Abstract
Hexose and pentose cofermentation is regarded as one of the chief obstacles impeding economical conversion of lignocellulosic biomass to biofuels. Over time, successful application of traditional metabolic engineering strategy has produced yeast strains capable of utilizing the pentose sugars (especially xylose and arabinose) as sole carbon sources, yet major difficulties still remain for engineering simultaneous, exogenous sugar metabolism. Beyond catabolic pathways, the focus must shift towards non-traditional aspects of cellular engineering such as host molecular transport capability, catabolite sensing and stress response mechanisms. This review highlights the need for an approach termed 'panmetabolic engineering', a new paradigm for integrating new carbon sources into host metabolic pathways. This approach will concurrently optimize the interdependent processes of transport and metabolism using novel combinatorial techniques and global cellular engineering. As a result, panmetabolic engineering is a whole pathway approach emphasizing better pathways, reduced glucose-induced repression and increased product tolerance. In this paper, recent publications are reviewed in light of this approach and their potential to expand metabolic engineering tools. Collectively, traditional approaches and panmetabolic engineering enable the reprogramming of extant biological complexity and incorporation of exogenous carbon catabolism.
Introduction
Conversion of lignocellulosic biomass into fuels and chemicals is an attractive, sustainable alternative to traditional petroleum refining [1]. Accomplishing this goal using fungal or microbial hosts requires optimized cellular systems [2]. Since its inception, metabolic engineering has served as an effective platform approach to optimize and control small molecule production in cells [3-8]. Moreover, recent developments in genetic approaches have advanced the metabolic engineering toolbox beyond overexpression and knockout techniques [9-13]. This progress is evinced by the wide variety of secondary metabolite and novel products engineered for production in cells [14-21]. It is anticipated that metabolic engineering approaches can help solve the challenges of biofuel production [22-24]. However, these approaches have all traditionally focused on rerouting cellular metabolism using standard native sugars as carbon sources.
Lignocellulosic biomass presents new challenges for the metabolic engineering paradigm [25,26]. Although biomass is chiefly composed of glucose, the pentose sugars D-xylose and L-arabinose can also constitute significant fractions (up to 20%) that must be converted [27]. Even though many organisms are capable of natively converting these sugars, the most commonly selected organism is baker's yeast (Saccharomyces cerevisiae). The genetic tractability, widespread industrial use and endogenous ethanol production capacity of yeast motivate its use [28]; however, baker's yeast must be engineered to convert xylose and arabinose. Traditional pathway engineering approaches have enabled xylose and arabinose catabolism in yeast, but continued optimization of these strains requires novel metabolic engineering tools and strategies. Specifically, novel approaches should target and exploit additional cellular mechanisms influencing metabolic pathways, such as molecular transport, catabolite sensing and cellular tolerances.
In this review, we propose simultaneous transport and metabolic engineering, including new global cellular metabolic engineering techniques, as a powerful approach for the development of an economical hexose and pentose cofermenting yeast. As this review will illustrate, molecular transport and metabolic pathway engineering have conventionally been studied in isolation despite their intricate interdependence. To efficiently arrive at an optimized strain, metabolic engineering tools must be expanded to modify multiple interdependent steps, an approach we term 'panmetabolic engineering'. Metabolic engineering tools must also expand to incorporate recent breakthroughs in modifying catabolite sensing and in increasing cellular tolerances to significantly affect biofuel-producing organisms. Once developed, these tools will enable the addition of other substrates to the yeast carbon source portfolio. Therefore, the scope of this review is to discuss recent literature concerning pentose transport and metabolism in yeast, and to suggest a path forward for engineering nonnative carbon source metabolism in organisms.
Engineering exogenous pentose sugar utilization in yeast
Pathway assembly by heterologous gene expression
Baker's yeast is an ideal industrial fermentation host; however, it possesses little to no xylose or arabinose metabolic capability. Interestingly, the S. cerevisiae genome is encoded with a putative xylose metabolic pathway, although the expression level of these genes are often too low to permit growth on xylose as the sole carbon source [29-31]. Recent work suggests that some strains of Saccharomyces may possess a latent oxidoreductase pathway with an active xylitol dehydrogenase [32]. However, this pathway was shown to be repressed by other putative xylitol dehydrogenases, illustrating unique and potentially trans-acting control mechanisms. Even so, yeast lack effective xylose and arabinose utilization pathways, and therefore require heterologous complementation or significant genetic modification.
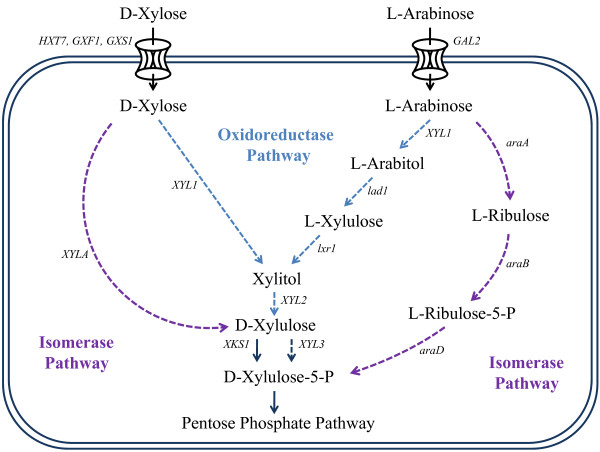
The advent of recombinant DNA technology enabled the transfer of genes from native pentose catabolizing organisms into baker's yeast, thus facilitating novel pentose catabolism [33-36]. Two types of pentose pathways have been constructed in yeast: the oxidoreductase pathway and the isomerase pathway (Figure 1). Both xylose and arabinose can be metabolized through each of these pathways, although arabinose assimilation involves additional steps in both cases [37]. All four possible pathway variants have been previously constructed [37-40], and all feed into native yeast metabolism via D-xylulose or D-xylulose-5-phosphate (P). Once converted to xylulose 5-P, these sugars are further metabolized through the native pentose phosphate pathway (PPP). The following sections give a brief review of these pathways. Combined with molecular transporter engineering, they form the foundation of the panmetabolic engineering described in more detail later.
Figure 1.
Successful exogenous transport and metabolic pathways introduced in S. cerevisiae. Reported improvements of pentose utilization in yeast (as described in the text) are depicted in this schematic. (A) Bacterial xylose isomerase pathway; (B) fungal xylose oxidoreductase pathway; (C) Fungal arabinose oxidoreductase pathway; (D) Bacterial arabinose isomerase pathway. The genes used to accomplish the enzymatic step are italicized. Heterologous steps are indicated by dashed lines.
