Abstract
Background
Vampire bat related rabies harms both livestock industry and public health sector in central Brazil. The geographical distributions of vampire bat-transmitted rabies virus variants are delimited by mountain chains. These findings were elucidated by analyzing a high conserved nucleoprotein gene. This study aims to elucidate the detailed epidemiological characters of vampire bat-transmitted rabies virus by phylogenetic methods based on 619-nt sequence including unconserved G-L intergenic region.
Findings
The vampire bat-transmitted rabies virus isolates divided into 8 phylogenetic lineages in the previous nucleoprotein gene analysis were divided into 10 phylogenetic lineages with significant bootstrap values. The distributions of most variants were reconfirmed to be delimited by mountain chains. Furthermore, variants in undulating areas have narrow distributions and are apparently separated by mountain ridges.
Conclusions
This study demonstrates that the 619-nt sequence including G-L intergenic region is more useful for a state-level phylogenetic analysis of rabies virus than the partial nucleoprotein gene, and simultaneously that the distribution of vampire bat-transmitted RABV variants tends to be separated not only by mountain chains but also by mountain ridges, thus suggesting that the diversity of vampire bat-transmitted RABV variants was delimited by geographical undulations.
Background
Rabies is a zoonosis that kills infected mammals, including humans, and is mainly transmitted by carnivores. In the Americas, chiropterans (insectivorous, frugivorous and hematophagous bat) are another reservoir of this disease. Although dog-transmitted rabies in central Brazil has been reduced by aggressive vaccination programs [1], chiroptera (particularly the common vampire bat, Desmodus rotundas)-transmitted rabies remains endemic in this region, and harms both the livestock industry and the public health sector [2,3].
To date, vampire bat-transmitted rabies in livestock has been controlled by reducing the population of vampire bats and by vaccinating livestock [3,4]. However, the depopulation of vampire bats has limitations and the effects are temporary, while vaccination of livestock is only carried out for some animals and is ineffective in decreasing rabies levels in vampire bats.
For the sustainable and effective control of vampire bat rabies, further knowledge of epidemiological features, such as vampire bat ecology and the dynamics of vampire bat-transmitted rabies, is necessary. Molecular epidemiological analysis of vampire bat-transmitted cattle rabies cases using the partial nucleoprotein gene, which is the most conserved gene in the rabies virus (RABV) genome, has suggested that the distribution of variants in Brazil is delimited by mountain chains and clustered in tens of thousands of square kilometers [5]. However, the vampire bats migrate several kilometers from their nests [6]. To elucidate a more detailed genetic clustering and geo-distribution of genetic clades of vampire bat-transmitted RABV, 204 isolates from Goiás, which includes the 185 isolates analyzed previously, were employed and analyzed by a phylogenetic method based on a nucleotide sequence encompassing the G-L intergenic region locating between glycoprotein (G) and polymerase (L) gene loci, which is the most divergent region in the RABV genome and is used for monitoring epidemiological changes in the evolution of RABV [7,8].
Results
Phylogenetic analysis
The 204 RABV isolates divided into 8 phylogenetic lineages in the previous nucleoprotein gene analysis were divided into 10 phylogenetic lineages with significant bootstrap values (Figure 1; details shown in Table 1). Isolates of the C-5 and C-6 lineages designated by Kobayashi et al. belonged to the A-lineage, while the B-lineage consisted of some isolates of C-5. The C-, D-, F- and J-lineages included isolates belonging to C-12, C-1, C-22 and C-3, respectively. Isolates of C-20 belonged to the I-lineage, and C-21 was divided into three lineages; G-, H- and I-lineages. Two isolates not belonging to any lineages in previous studies were assigned to the E-lineage.
Figure 1.
