doi: 10.1016/j.ijrobp.2004.06.205. [DOI] [PubMed] [Google Scholar]
- 3.Bybel B, Brunken RC, DiFilippo FP, et al. SPECT/CT imaging: clinical utility of an emerging technology. Radiographics. 2008;28:1097–1113. doi: 10.1148/rg.284075203. [DOI] [PubMed] [Google Scholar]
- 4.von Schulthess G. Molecular anatomic imaging: PET-CT and SPECT-CT integrated modality imaging. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. [Google Scholar]
- 5.Seltzer SE, Getty DJ, Pickett RM, et al. Multimodality diagnosis of liver tumors: feature analysis with CT, liver-specific and contrast-enhanced MR, and a computer model. Acad Radiol. 2002;9:256–269. doi: 10.1016/s1076-6332(03)80368-9. [DOI] [PubMed] [Google Scholar]
- 6.Kennel SJ, Davis IA, Branning J, et al. High resolution computed tomography and MRI for monitoring lung tumor growth in mice undergoing radioimmunotherapy: correlation with histology. Med Phys. 2000;27:1101–1107. doi: 10.1118/1.598974. [DOI] [PubMed] [Google Scholar]
- 7.Chan R, He Y, Haque A, et al. Computed tomographic-pathologic correlation of gross tumor volume and clinical target volume in non-small cell lung cancer: a pilot experience. Arch Pathol Lab Med. 2001;125:1469–1472. doi: 10.5858/2001-125-1469-CTPCOG. [DOI] [PubMed] [Google Scholar]
- 8.Clarke SE, Hammond RR, Mitchell JR, et al. Quantitative assessment of carotid plaque composition using multicontrast MRI and registered histology. Magn Reson Med. 2003;50:1199–1208. doi: 10.1002/mrm.10618. [DOI] [PubMed] [Google Scholar]
- 9.Namati E, De Ryk J, Thiesse J, et al. Large image microscope array for the compilation of multimodality whole organ image databases. Anat Rec (Hoboken) 2007;290:1377–1387. doi: 10.1002/ar.20600. [DOI] [PubMed] [Google Scholar]
- 10.de Ryk J, Namati E, Reinhardt J, et al. SPIE: Three-dimensional and multidimensional microscopy. San Diego, CA: 2004. A whole organ serial sectioning and imaging system for correlation of pathology to computed tomography; pp. 224–234. [Google Scholar]
- 11.de Ryk J, Thiesse J, Reinhardt J, et al. SPIE: Medical imaging. San Diego, CA: 2006. Establishing multimodality datasets with incorporation of 3D histopathology for soft tissue classification; pp. 1–8. [Google Scholar]
- 12.Thiesse J, Namati E, De Ryk J, et al. SPIE: Three-dimensional and multidimensional microscopy. San Diego, CA: 2004. Bright field segmentation tomography (BFST) for use as surface identification in stereomicroscopy; pp. 216–224. [Google Scholar]
- 13.Heitzman R. The Lung. 2nd ed. New York, NY: The C.V. Mosby Company; 1984. Radiologic-pathologic correlations. [Google Scholar]
- 14.Korobkin M, Brodeur FJ, Francis IR, et al. Delayed enhanced CT for differentiation of benign from malignant adrenal masses. Radiology. 1996;200:737–742. doi: 10.1148/radiology.200.3.8756924. [DOI] [PubMed] [Google Scholar]
- 15.Goshtasby A. 2D and 3D image registration for medical, remote sensing, and industrial applications. Hoboken, NJ: John Wiley and Sons; 2005. [Google Scholar]
- 16.Fitzpatrick JM, West JB. The distribution of target registration error in rigid-body point-based registration. IN: IEEE Trans Med Imaging; 2001. pp. 917–927. [DOI] [PubMed] [Google Scholar]
- 17.Fitzpatrick JM, West JB, Maurer CRJ. Predicting error in rigid-body point-based registration. IN: IEEE Trans Med Imaging; 1998. pp. 694–702. [DOI] [PubMed] [Google Scholar]
- 18.Bookstein F. Principal warps: thin-plate splines and the decomposition of deformations. IEEE Transactions on pattern analysis and machine intelligence. 1989;11:567–585. [Google Scholar]