Oxidoreductase pathways
The pentose oxidoreductase pathways are conserved between certain species of native fungi, and employ common enzymes and redox cofactors to catalyze substrate conversion. The xylose oxidoreductase pathway was the first heterologous pentose pathway constructed in baker's yeast [38]. In this pathway, depicted in Figure 1, xylose is reduced to xylitol by an aldose reductase (AR), then xylitol is oxidized to xylulose by xylitol dehydrogenase (XDH). The AR most commonly used is encoded by Pichia stipitis XYL1 (xylose reductase, XR) which prefers the cofactor NADPH over NADH. The XDH is encoded by P. stipitis XYL2, which is NAD+ dependent [41].
The L-arabinose oxidoreductase pathway was also the first variant constructed in baker's yeast for arabinose conversion [42]. Figure 1 illustrates that the two enzymes from the xylose oxidoreductase pathway AR and XDH serve as catalysts of the first and last reactions, respectively. The remaining two steps have recently been constructed using two genes from the fungus Trichoderma reesei (Hypocrea jecorina). The first gene, lad1, encodes an arabitol 4-dehydrogenase (ADH) which reduces arabitol to L-xylulose using NADPH as a cofactor [43]. The second is an L-xylulose reductase (XR) encoded by lxr1, which reduces L-xylulose to D-xylulose using NAD+ as a cofactor [42].
The D-xylulose from both the xylose and arabinose variants is then utilized by native metabolic steps, culminating in the production of biomass, carbon dioxide and ethanol.
It is important to recognize the cofactor imbalances of the arabinose and xylose oxidoreductase pathways. This imbalance is thought to limit theoretical and actual pentose conversion by yeast [25,44]. Whereas the enzymes discussed above alone are sufficient to enable xylose or arabinose metabolism in yeast, the xylulokinase gene from P. stipitis (XYL3) is often complemented to create a complete heterologous pathway and further reduce xylitol production [28,45,46].
Isomerase pathways
In contrast to the pentose oxidoreductase pathway, the isomerase pathway variants require no cofactors. The pathway is native to bacterial species and to rare yeasts. The heterologous xylose isomerase pathway minimally consists of one enzyme, xylose isomerase (XI), which directly converts xylose to xylulose (Figure 1). Because most XIs are native to bacteria, difficulties for heterologous expression in yeast exist [47], yet recent work has demonstrated functional bacterial XI pathways [39,48]. In addition, bioprospecting has yielded functional heterologous XIs isolated from rare fungi [49,50]. As with the oxidoreductase pathway, the complementation of a xylulokinase can further improve yields and assimilation rates. This pathway has improved ethanol conversion yields over the oxidoreductase pathway; however, strains have lower growth rates and sugar uptake rates, as discussed later. Nevertheless, this pathway is attractive because of its lack of cofactor imbalance.
Whereas the xylose isomerase pathway involves one step, the arabinose isomerase pathway consists of three. First, an arabinose isomerase encoded by araA converts arabinose to ribulose. Second, a ribulokinase encoded by araB phosphorylates ribulose to ribulose 5-P. Third, ribulose-5-P-4-epimerase, encoded by araD, catalyzes the final epimerization of ribulose 5-P to D-xylulose-5-P, an intermediate in the PPP. Two variations of this pathway have been constructed in yeast using distinct sets of heterologous genes. The first set uses Bacillus subtilis araA along with Escherichia coli araB and araD [40]. The second relies on the expression of Lactobacillus plantarum araA, araB and araD [51]. However, unlike the other pathways described above, evolutionary engineering was needed in both cases to isolate a yeast strain with an active arabinose isomerase pathway. Thus, only after mutations is a functional arabinose isomerase pathway in yeast possible.
Optimization of limiting steps
The work described above establishes viable pentose utilization pathways in baker's yeast, enabling xylose and arabinose to be used as a sole carbon source. However, reported growth rates remain suboptimal for economical production of biofuels from lignocellulosic biomass [2]. This limitation has driven further improvements of the heterologous pathways by both traditional metabolic engineering approaches and heterologous molecular transporter expression.
The following sections review individual efforts to improve internal pathways and transport in yeast. Through this work, limiting steps have been identified and partially alleviated. However, pathway optimization and transporter engineering in isolation has not yet produced an optimal organism. An approach integrating both pathway and transport engineering may produce more industrially viable results.
Internal pathway
When the xylose oxidoreductase pathway is expressed in S. cerevisiae, several factors limit ethanol production, with the primary one being cofactor imbalance. XR has a higher specificity for NADPH than for NADH (Km = 3.2 μmol/l for NADPH and Km = 40 μmol/l for NADH) and XDH uses only NAD+. In yeast, there is a disparity between the amount and accessibility of intracellular NADPH and NADH for the xylose pathway enzymes [52,53]. This disparity could be due to competition for NAD+ by other endogenous metabolic enzymes or to inefficient xylose pathway enzymes. As a result, high amounts of xylitol may be produced [41]. A severe cofactor imbalance (with two NADPH and two NAD+ required) limits the arabinose oxidoreductase pathway [54]. As a result, this pathway exhibits poor cell growth or little ethanol production, even though the enzymes are actively expressed [36,54].
Modifying the affinity or expression level of XR and XDH in the xylose oxidoreductase pathway has been shown to decrease xylitol production and increase ethanol yield [37,55-57]. However, even with adjusting cofactor preferences, a significant increase in ethanol production has not been achieved [25,56]. Xylitol formation may not be solely a consequence of inappropriate redox balance; some additional metabolic factors in the host may cause its production. Supporting this hypothesis, native P. stipitis does not produce significant amounts of xylitol, whereas recombinant S. cerevisiae expressing the same genes produces abundant xylitol [53]. Xylitol is still seen in strains expressing a complete XYL1/2/3 pathway from P. stipitis. Therefore, balancing mechanisms eliminating excess xylitol in P. stipitis may not be present in baker's yeast.
In the xylose isomerase pathway, xylose is converted to xylulose through a one step enzyme reaction catalyzed by xylose isomerase [41]. In this redox-neutral process, cofactor imbalance is avoided, no xylitol is produced, and the ethanol yield is much higher than in the oxidoreductase pathway [25,49,50]. However, strains expressing xylose isomerase have low anaerobic xylose consumption and growth rates due to insufficient activity of the heterologous xylose isomerase [35,46,50,53,58]. Moreover, at equilibrium, the isomerization reaction is more favorable for xylose than xylulose formation (ratio of 80:20) [59,60]. Beyond genetic approaches, the equilibrium of the xylose isomerase reaction can be shifted to favor xylulose formation by supplementing with chemicals such as borate [61].
These facts emphasize the need for either increased bioprospecting for more active xylose isomerases or protein engineering to improve performance in yeast. Optimizing expression is also important, and coregulating a xylose isomerase with downstream enzymes may prove effective. Because the xylose isomerase pathway has only recently been engineered in S. cerevisiae, an improved xylose isomerase with satisfactory activity may still be discovered or engineered.