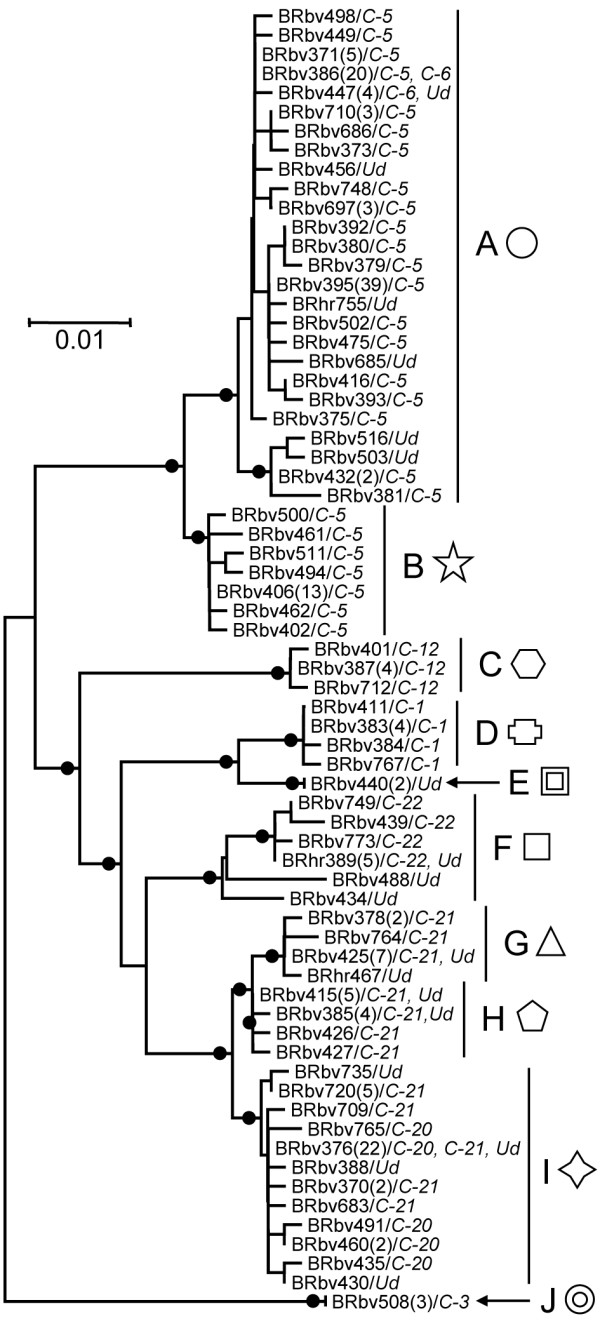
NJ tree based on 619-nt sequence of the partial G and the G-L intergenic region. Full circles on internal branches indicate significant bootstrap values (>70%). The number of isolates retaining 100% nucleotide similarity with the representative isolate is shown in parentheses. Lineages of the isolates defined in a previous study [5] are shown in italics after a diagonal (Ud: undefined).
Table 1.
Isolates from Goiás
| Grouping | Accession No. | ||||||
|---|---|---|---|---|---|---|---|
| Sample | Species | Location | Year | This study | Previous studyc | N203-nt | Partial G & GL |
| BRbv371 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307182 | AB544082 |
| BRbv372 | Cattle | Água Limpa | 2002 | A | C-5 | AB307183 | AB544083 |
| BRbv373 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307184 | AB544084 |
| BRbv375 | Cattle | Santa Cruz de Goias | 2002 | A | C-5 | AB307186 | AB544085 |
| BRbv379 | Cattle | Corumbaíba | 2002 | A | C-5 | AB307190 | AB544089 |
| BRbv380 | Cattle | Corumbaíba | 2002 | A | C-5 | AB307191 | AB544090 |
| BRbv381 | Cattle | Cristalina | 2002 | A | C-5 | AB307192 | AB544091 |
| BRbv386 | Cattle | Ipameri | 2002 | A | C-5 | AB307197 | AB544095 |
| BRbv391 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307201 | AB544099 |
| BRbv392 | Cattle | Água Limpa | 2002 | A | C-5 | AB307202 | AB544100 |
| BRbv393 | Cattle | Itumbiara | 2002 | A | C-5 | AB307203 | AB544101 |
| BRbv395 | Cattle | Itumbiara | 2002 | A | C-5 | AB307205 | AB544102 |
| BRbv396 | Cattle | Itumbiara | 2002 | A | C-5 | AB307206 | AB544103 |
| BRbv397 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307207 | AB544104 |
| BRbv398 | Cattle | Itumbiara | 2002 | A | C-5 | AB307208 | AB544105 |
| BRbv400 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307210 | AB544107 |
| BRbv404 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307214 | AB544111 |
| BRbv405 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307215 | AB544112 |
| BRbv407 | Cattle | Goiandira | 2002 | A | C-5 | AB307217 | AB544114 |
| BRbv412 | Cattle | Itapaci | 2002 | A | C-5 | AB307222 | AB544118 |
| BRbv413 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307223 | AB544119 |
| BRbv416 | Cattle | Morrinhos | 2002 | A | C-5 | AB307226 | AB544122 |
| BRbv421 | Cattle | Nova Aurora | 2002 | A | C-5 | AB307231 | AB544124 |
| BRbv429 | Cattle | Corumbaíba | 2002 | A | Uda | AB307238 | AB544129 |
| BRbv432 | Cattle | Ipameri | 2002 | A | C-5 | AB307241 | AB544132 |
| BRbv438 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307247 | AB544137 |
| BRhr441 | Horse | Buriti Alegre | 2002 | A | Ud | AB307251 | AB544140 |