- 19.Rohr K, Stiehl HS, Sprengel R, et al. Landmark-based elastic registration using approximating thin-plate splines. IEEE Trans Med Imaging. 2001:526–534. doi: 10.1109/42.929618. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Nikolova S, Bartha R. Registration of in vivo magnetic resonance T1-weighted brain images to triphenyltetrazolium chloride stained sections in small animals. J Neurosci Methods. 2006;156:368–375. doi: 10.1016/j.jneumeth.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Studholme C, Hill D, Hawkes D. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32:71–86. [Google Scholar]
- 22.Travis WD, Garg K, Franklin WA, et al. Evolving concepts in the pathology and computed tomography imaging of lung adenocarcinoma and bronchioloalveolar carcinoma. J Clin Oncol. 2005;23:3279–3287. doi: 10.1200/JCO.2005.15.776. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki K, Yokose T, Yoshida J, et al. Prognostic significance of the size of central fibrosis in peripheral adenocarcinoma of the lung. Ann Thorac Surg. 2000;69:893–897. doi: 10.1016/s0003-4975(99)01331-4. [DOI] [PubMed] [Google Scholar]
- 24.Okudera K, Kamata Y, Takanashi S, et al. Small adenocarcinoma of the lung: prognostic significance of central fibrosis chiefly because of its association with angiogenesis and lymphangiogenesis. Pathol Int. 2006;56:494–502. doi: 10.1111/j.1440-1827.2006.01997.x. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi M, Morikawa A, Kawasaki M, et al. Small adenocarcinoma of the lung. Histologic characteristics and prognosis. Cancer. 1995;75:2844–2852. doi: 10.1002/1097-0142(19950615)75:12<2844::aid-cncr2820751209>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 26.Olcott CR. Cell types and histologic patterns in carcinoma of the lung; observations on the significance of tumors containing more than one type of cell. Am J Pathol. 1955;31:975–995. [PMC free article] [PubMed] [Google Scholar]
- 27.Shah SK, McNitt-Gray MF, Rogers SR, et al. Computer aided characterization of the solitary pulmonary nodule using volumetric and contrast enhancement features. Acad Radiol. 2005;12:1310–1319. doi: 10.1016/j.acra.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Way TW, Hadjiiski LM, Sahiner B, et al. Computer-aided diagnosis of pulmonary nodules on CT scans: segmentation and classification using 3D active contours. Med Phys. 2006;33:2323–2337. doi: 10.1118/1.2207129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs MA, Windham JP, Soltanian-Zadeh H, et al. Registration and warping of magnetic resonance images to histological sections. Med Phys. 1999;26:1568–1578. doi: 10.1118/1.598671. [DOI] [PubMed] [Google Scholar]
- 30.Pelizzari CA, Chen GT, Spelbring DR, et al. Accurate three-dimensional registration of CT, PET, and/or MR images of the brain. J Comput Assist Tomogr. 1989;13:20–26. doi: 10.1097/00004728-198901000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Dodd LE, Wagner RF, Armato SG, 3rd, et al. Assessment methodologies and statistical issues for computer-aided diagnosis of lung nodules in computed tomography: contemporary research topics relevant to the lung image database consortium. Acad Radiol. 2004;11:462–475. doi: 10.1016/s1076-6332(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 32.Wormans D, Beyer F, Diederich S, et al. Diagnostic performance of a commercially available computer-aided diagnosis system for automatic detection of pulmonary nodules: comparison with single and double reading. Fortschr Rontgenstr. 2004;176:953–958. doi: 10.1055/s-2004-813251. [DOI] [PubMed] [Google Scholar]
- 33.Awai K, Murao K, Ozawa A, et al. Pulmonary nodules at chest CT: effect of computer-aided diagnosis on radiologists’ detection performance. Radiology. 2004;230:347–352. doi: 10.1148/radiol.2302030049. [DOI] [PubMed] [Google Scholar]
- 34.McLennan G, Clarke L, Hohl R. Imaging as a biomarker for therapy response: cancer as a prototype for the creation of research resources. Clin Pharmacol Ther. 2008;84:433–436. doi: 10.1038/clpt.2008.171. [DOI] [PubMed] [Google Scholar]