Consistent with the xylose isomerase pathway, the activity of arabinose isomerase is insufficient, and the isomerization of arabinose to ribulose is unfavorable [54,62]. In particular, insufficient cell growth and lower ethanol production rates are seen with cells grown on arabinose compared with glucose or even xylose [54]. Therefore, metabolic and protein engineering solutions for increasing the growth rate and shifting the driving force from aldose to ketose could improve both xylose and arabinose assimilation in recombinant S. cerevisiae.
Beyond the recombinant pathway steps, additional work in improving pentose utilization has focused on the native PPP. The enzymes xylulokinase (XKS1 gene), transaldolase (TAL1) and transketolase (TKL1) have been supplemented by heterologous or overexpressed native genes. These heterologous genes have typically been imported from P. stipitis;, for example, PsTAL1 may be expressed to supplement ScTAL1[45]. In the case of transaldolase, the Pichia version of the enzyme had more advantageous enzyme kinetics than the Saccharomyces version. This observation raises the possibility that additional superior PPP enzyme may exist in native xylose consumers. Bioprospecting provides a means of identifying such enzymes.
Taken together, metabolic engineering has been able to construct and improve the pentose utilization pathways in yeast. However, these improvements have yet to produce a strain that allows widespread lignocellulosic biomass conversion to biofuels. Explanations may include host genotype fitness; for example, the putative xylitol dehydrogenases may actually be detrimental to xylose metabolism [32,45]. These and other potential regulatory mechanisms such as protein acetylation patterns or other unknown regulatory factors may be inhibiting pentose utilization [63]. Therefore, more targets and studies are required to advance the field. One of the unique primary targets being examined is the optimization of pentose transport in yeasts.
Transport
Sugar transport across the membrane does not significantly limit the endogenous metabolism of sugars such as glucose, although it may limit exogenous pentose metabolism. Without any genetic modifications, baker's yeast will transport pentoses across the cell membrane through one of many native hexose transport proteins: the S. cerevisiae proteins Hxt7p, Hxt5p, Hxt4p, Hxt2p, Hxt1p and Gal2p [40,64-66]. However, these proteins have a much higher affinity for their native hexose substrates, which may create unfavorable competitive inhibition and lead to diauxic growth in a hexose-pentose cofermentation. Therefore, dedicated pentose transport provides an opportunity to improve the simultaneous use of hexoses and pentoses.
It has been demonstrated or inferred in several cases that pentose transport accounts for significant pathway flux control [67-69]. Gardonyi et al. reported a flux control coefficient of 0.2 for xylose transport in S. cerevisiae TMB 3001 (CEN.PK-2 XR/XDH/XKS) regardless of xylose concentration, and rising to 0.5 in S. cerevisiae TMB 3206 (TMB 3001 with overexpressed XR) at xylose concentrations below 0.6 g/l [67]. Recently, an evolutionary engineering experiment (aided by continuous culturing in xylose) was performed using an optimized S. cerevisiae strain overexpressing six key xylose metabolic enzymes (including XI, XKS, TAL and TKL) [68]. Accumulated mutations over time resulted in greatly altered xylose transport kinetics, doubling Vmax (15.8 to 32 mmol per dry weight per hour) and reducing Km by 25% (132 to 99 mmol/lM). From this result, it may be inferred that xylose transport is a limiting step, especially in metabolically optimized strains with higher downstream flux capacity. An additional evolutionary engineering experiment with recombinant S. cerevisiae found an increase in expression of the hexose transporter gene HXT5 [69], showing that cells choose to increase transport activity when selected on xylose. In terms of L-arabinose transport, it has been demonstrated that the native hexose transporter gene GAL2 is essential for pathway function [40], thus implicating the lack of a specific arabinose transporter protein.
The above findings underpin recent work focusing on heterologous transporter protein identification and expression to improve xylose uptake. Heterologous transport proteins from plant, bacteria and other yeasts have been cloned and expressed in recombinant S. cerevisiae. However, only the class of transporters native to yeast, the major facilitator superfamily (MFS) [70], has been investigated. Other classes, such as the ATP binding cassette (ABC) transporters and the bacterial phosphoenolpyruvate (PEP) dependent transporters, are unlikely to be effective, because of expression difficulty and high relative energy requirements [71]. Despite ongoing work and bioprospecting, researchers have been unable to isolate or evolve a transport protein with xylose as its highest affinity substrate. Such a transporter would greatly enhance the prospects of a glucose-xylose cofermentation.
The collective results from heterologous pentose transporter expression studies are summarized in Table 1. To date, no heterologous arabinose transport experiments have been conducted, in part because of the recent reconstruction of an arabinose metabolic pathway in yeast. The most successful improvement of xylose transport to date is the expression of Candida intermedia PYCC 4715 transport proteins [72]. Both C. intermedia GXF1 and GXS1 conferred significant improved growth phenotypes in recombinant S. cerevisiae when glucose and xylose were used as sole carbon sources. Subsequent work has evaluated coexpression of the two proteins [73] as well as more in-depth fermentation analysis [74]. Both of these transporters are efficient xylose transporters, yet also have a high affinity for glucose. Therefore, they remain primarily hexose transporters, despite the excellent xylose transport characteristics.
Table 1.
Transport proteins studied for xylose uptake.