| BRbv442 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307252 | AB544141 |
| BRbv447 | Cattle | Urutaí | 2002 | A | C-6 | AB307255 | AB544143 |
| BRbv449 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307256 | AB544145 |
| BRbv451 | Cattle | Ipameri | 2002 | A | C-5 | AB307258 | AB544147 |
| BRbv453 | Cattle | São Luis de Montes Belos | 2002 | A | C-5 | AB307260 | AB544149 |
| BRbv456 | Cattle | Orizona | 2002 | A | Ud | AB307263 | AB544150 |
| BRbv457 | Cattle | Água Limpa | 2002 | A | C-5 | AB307264 | AB544151 |
| BRbv458 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307265 | AB544152 |
| BRbv466 | Cattle | Itumbiara | 2002 | A | C-5 | AB307273 | AB544157 |
| BRbv469 | Cattle | Ipameri | 2002 | A | C-6 | AB307275 | AB544159 |
| BRbv471 | Cattle | Santa Cruz de Goias | 2002 | A | C-6 | AB307277 | AB544161 |
| BRbv472 | Cattle | Ipameri | 2002 | A | C-6 | AB307278 | AB544162 |
| BRbv473 | Cattle | Itumbiara | 2002 | A | C-5 | AB307279 | AB544163 |
| BRbv475 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307281 | AB544165 |
| BRbv477 | Cattle | Itumbiara | 2002 | A | C-5 | AB307283 | AB544166 |
| BRbv478 | Cattle | Itumbiara | 2002 | A | C-5 | AB307284 | AB544167 |
| BRbv482 | Cattle | Panamá | 2002 | A | C-5 | AB307288 | AB544170 |
| BRhr483 | Horse | Panamá | 2002 | A | Ud | AB307291 | AB544171 |
| BRbv486 | Cattle | Itumbiara | 2002 | A | C-5 | AB307293 | AB544172 |
| BRbv489 | Cattle | Itumbiara | 2002 | A | Ud | AB307295 | AB544174 |
| BRbv493 | Cattle | Ipameri | 2002 | A | C-6 | AB307298 | AB544177 |
| BRbv495 | Cattle | Panamá | 2002 | A | C-5 | AB307300 | AB544179 |
| BRbv496 | Cattle | Panamá | 2002 | A | C-5 | AB307301 | AB544180 |
| BRbv497 | Cattle | Itumbiara | 2002 | A | C-5 | AB307302 | AB544181 |
| BRbv498 | Cattle | Bela Vista de Goiás | 2002 | A | C-5 | AB307303 | AB544182 |
| BRbv502 | Cattle | Itumbiara | 2002 | A | C-5 | AB307306 | AB544186 |
| BRbv503 | Cattle | Orizona | 2002 | A | Ud | AB307307 | AB544187 |
| BRbv509 | Cattle | Buriti Alegre | 2002 | A | C-5 | AB307311 | AB544192 |
| BRbv514 | Cattle | Panamá | 2002 | A | C-5 | AB307316 | AB544196 |
| BRbv516 | Cattle | Orizona | 2002 | A | Ud | AB307318 | AB544198 |
| BRbv521 | Cattle | Urutaí | 2002 | A | Ndb | Nd | AB544201 |
| BRbv524 | Cattle | Urutaí | 2002 | A | C-6 | AB307323 | AB544204 |
| BRbv526 | Cattle | Panamá | 2002 | A | C-5 | AB307325 | AB544206 |
| BRbv527 | Cattle | Campos Belos | 2002 | A | C-5 | AB307326 | AB544207 |
| BRbv684 | Cattle | Ipameri | 2001 | A | C-5 | AB307412 | AB544211 |
| BRbv685 | Cattle | Corumbaíba | 2001 | A | Nd | Nd | AB544212 |