| Genus | Species | Gene or DNA Library | UniProt | Plasmid | Study | Strain | Xylose transport phenotype |
|---|---|---|---|---|---|---|---|
| Arabidopsis | thaliana | Stp2 | Q9LNV3 | p4H7 | [64] | TMB3201 | - |
| Arabidopsis | thaliana | Stp3 | Q8L7R8 | p4H7 | [64] | TMB3201 | - |
| Arabidopsis | thaliana | At5g59250 | Q0WWW9 | p4H7 | [64] | TMB3201 | - |
| Arabidopsis | thaliana | At5g59250 | Q0WWW9 | pRH145 | [75] | BY4727 | + |
| Arabidopsis | thaliana | At5g17010 | Q6AWX0 | pRH145 | [75] | BY4727 | + |
| Arabidopsis | thaliana | Seedling cDNA [104] | - | pFL61 | [64] | TMB3201 | - |
| Candida | intermedia | Gxf1 | Q2MDH1 | YEplac195 | [72] | TMB3201 | + |
| Candida | intermedia | Gxs1 | Q2MEV7 | YEplac195 | [72] | TMB3201 | + |
| Chlorella | kessleri | Hup1 | P15686 | p4H7 | [64] | TMB3201 | - |
| Escherichia | coli | XylE | P0AGF4 | p4H7 | [64] | TMB3201 | - |
| Pichia | stipitis | SUT1 | O94155 | YEp24 | [105] | RE700 | - |
| Pichia | stipitis | SUT2 | O94151 | YEp24 | [105] | RE700 | - |
| Pichia | stipitis | SUT3 | Q9UWF5 | YEp24 | [105] | RE700 | - |
| Pichia | stipitis | Genomic library [105] | - | YEp24 | [64] | TMB3201 | - |
| Trichoderma | reesei | Xlt1 | Q1EG32 | pAJ401 | [66] | H2219 | +* |
| Saccharomyces | cerevisiae | HXT1 | P32465 | pYX212 | [66] | H2219 | + |
| Saccharomyces | cerevisiae | HXT2 | P23585 | pYX212 | [66] | H2219 | + |
| Saccharomyces | cerevisiae | HXT4 | P32467 | pYX212 | [66] | H2219 | + |
| Saccharomyces | cerevisiae | HXT7 | P39004 | pYX212 | [66] | H2219 | + |
| Saccharomyces | cerevisiae | HXT1 | P32465 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT3 | P32466 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT4 | P32467 | p4H7 | [64] | TMB3201 | + |
| Saccharomyces | cerevisiae | HXT5 | P38695 | p4H7 | [64] | TMB3201 | + |
| Saccharomyces | cerevisiae | HXT7 | P39004 | YEpkHXT7 | [64] | TMB3201 | + |
| Saccharomyces | cerevisiae | HXT8 | P40886 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT9 | P40885 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT10 | P43581 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT11 | P54862 | p426MET25 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT13 | P39924 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT14 | P42833 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | HXT15 | P54854 | p4H7 | [64] | TMB3201 | - |
| Saccharomyces | cerevisiae | GAL2 | P13181 | pHL125 | [64] | TMB3201 | + |
Several previous studies have evaluated the capacity of selected transport proteins to uptake xylose in yeast.
The strain genotype (described in Table 2) used and the identified xylose phenotype is listed.
*Indicates that mutations (either in the gene or in the host cell) were required for the phenotype to appear.
Heterologous xylose transport phenotypes were observed in yeast expressing A. thaliana At5g59250 and At5g17010 [75]. However, this observation was not supported by a second study in which At5g59250 expression did not confer growth on xylose [64]. These studies, as Table 2 clarifies, did not use the same host strain. In the first study with At5g59250, a standard strain of yeast was used whereas in the second, a strain lacking the HXT family of proteins was used. Interactions between membrane proteins, such as those demonstrated above with GXF1 and GXS1 [73], may explain this discrepancy. In this regard, the A. thaliana proteins may potentially act as sensors or activators of HXT family transporters. These results illustrate the importance of genotype on transporter characterization studies. Moreover, they emphasize the need for simultaneous optimization of both transport processes and metabolic pathways.
Table 2.
Various host strains used to analyze transporter function.
| Strain | Relevant genotype | Other plasmids (promoter and gene) |
|---|---|---|
| TMB3201 | Δhxt1-17 Δgal2 Δstl1 Δagt1 Δmph2 Δmph3 his3-Δ1::YIpXR/XDH/XK | - |
| BY4727 | MATα his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | PADH1-PsXYL2, PPGK1-PsXYL1, PHXT7-ScXKS1-THXT7 |
| H2219 | Δhxt1-7 Δgal2 ura3-Δ1::XR/XDH his3-Δ1::XK | - |
| RE700 | Δhxt1-7 | - |
The basic genotype of strains used in prior transporter assay studies is provided.
Importantly, research has been unable to experimentally identify the highly efficient pentose transport system of P. stipitis, even though it is the source organism for many pentose metabolic genes. Specifically, a P. stipitis genomic DNA library transformed into a transporter-null strain of S. cerevisiae yielded no transformants able to grow on xylose [64]. Further investigation is needed because P. stipitis is known to have an excellent xylose transport system, yet to date no high-affinity xylose transporters have been isolated. The recently published genome sequence of this organism will certainly aid future research in this area [76], although most transporter proteins in the genome are currently annotated by homology and have not been characterized.
All of these studies make up a large body of work concerning heterologous transport protein expression in S. cerevisiae, and motivate further exploration for dedicated xylose transporters. However, future studies in this area must proceed with caution. Owing to the native transport characteristics of yeast, efficient transporters may need to be discovered through the use HXT family knockout strains [77]. It is also unknown how transporter proteins interact with each other to produce certain phenotypes. Specifically, P. stipitis xylose transport may involve transporter cooperation to achieve a high rate of xylose influx. Evidence of such transporter cooperation has already been demonstrated when coexpressing C. intermedia GXS1 and GXF1 [73]. Additionally, many transmembrane proteins act as sensors; for example, RGT2 and can control transporter expression [78]. Furthermore, the internal xylose catabolism pathway used in conjunction with heterologous or engineered xylose transporters can determine the level of flux control exhibited by the transport step [67]. Naturally, as the internal metabolic pathway is improved, pentose transport will become a greater limitation. Therefore, novel tools and approaches must accomplish simultaneous optimization of transport and metabolism - the tools and approaches of panmetabolic engineering.
Panmetabolic engineering: the path forward for pentose utilization in yeast
As demonstrated above, pathway and transport optimization strategies have had little crossover. Pathway optimization targets were shown to have moved beyond merely expressing initial pathway genes to include those encoding proteins in the pentose phosphate pathway. These modifications greatly improved xylose assimilation and fermentation rates, yet, the most advanced strains used in the transporter expression studies (TMB3201 and H2219) only contained simple complementation of the three-enzyme xylose oxidoreductase pathway. We propose that this dichotomy exists because pathway-engineering approaches begin with the assumption that internal cellular pathways limit maximum production rates. As a result, approaches focus on importing heterologous pathways, and subsequently removing intracellular flux bottlenecks by replacing low capacity enzymes with overexpressed or optimized versions [28,79]. Despite this, strains with these improvements still exhibit suboptimal growth rates on xylose and arabinose [45,51]. Regardless, these strictly intracellular pathway engineering approaches will fail when the assumption is incorrect; that is, when molecular transport limits maximum production rates. This premise is the motivation for heterologous transporter expression. However, the vast majority of transporter studies have only investigated how to obtain improved transport through heterologous transporter expression; pathway fitness is generally not considered. From these points of view, the inter-related problems of pathway inadequacy and transport limitation have been studied in isolation. Therefore, to achieve the optimal biofuel-producing cell, novel pathway engineering and molecular transport engineering approaches must be combined and simultaneously optimized.
Therefore, the field seems poised for a new development: panmetabolic engineering. In this paradigm, the transport and pathway steps comprising the whole utilization pathway will be optimized concurrently, and may result in leaps forward in biofuel production from lignocellulosic biomass. In this regard, the expression and activity profiles across the entire pathway will be coordinated for maximal flux.
To implement panmetabolic engineering, new tools will be necessary. These tools must be able to simultaneously evaluate the effectiveness of individual mutant proteins while balancing the expression of transporters and multiple pathway genes. At the moment, nearly all protein and metabolic engineering strategies proceed in a stepwise manner to improve pathways. A recent example of complex, multistep pathway optimization is seen with improving levopimaradiene production in E. coli [80]. As a result, high-throughput methods involving combinatorial engineering of transporters and downstream enzymes are necessary to accelerate pathway improvement. One such approach could be achieved by adapting existing tools such as DNA assembler [10]. This tool takes advantage of yeast homologous recombination to build multiplex gene assembly and plasmid construction without restriction enzyme cloning, and can be used to randomize genes and promoters. Thus, mixtures of transporter and pathway enzyme mutants along with expression elements could be transformed into yeast to allow for simultaneous gene and expression level selection. Other approaches are certainly possible, and will largely be informed by the ever-increasing list of combinatorial pathway modification strategies [81]. Particular challenges with such approaches include search space reduction and improved selection strategies.
Bioinformatic tools can help pre-select (or potentially, may one day design de novo) enzymes of interest for the desired pathway. Protein structural information can help limit search spaces for directed evolution or rational design strategies. Resources such as the Basic Local Alignment Search Tool (BLAST) draw upon the success of genomic sequencing projects [76,82], allowing access to the extant biodiversity in other organisms. An example of the power of structural information and homology was shown in the recent engineering of levopimaradiene synthase [80]. By combining protein engineering with upstream pathway modifications, a 2,600-fold increase in levopimaradiene production was observed [80]. In terms of pentose utilization, enzyme sequence homology among organisms was used to assemble the L. plantarum arabinose pathway in yeast [51], to discover a functional heterologous bacterial XI [48], and to find likely candidates for pentose transport capability. Increasing sequence information from organisms relevant to lignocellulosic biomass utilization (including organism such as P. stipitis and T. reesei) can advance the panmetabolic engineering approach by providing more diversity of starting pathway proteins.
However, a high-throughput method will eventually be necessary to experimentally validate the functions of sequenced genes. Screening strategies (often employed to enable directed evolution) may be useful, but a high-throughput genotype to phenotype mapping method such as multiscale analysis of library enrichment (SCALEs) [83] may work best. With this approach, individual enzymes or whole operons that confer beneficial phenotypes may be discovered. With advancements in microarray technology, an adapted form of SCALEs could be used to screen a genomic library in yeast for transporter, pathway and regulatory enzymes. Such an approach could identify novel enzymes that confer beneficial phenotypes but do not yet have an annotated 'relative' and therefore are not identifiable using in silico search strategies such as BLAST. High-throughput phenotyping and analysis will not only be useful for bioprospecting efforts, but for screening the large libraries created from the large-scale combinatorial techniques mentioned above.
Classic strain engineering methods such as evolutionary engineering have been, and will probably continue to be, effective in bringing about efficient pentose metabolism in S. cerevisiae. Thus, the role of evolutionary engineering combined with panmetabolic engineering must be considered. Evolutionary engineering has been intensively used in industry for obtaining microorganisms with desired properties, such as efficient utilization of substrate and higher product tolerance [84]. Recently, evolutionary adaptation of engineered S. cerevisiae showed significant improvements in growth rate, utilization of pentose sugars and ethanol production [61,68,85-88]. However, this process should be directed and accelerated to more robustly harness the power of natural selection to evolve S. cerevisiae strains with the desired properties for ethanol production from lignocellulosic biomass. Additionally, advances in high-throughput genome sequencing and detection of single nucleotide polymorphisms have begun to enable the resequencing of evolved strains. Such capability would speed the development of rational techniques for strain improvement by enabling a link between genotype and phenotype. Already, combining the power of evolution with rational bioinformatics and systems biology has produced iterative strain engineering [89]. As these techniques develop, combining evolutionary engineering with pathway construction can further increase yields and efficiencies. However, computational paradigms will be necessary to draw the complete link between mutant genome sequence and functional phenotype.
Directed evolution of proteins has been used for redesigning and optimizing target enzymes and has obtained significant improvements in stability, tolerance, substrate specificity and product selectivity [90], although this technique has been underused in engineering transmembrane proteins [91]. For successful biofuel production with pentose sugars, directed evolution could be used at several points in the pentose assimilation process: for optimizing enzyme properties (such as higher activity of XI, low selectivity of XR to NADPH and optimal activity of XKS), for increasing transporter specificity towards xylose and arabinose, and for improving tolerance to ethanol. By combining directed evolution with more global evolutionary engineering strategies, it may be possible to simultaneously co-evolve multiple proteins in this process. Thus, this vision of panmetabolic engineering would prove powerful for improving metabolic flux in the context of lignocellulosic biomass utilization.
Global cellular tools for improving non-pathway based phenotypes
Global approaches emerging from synthetic and systems biology may also have a role in improving pentose pathways [92]. Particularly, enhanced understanding of cellular signaling and synthetic circuitry could play a large role in optimization of pentose utilization. Lessons can be learned from other successful metabolic synthetic biology projects [93], yet the singular problem of exogenous sugar utilization will necessitate panmetabolic engineering along with global, synthetic cellular engineering.
Catabolite sensing may limit pentose metabolism, although mechanisms could be optimized to improve the fermentation characteristics of modified yeasts. Initial work has begun on understanding the regulatory effects of heterologous pathway expression [78], but further investigation must be undertaken. Glucose sensing and repression plays a large role in the metabolic response of yeast [94-98], including regulation of the hexokinase HXK2 in response to glucose [99]. Additionally, the yeast hexose transporters are subject to glucose regulation through a variety of mechanisms [100]. Furthermore, recent discoveries point to additional regulatory mechanisms triggered by glucose, such as cytosolic pH, influencing global cellular metabolic phenotypes [101]. Finally, transmembrane sensor proteins are thought to initiate several responses to extracellular metabolites. In many cases, these are major facilitator superfamily proteins with strong similarity to transport-capable transmembrane proteins. Future work must investigate the differences between a sensor and a transporter.
All of these regulatory mechanisms may play a role in the diauxic growth phenotype exhibited when yeast are presented with two carbon sources. Indeed, glucose-activated repression of the PPP may contribute to the low flux phenotypes observed in hexose and pentose cofermentations [78]. Optimization of this regulatory network must take place for panmetabolic engineering to succeed. For direction, reprogramming motility and toxin degradation in bacteria [102] demonstrates the feasibility of regulatory reprogramming, which may permit more efficient hexose and pentose consumption.