| BRbv686 | Cattle | Ipameri | 2001 | A | C-5 | AB307413 | AB544213 |
| BRbv687 | Cattle | Corumbaíba | 2001 | A | C-5 | AB307414 | AB544214 |
| BRbv690 | Cattle | Ipameri | 2001 | A | C-5 | AB307417 | AB544215 |
| BRbv691 | Cattle | Ipameri | 2001 | A | C-5 | AB307418 | AB544216 |
| BRbv694 | Cattle | Iporá | 2001 | A | Nd | Nd | AB544219 |
| BRbv697 | Cattle | Campo Alegre de Goiás | 2001 | A | C-5 | AB307423 | AB544221 |
| BRbv698 | Cattle | Campo Alegre de Goiás | 2001 | A | C-5 | AB307424 | AB544222 |
| BRbv699 | Cattle | Campo Alegre de Goiás | 2001 | A | C-5 | AB307425 | AB544223 |
| BRbv700 | Cattle | Caldas Novas | 2001 | A | Nd | Nd | AB544224 |
| BRbv701 | Cattle | Caldas Novas | 2001 | A | C-5 | AB307426 | AB544225 |
| BRhr704 | Horse | Corumbaíba | 2001 | A | Nd | Nd | AB544227 |
| BRbv705 | Cattle | Ipameri | 2001 | A | C-5 | AB307428 | AB544228 |
| BRbv707 | Cattle | Corumbaíba | 2001 | A | Nd | Nd | AB544229 |
| BRsp708 | Sheep | Ouvidor | 2001 | A | Nd | Nd | AB544230 |
| BRbv710 | Cattle | Morrinhos | 2001 | A | C-5 | AB307431 | AB544232 |
| BRbv711 | Cattle | Morrinhos | 2001 | A | C-5 | AB307432 | AB544233 |
| BRbv717 | Cattle | Ipameri | 2001 | A | C-5 | AB307438 | AB544235 |
| BRbv718 | Cattle | Ipameri | 2001 | A | C-5 | AB307439 | AB544236 |
| BRbv741 | Cattle | Marzagão | 2001 | A | C-5 | AB307450 | AB544244 |
| BRbv742 | Cattle | Ipameri | 2001 | A | C-5 | AB307451 | AB544245 |
| BRbv745 | Cattle | Buriti Alegre | 2001 | A | C-5 | AB307454 | AB544248 |
| BRbv747 | Cattle | Corumbaíba | 2001 | A | Ud | AB307456 | AB544250 |
| BRbv748 | Cattle | Campo Alegre de Goiás | 2001 | A | C-5 | AB307457 | AB544251 |
| BRbv752 | Cattle | Catalão | 2001 | A | Ud | AB307461 | AB544254 |
| BRhr755 | Horse | Cumari | 2001 | A | Nd | Nd | AB544257 |
| BRbv770 | Cattle | Ipameri | 2002 | A | C-5 | AB307476 | AB544269 |
| BRbv771 | Cattle | Caldas Novas | 2002 | A | C-5 | AB307477 | AB544270 |
| BRbv780 | Cattle | Nova Crixás | 2002 | A | C-5 | AB307484 | AB544274 |
| BRbv785 | Cattle | Corumbaíba | 2002 | A | C-5 | AB307486 | AB544277 |
| BRbv786 | Cattle | Buriti Alegre | 2002 | A | Ud | AB307487 | AB544278 |
| BRbv796 | Cattle | Pires do Rio | 2002 | A | C-6 | AB307495 | AB544283 |
| BRbv797 | Cattle | Cristalina | 2002 | A | C-6 | AB307496 | AB544284 |
| BRbv402 | Cattle | Divinópolis de Goiás | 2002 | B | C-5 | AB307212 | AB544109 |
| BRbv406 | Cattle | Posse | 2002 | B | C-5 | AB307216 | AB544113 |
| BRbv459 | Cattle | Monte Alegre de Goiás | 2002 | B | C-5 | AB307266 | AB544153 |
| BRbv461 | Cattle | Monte Alegre de Goiás | 2002 | B | C-5 | AB307268 | AB544155 |
| BRbv462 | Cattle | Monte Alegre de Goiás | 2002 | B | C-5 | AB307269 | AB544156 |