In addition to catabolite sensing and response, other global cellular phenotypes such as product tolerance have a large influence on the success of the fermentation. Importantly, improving cellular tolerances is possible, although underused [103]. In both of these applications, next generation metabolic engineering approaches such as global transcription machinery engineering (gTME) [103] may be able to alter global characteristics of S. cerevisiae. Without relying on a priori knowledge, gTME applies the success of directed evolution strategies to transcription factors. The major premise of this approach is that introducing dominant mutant alleles of generic transcription-related proteins can reprogram gene networks and cellular metabolism. In particular, key transcription factors (such as the TATA binding protein in yeast) are mutated to create diverse libraries. These mutants are selected on the basis of their ability to improve cellular phenotypes. By doing so, it is possible to alter complex phenotypes such as ethanol tolerance in yeast [103]. Using this method, transcription factors may also be engineered to optimize cellular systems for improved pentose fermentation characteristics, because the metabolism of sugars is tightly regulated [96,98]. This approach has the ability to improve several of the phenotypes related to pentose utilization, including high sugar tolerance and regulatory issues. Collectively, whole cell approaches may integrate the utilization pathway more fully into the host, and may alleviate difficulties such as cofactor imbalances and regulation.
Conclusion
Metabolic engineering approaches must be adapted to address the challenges for pathway and global cellular optimization that currently limit the construction of an integrated, efficient pentose pathway. Although much progress has indeed been made in the past 20 years towards lignocellulosic biomass conversion by yeasts, the iterative approach of identifying limiting steps, optimizing and identifying limiting steps, ad infinitum, must accelerate. More emphasis on host genome and regulatory structure must occur in future projects to understand the full effect of biological complexity on a pathway. In this regard, classic approaches combined with next-generation technologies may be combined to allow simultaneous optimization of all steps in a pathway. The idealized approach combines the power of many approaches including pathway engineering, directed evolution, evolutionary engineering, and combinatorial genetics to harness this cellular complexity. To date, many of these approaches have been performed in isolation for creating pentose-utilizing yeast. In summary, the field stands ready for the paradigm shift to panmetabolic engineering, necessarily including novel global cellular engineering tools.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
EY, SL and HA conceptualized the manuscript. EY prepared the sections on xylose transport and SL prepared the sections on catabolic pathways. All authors contributed to the introduction, conclusions and panmetabolic engineering sections. HA provided crucial review and contributed to the writing and editing of all sections. All authors read and approved the final manuscript.
Contributor Information
Eric Young, Email: young@che.utexas.edu.
Sun-Mi Lee, Email: sunmilee@che.utexas.edu.
Hal Alper, Email: halper@che.utexas.edu.
Acknowledgements
We acknowledge support from the Camille and Henry Dreyfus New Faculty Award. EY is supported by a National Science Foundation Graduate Research Fellowship.
References
- Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM. Ethanol can contribute to energy and environmental goals. Science. 2006;311(5760):506–508. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nature Reviews Microbiology. 2009;7(10):715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- Bailey JE. Toward a science of metabolic engineering. Science. 1991;252(5013):1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G, Vallino JJ. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252(5013):1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- Wiechert W. C-13 metabolic flux analysis. Metabolic Engineering. 2001;3(3):195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- Shintani D, DellaPenna D. Elevating the vitamin E content of plants through metabolic engineering. Science. 1998;282(5396):2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiology and Molecular Biology Reviews. 1999;63(1):21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Palsson BO. Metabolic flux balancing - basic concepts, scientific and practical use. Bio-Technology. 1994;12(10):994–998. [Google Scholar]
- Martin CH, Nielsen DR, Solomon KV, Prather KLJ. Synthetic metabolism: engineering biology at the protein and pathway scales. Chemistry & Biology. 2009;16(3):277–286. doi: 10.1016/j.chembiol.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Shao ZY, Zhao H, Zhao HM. DNA assembler: an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Research. 2009;37(2):e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair NU, Zhao HM. Mutagenic inverted repeat assisted genome engineering (MIRAGE) Nucleic Acids Research. 2009;37(1) doi: 10.1093/nar/gkn943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Computational Biology. 2009;5(4):e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine EL, Arnold FH. Implications of rewiring bacterial quorum sensing. Applide and Environmental Microbiology. 2008;74(2):437–445. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Lee PC, Schmidt-Dannert C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Applied Microbiology and Biotechnology. 2002;60(1-2):1–11. doi: 10.1007/s00253-002-1101-x. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nature Biotechnology. 2000;18(7):750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metabolic Engineering. 2009;11(1):13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nature Chemical Biology. 2008;4(9):564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJY, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metabolic Engineering. 2008;10(6):305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Yu H, Tyo K, Alper H, Klein-Marcuschamer D, Stephanopoulos G. A high-throughput screen for hyaluronic acid accumulation in recombinant Escherichia coli transformed by libraries of engineered sigma factors. Biotechnology and Bioengineering. 2008;101(4):788–796. doi: 10.1002/bit.21947. [DOI] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G. Uncovering the gene knockout landscape for improved lycopene production in E. coli. Applied Microbiology and Biotechnology. 2008;78(5):801–810. doi: 10.1007/s00253-008-1373-x. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315(5813):801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Liao JC. Metabolic engineering for advanced biofuels production from Escherichia coli. Current Opinion in Biotechnology. 2008;19(5):414–419. doi: 10.1016/j.copbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Current Opinion in Biotechnology. 2008;19(6):556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Van Vleet JH, Jeffries TW. Yeast metabolic engineering for hemicellulosic ethanol production. Current Opinion in Biotechnology. 2009;20(3):300–306. doi: 10.1016/j.copbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Applied Microbiology and Biotechnology. 2007;74(5):937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G. Bio-ethanol - the fuel of tomorrow from the residues of today. Trends in Biotechnology. 2006;24(12):549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Ho N, Chen Z, Brainard A. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Applied Environmental Microbiology. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Toivari MH, Penttilä M. Evidence that the gene YLR070c of Saccharomyces cerevisiae encodes a xylitol dehydrogenase. FEBS Letters. 1999;457(1):135–138. doi: 10.1016/S0014-5793(99)01016-9. [DOI] [PubMed] [Google Scholar]
- Traff K, Jonsson L, Hahn-Hagerdal B. Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast. 2002;19:1233–1241. doi: 10.1002/yea.913. [DOI] [PubMed] [Google Scholar]
- Richard P, Toivari MH, Penttilä M. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiology Letters. 2000;190(1):39–43. doi: 10.1111/j.1574-6968.2000.tb09259.x. [DOI] [PubMed] [Google Scholar]
- Wenger JW, Schwartz K, Sherlock G. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genetics. p. e1000942. [DOI] [PMC free article] [PubMed]
- Matsushika A, Inoue H, Kodaki T, Sawayama S. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Applied Microbiology and Biotechnology. 2009;84(1):37–53. doi: 10.1007/s00253-009-2101-x. [DOI] [PubMed] [Google Scholar]
- van Maris A, Winkler A, Kuyper M, de Laat W, van Dijken J, Pronk J. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Advances in Biochemical Engineering/Biotechnology. 2007. pp. 179–204. full_text. [DOI] [PubMed]
- Jeffries TW. Engineering yeasts for xylose metabolism. Current Opinion in Biotechnology. 2006;17(3):320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund M. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Advances in Biochemical Engineering/Biotechnology. 2007;108:147–177. doi: 10.1007/10_2007_062. full_text. [DOI] [PubMed] [Google Scholar]
- Richard P, Verho R, Putkonen M, Londesborough J, Penttila M. Production of ethanol from L-arabinose by Saccharomyces cerevisiae containing a fungal L-arabinose pathway. FEMS Yeast Research. 2003;3(2):185–189. doi: 10.1016/S1567-1356(02)00184-8. [DOI] [PubMed] [Google Scholar]
- Kotter P, Amore R, Hollenberg CP, Ciriacy M. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Current Genetics. 1990;18(6):493–500. doi: 10.1007/BF00327019. [DOI] [PubMed] [Google Scholar]
- Walfridsson M, Bao XM, Anderlund M, Lilius G, Bulow L, Hahn-Hagerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Applied and Environmental Microbiology. 1996;62(12):4648–4651. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Applied and Environmental Microbiology. 2003;69(7):4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 2008;72(3):379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Putkonen M, Vaananen R, Londesborough J, Penttila M. The missing link in the fungal L-arabinose catabolic pathway, identification of the L-xylulose reductase gene. Biochemistry. 2002;41(20):6432–6437. doi: 10.1021/bi025529i. [DOI] [PubMed] [Google Scholar]
- Richard P, Londesborough J, Putkonen M, Kalkkinen N, Penttila M. Cloning and expression of a fungal L-arabinitol 4-dehydrogenase gene. Journal of Biological Chemistry. 2001;276(44):40631–40637. doi: 10.1074/jbc.M104022200. [DOI] [PubMed] [Google Scholar]
- van Maris A, Abbott D, Bellissimi E, van den Brink J, Kuyper M, Luttik M, Wisselink H, Scheffers W, van Dijken J, Pronk J. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie van Leeuwenhoek. 2006;90(4):391–418. doi: 10.1007/s10482-006-9085-7. [DOI] [PubMed] [Google Scholar]
- Jin YS, Alper H, Yang YT, Stephanopoulos G. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Applied and Environmental Microbiology. 2005;71(12):8249–8256. doi: 10.1128/AEM.71.12.8249-8256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettiga M, Hahn-Hagerdal B, Gorwa-Grauslund M. Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnology for Biofuels. 2008;1(1):16. doi: 10.1186/1754-6834-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardonyi M, Hahn-Hagerdal B. The Streptomyces rubiginosus xylose isomerase is misfolded when expressed in Saccharomyces cerevisiae. Enzyme and Microbial Technology. 2003;32(2):252–259. doi: 10.1016/S0141-0229(02)00285-5. [DOI] [Google Scholar]
- Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2009;75(8):2304–2311. doi: 10.1128/AEM.02522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MSM, de Laat W, den Ridder JJJ, Op den Camp HJM, van Dijken JP, Pronk JT. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Research. 2003;4(1):69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Madhavan A, Tamalampudi S, Ushida K, Kanai D, Katahira S, Srivastava A, Fukuda H, Bisaria VS, Kondo A. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Applied Microbiology and Biotechnology. 2009;82(6):1067–1078. doi: 10.1007/s00253-008-1794-6. [DOI] [PubMed] [Google Scholar]
- Wisselink H, Toirkens M, del Rosario Franco Berriel M, Winkler A, van Dijken J, Pronk J, van Maris A. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Applied and Environmental Microbiology. 2007;73:4881–4891. doi: 10.1128/AEM.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos dos MM, Raghevendran V, Kötter P, Olsson L, Nielsen J. Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metabolic Engineering. 2004;6(4):352–363. doi: 10.1016/j.ymben.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Jin YS. Metabolic engineering for improved fermentation of pentoses by yeasts. Applied Microbiology and Biotechnology. 2004;63(5):495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- Wiedemann B, Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2008;74(7):2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal B, Gorwa-Grauslund M. The expression of a Pichia stipitis xylose reductase mutant with higher Km for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnology and Bioengineering. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Abu Saleh A, Pack SP, Annaluru N, Kodaki T, Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007;153(9):3044–3054. doi: 10.1099/mic.0.2007/007856-0. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Fromanger R, Hahn-Hagerdal B, Gorwa-Grauslund M. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 2007;73:1039–1046. doi: 10.1007/s00253-006-0575-3. [DOI] [PubMed] [Google Scholar]
- Bellissimi E, Dijken JP, Pronk JT, Maris AJA. Effects of acetic acid on the kinetics of xylose fermentation by an engineered, xylose-isomerase-based Saccharomyces cerevisiae strain. FEMS Yeast Research. 2009;9(3):358–364. doi: 10.1111/j.1567-1364.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- Chiang LC, Hsiao HY, Ueng PP, Tsao GT. Enzymatic and microbial preparation of D-xylulose from D-xylose. Applied and Environmental Microbiology. 1981;42(1):66–69. doi: 10.1128/aem.42.1.66-69.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries T. Utilization of xylose by bacteria, yeasts, and fungi. Advances in Biochemical Engineering/Biotechnology. 1983;27:1–32. doi: 10.1007/BFb0009101. full_text. [DOI] [PubMed] [Google Scholar]
- Madhavan A, Tamalampudi S, Srivastava A, Fukuda H, Bisaria V, Kondo A. Alcoholic fermentation of xylose and mixed sugars using recombinant Saccharomyces cerevisiae engineered for xylose utilization. Applied Microbiology and Biotechnology. 2009;82(6):1037–1047. doi: 10.1007/s00253-008-1818-2. [DOI] [PubMed] [Google Scholar]
- Heath EC, Horecker BL, Smyrniotis PZ, Takagi Y. Pentose fermentation by Lactobacillus planarum. Journal of Biological Chemistry. 1958;231(2):1031–1037. [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y. et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher T, Becker J, Gardonyi M, Hahn-Hagerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148(Pt 9):2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- Sedlak M, Ho NWY. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast. 2004;21(8):671–684. doi: 10.1002/yea.1060. [DOI] [PubMed] [Google Scholar]