| BRbv479 | Cattle | Colinas do Sul | 2002 | B | C-5 | AB307285 | AB544168 |
| BRbv481 | Cattle | Serranópolis | 2002 | B | C-5 | AB307287 | AB544169 |
| BRbv490 | Cattle | Colinas do Sul | 2002 | B | C-5 | AB307296 | AB544175 |
| BRbv494 | Cattle | Divinópolis de Goiás | 2002 | B | C-5 | AB307299 | AB544178 |
| BRbv500 | Cattle | Colinas do Sul | 2002 | B | C-5 | AB307304 | AB544184 |
| BRbv505 | Cattle | Colinas do Sul | 2002 | B | C-5 | AB307309 | AB544188 |
| BRhr506 | Horse | Colinas do Sul | 2002 | B | Nd | Nd | AB544189 |
| BRhr507 | Horse | Colinas do Sul | 2002 | B | Nd | Nd | AB544190 |
| BRbv510 | Cattle | Monte Alegre de Goiás | 2002 | B | C-5 | AB307312 | AB544193 |
| BRbv511 | Cattle | Divinópolis de Goiás | 2002 | B | C-5 | AB307313 | AB544194 |
| BRbv518 | Cattle | Campos Belos | 2002 | B | C-5 | AB307319 | AB544199 |
| BRbv525 | Cattle | Panamá | 2002 | B | C-5 | AB307324 | AB544205 |
| BRbv753 | Cattle | Monte Alegre de Goiás | 2001 | B | C-5 | AB307462 | AB544255 |
| BRbv762 | Cattle | Divinópolis de Goiás | 2001 | B | C-5 | AB307470 | AB544263 |
| BRbv387 | Cattle | Doverlândia | 2002 | C | C-12 | AB307198 | AB544096 |
| BRbv401 | Cattle | Mineiros | 2002 | C | C-12 | AB307211 | AB544108 |
| BRbv424 | Cattle | Doverlândia | 2002 | C | C-12 | AB307234 | AB544125 |
| BRbv692 | Cattle | Mineiros | 2001 | C | C-12 | AB307419 | AB544217 |
| BRbv712 | Cattle | Doverlândia | 2001 | C | C-12 | AB307433 | AB544234 |
| BRbv751 | Cattle | Mineiros | 2001 | C | C-12 | AB307460 | AB544253 |
| BRbv383 | Cattle | Nova América | 2002 | D | C-1 | AB307194 | AB544092 |
| BRbv384 | Cattle | Nova América | 2002 | D | C-1 | AB307195 | AB544093 |
| BRbv409 | Cattle | Rubiataba | 2002 | D | C-1 | AB307219 | AB544115 |
| BRbv411 | Cattle | Morrinhos | 2002 | D | C-1 | AB307221 | AB544117 |
| BRbv452 | Cattle | Rubiataba | 2002 | D | C-1 | AB307259 | AB544148 |
| BRbv767 | Cattle | Rubiataba | 2002 | D | C-1 | AB307474 | AB544267 |
| BRbv793 | Cattle | Urutaí | 2002 | D | C-1 | AB307493 | AB544282 |
| BRbv440 | Cattle | Nova Crixás | 2002 | E | Ud | AB307249 | AB544139 |
| BRbv519 | Cattle | Mundo Novo | 2002 | E | Ud | AB307320 | AB544200 |
| BRhr389 | Horse | Nova Crixás | 2002 | F | Nd | Nd | AB544098 |
| BRbv434 | Cattle | Itapuranga | 2002 | F | Ud | AB307243 | AB544133 |
| BRbv439 | Cattle | Mundo Novo | 2002 | F | C-22 | AB307248 | AB544138 |
| BRbv488 | Cattle | Carmo do Rio Verde | 2002 | F | Ud | AB307294 | AB544173 |
| BRbv749 | Cattle | Nova Crixás | 2001 | F | C-22 | AB307458 | AB544252 |
| BRbv757 | Cattle | Monte Alegre de Goiás | 2001 | F | C-22 | AB307465 | AB544259 |