- Saloheimo A, Rauta J, Stasyk OV, Sibirny AA, Penttila M, Ruohonen L. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Applied Microbiology and Biotechnology. 2007;74(5):1041–1052. doi: 10.1007/s00253-006-0747-1. [DOI] [PubMed] [Google Scholar]
- Gardonyi M, Jeppsson M, Liden G, Gorwa-Grausland MF, Hahn-Hagerdal B. Control of xylose consumption by xylose transport in recombinant Saccharomyces cerevisiae. Biotechnology and Bioengineering. 2003;82(7):818–824. doi: 10.1002/bit.10631. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Research. 2005;5(10):925–934. doi: 10.1016/j.femsyr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Wahlbom CF, Otero RRC, van Zyl WH, Hahn-Hagerdal B, Jonsson LJ. Molecular analysis of a Saccharomyces cerevisiae mutant with improved ability to utilize xylose shows enhanced expression of proteins involved in transport, initial xylose metabolism, and the pentose phosphate pathway. Applied and Environmental Microbiology. 2003;69(2):740–746. doi: 10.1128/AEM.69.2.740-746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiology and Molecular Biology Reviews. 1998;62(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandro MJ, Fonseca C, Goncalves P. Hexose and pentose transport in ascomycetous yeasts: an overview. FEMS Yeast Research. 2009;9(4):511–525. doi: 10.1111/j.1567-1364.2009.00509.x. [DOI] [PubMed] [Google Scholar]
- Leandro MJ, Goncalves P, Spencer-Martins I. Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochemical Journal. 2006;395:543–549. doi: 10.1042/BJ20051465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandro MJ, Spencer-Martins I, Goncalves P. The expression in Saccharomyces cerevisiae of a glucose/xylose symporter from Candida intermedia is affected by the presence of a glucose/xylose facilitator. Microbiology. 2008;154(Pt 6):1646–1655. doi: 10.1099/mic.0.2007/015511-0. [DOI] [PubMed] [Google Scholar]
- Runquist D, Fonseca C, Radstrom P, Spencer-Martins I, Hahn-Hagerdal B. Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 2009;82(1):123–130. doi: 10.1007/s00253-008-1773-y. [DOI] [PubMed] [Google Scholar]
- Hector RE, Qureshi N, Hughes SR, Cotta MA. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Applied Microbiology and Biotechnology. 2008;80(4):675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H. et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nature Biotechnology. 2007;25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Letters. 1999;464(3):123–128. doi: 10.1016/S0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- Salusjarvi L, Kankainen M, Soliymani R, Pitkanen JP, Penttila M, Ruohonen L. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microbial Cell Factories. 2008;7:16. doi: 10.1186/1475-2859-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 1993;38(6):776–783. doi: 10.1007/BF00167144. [DOI] [Google Scholar]
- Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KLJ. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(31):13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CNS, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Current Opinion in Chemical Biology. 2008;12(2):168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D. et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nature Biotechnology. 2008;26(5):553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Lynch MD, Warnecke T, Gill RT. SCALEs: multiscale analysis of library enrichment. Nature Methods. 2007;4(1):87–93. doi: 10.1038/nmeth946. [DOI] [PubMed] [Google Scholar]
- Wisselink HW, Toirkens MJ, Wu Q, Pronk JT, van Maris AJA. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Applied and Environmental Microbiology. 2009;75(4):907–914. doi: 10.1128/AEM.02268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlbom C, van Zyl W, Jonsson L, Hahn-Hagerdal B, Otero R. Generation of the improved recombinant xylose-utilizing Saccharomyces cerevisiae TMB 3400 by random mutagenesis and physiological comparison with Pichia stipitis CBS 6054. FEMS Yeast Res. 2003;3:319–326. doi: 10.1016/S1567-1356(02)00206-4. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Hartog MMP, Toirkens MJ, Almering MJH, Winkler AA, van Dijken JP, Pronk JT. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Research. 2005;5(4-5):399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Oguri E, Sawayama S. Evolutionary adaptation of recombinant shochu yeast for improved xylose utilization. Journal of Bioscience and Bioengineering. 2010;110(1):102–105. doi: 10.1016/j.jbiosc.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Garcia Sanchez R, Karhumaa K, Fonseca C, Sanchez Nogue V, Almeida J, Larsson C, Bengtsson O, Bettiga M, Hahn-Hagerdal B, Gorwa-Grauslund M. Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnology for Biofuels. 2010;3(1):13. doi: 10.1186/1754-6834-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo KEJ, Kocharin K, Nielsen J. Toward design-based engineering of industrial microbes. Current Opinion in Microbiology. 2010;13(3):255–262. doi: 10.1016/j.mib.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäckel C, Kast P, Hilvert D. Protein design by directed evolution. Annual Review of Biophysics. 2008;37(1):153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- Bayley H, Jayasinghe L. Functional engineered channels and pores. Molecular Membrane Biology. 2004;21(4):209–220. doi: 10.1080/09687680410001716853. [DOI] [PubMed] [Google Scholar]
- Young E, Alper H. Synthetic biology: tools to design, build, and optimize cellular processes. Journal of Biomedicine and Biotechnology. 2010. p. 130781. [DOI] [PMC free article] [PubMed]
- Cantone I, Marucci L, Iorio F, Ricci MA, Belcastro V, Bansal M, Santini S, di Bernardo M, di Bernardo D, Cosma MP. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell. 2009;137(1):172–181. doi: 10.1016/j.cell.2009.01.055. [DOI] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting - glucose sensing in yeast and other cells. Trends in Genetics. 1999;15(1):29–33. doi: 10.1016/S0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Research. 2002;2(2):183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 2006;70(1):253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard SL, Oliveira AP, Bro C, Olsson L, Nielsen J. A systems biology approach to study glucose repression in the yeast Saccharomyces cerevisiae. Biotechnology and Bioengineering. 2007;96(1):134–145. doi: 10.1002/bit.21135. [DOI] [PubMed] [Google Scholar]
- Gancedo JM. The early steps of glucose signalling in yeast. FEMS Microbiology Reviews. 2008;32(4):673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- Palomino A, Herrero P, Moreno F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Research. 2006;34(5):1427–1438. doi: 10.1093/nar/gkl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiology and Molecular Biology Reviews. 1999;63(3):554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. The EMBO Journa. 2010;29(15):2515–2526. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nature Chemical Biology. pp. 464–470. [DOI] [PMC free article] [PubMed] [Retracted]
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314(5805):1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. The Plant Journal. 1992;2(3):417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Weierstall T, Hollenberg CP, Boles E. Cloning and characterization of three genes (SUT1-3) encoding glucose transporters of the yeast Pichia stipitis. Molecular Microbiology. 1999;31(3):871–883. doi: 10.1046/j.1365-2958.1999.01224.x. [DOI] [PubMed] [Google Scholar]