| BRbv763 | Cattle | Aruanã | 2002 | F | C-22 | AB307471 | AB544264 |
| BRbv773 | Cattle | Aruanã | 2002 | F | C-22 | AB307479 | AB544272 |
| BRbv774 | Cattle | Mozarlândia | 2002 | F | C-22 | AB307480 | AB544273 |
| BRsp787 | Sheep | Ipameri | 2002 | F | Nd | Nd | AB544279 |
| BRbv378 | Cattle | Novo Brasil | 2002 | G | C-21 | AB307189 | AB544088 |
| BRbv425 | Cattle | Novo Brasil | 2002 | G | C-21 | AB307235 | AB544126 |
| BRbv445 | Cattle | Morrinhos | 2002 | G | Ud | AB307254 | AB544142 |
| BRbv448 | Cattle | Buriti de Goiás | 2002 | G | C-21 | AB307256 | AB544144 |
| BRhr467 | Horse | Buriti de Goiás | 2002 | G | Nd | Nd | AB544158 |
| BRbv474 | Cattle | Moiporá | 2002 | G | C-21 | AB307280 | AB544164 |
| BRbv501 | Cattle | Anicuns | 2002 | G | C-21 | AB307305 | AB544185 |
| BRbv515 | Cattle | Córrego do Ouro | 2002 | G | C-21 | AB307317 | AB544197 |
| BRbv522 | Cattle | Buriti de Goiás | 2002 | G | Nd | Nd | AB544202 |
| BRbv523 | Cattle | Córrego do Ouro | 2002 | G | C-21 | AB307322 | AB544203 |
| BRbv764 | Cattle | Córrego do Ouro | 2002 | G | C-21 | AB307472 | AB544265 |
| BRbv385 | Cattle | Ivolândia | 2002 | H | C-21 | AB307196 | AB544094 |
| BRbv414 | Cattle | Ivolândia | 2002 | H | C-21 | AB307224 | AB544120 |
| BRbv415 | Cattle | Ivolândia | 2002 | H | C-21 | AB307225 | AB544121 |
| BRbv426 | Cattle | Amorinópolis | 2002 | H | C-21 | AB307236 | AB544127 |
| BRbv427 | Cattle | Ivolândia | 2002 | H | C-21 | AB307237 | AB544128 |
| BRbv431 | Cattle | Palestina de Goiás | 2002 | H | Ud | AB307240 | AB544131 |
| BRbv437 | Cattle | Goiandira | 2002 | H | Ud | AB307246 | AB544136 |
| BRbv693 | Cattle | Amorinópolis | 2001 | H | C-21 | AB307420 | AB544218 |
| BRhr703 | Horse | Ivolândia | 2001 | H | Nd | Nd | AB544226 |
| BRbv736 | Cattle | Amorinópolis | 2001 | H | C-21 | AB307446 | AB544242 |
| BRbv744 | Cattle | Amorinópolis | 2001 | H | C-21 | AB307453 | AB544247 |
| BRbv370 | Cattle | Caiapônia | 2002 | I | C-21 | AB307181 | AB544081 |
| BRbv376 | Cattle | Caiapônia | 2002 | I | C-21 | AB307187 | AB544086 |
| BRbv377 | Cattle | Bom Jardim de Goiás | 2002 | I | C-21 | AB307188 | AB544087 |
| BRbv388 | Cattle | Piranhas | 2002 | I | Ud | AB307199 | AB544097 |
| BRbv399 | Cattle | Rio Verde | 2002 | I | C-21 | AB307209 | AB544106 |
| BRbv403 | Cattle | Piranhas | 2002 | I | C-20 | AB307213 | AB544110 |
| BRbv410 | Cattle | Piranhas | 2002 | I | C-21 | AB307220 | AB544116 |
| BRbv420 | Cattle | Piranhas | 2002 | I | C-21 | AB307230 | AB544123 |
| BRbv430 | Cattle | Buriti Alegre | 2002 | I | Ud | AB307239 | AB544130 |
| BRbv435 | Cattle | Bom Jardim de Goiás | 2002 | I | C-20 | AB307244 | AB544134 |
| BRbv436 | Cattle | Piranhas | 2002 | I | Ud | AB307245 | AB544135 |
| BRbv450 | Cattle | Palestina de Goiás | 2002 | I | C-21 | AB307257 | AB544146 |
| BRbv460 | Cattle | Caiapônia | 2002 | I | C-20 | AB307267 | AB544154 |
| BRbv470 | Cattle | Piranhas | 2002 | I | C-20 | AB307276 | AB544160 |
| BRbv491 | Cattle | Caiapônia | 2002 | I | C-20 | AB307297 | AB544176 |
| BRbv499 | Cattle | Bom Jardim de Goiás | 2002 | I | Nd | Nd | AB544183 |
| BRbv513 | Cattle | Caiapônia | 2002 | I | C-20 | AB307315 | AB544195 |
| BRbv681 | Cattle | Piranhas | 2001 | I | Nd | Nd | AB544208 |
| BRbv682 | Cattle | Piranhas | 2001 | I | Nd | Nd | AB544209 |
| BRbv683 | Cattle | Piranhas | 2001 | I | C-21 | AB307411 | AB544210 |
| BRbv696 | Cattle | Piranhas | 2001 | I | C-21 | AB307422 | AB544220 |
| BRbv709 | Cattle | Piranhas | 2001 | I | C-21 | AB307430 | AB544231 |
| BRbv720 | Cattle | Arenópolis | 2001 | I | C-21 | AB307440 | AB544237 |
| BRbv721 | Cattle | Arenópolis | 2001 | I | C-21 | AB307441 | AB544238 |
| BRbv722 | Cattle | Arenópolis | 2001 | I | C-21 | AB307442 | AB544239 |
| BRbv734 | Cattle | Arenópolis | 2001 | I | C-21 | AB307444 | AB544240 |
| BRbv735 | Cattle | Arenópolis | 2001 | I | Ud | AB307445 | AB544241 |
| BRbv738 | Cattle | Piranhas | 2001 | I | C-20 | AB307448 | AB544243 |
| BRbv743 | Cattle | Marzagão | 2001 | I | C-20 | AB307452 | AB544246 |
| BRbv746 | Cattle | Caiapônia | 2001 | I | C-20 | AB307455 | AB544249 |
| BRbv756 | Cattle | Arenópolis | 2001 | I | C-21 | AB307464 | AB544258 |
| BRbv758 | Cattle | Iporá | 2001 | I | C-21 | AB307466 | AB544260 |
| BRbv759 | Cattle | Diorama | 2001 | I | C-21 | AB307467 | AB544261 |
| BRbv760 | Cattle | Palestina de Goiás | 2001 | I | C-21 | AB307468 | AB544262 |
| BRbv765 | Cattle | Caiapônia | 2002 | I | C-20 | AB307473 | AB544266 |
| BRbv769 | Cattle | Palestina de Goiás | 2002 | I | C-21 | AB307475 | AB544268 |
| BRbv772 | Cattle | Arenópolis | 2002 | I | C-21 | AB307478 | AB544271 |
| BRhr782 | Horse | Palestina de Goiás | 2002 | I | Nd | Nd | AB544275 |
| BRbv784 | Cattle | Piranhas | 2002 | I | C-20 | AB307485 | AB544276 |
| BRbv790 | Cattle | Pires do Rio | 2002 | I | C-21 | AB307490 | AB544280 |
| BRbv508 | Cattle | Porangatatu | 2002 | J | C-3 | AB307310 | AB544191 |
| BRbv754 | Cattle | Alvorada do Norte | 2001 | J | C-3 | AB307463 | AB544256 |
| BRbv792 | Cattle | Morrinhos | 2002 | J | C-3 | AB307492 | AB544281 |
aUndefined; bNo data; cKobayashi et al. (2008)
Geographical plotting
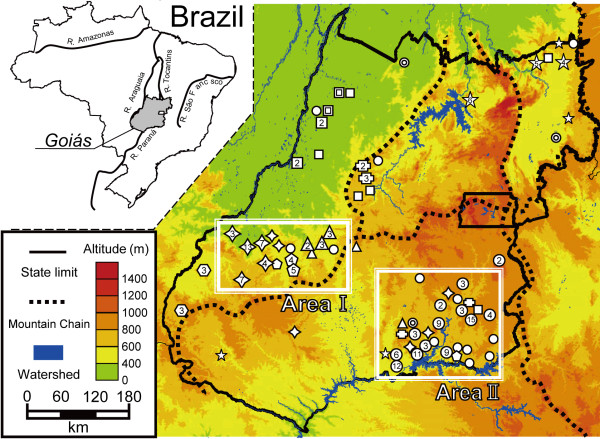
In this study, geographical areas in Goiás were divided by mountain chains into the Northwest, North central, Northeast and South regions (Figure 2). Most isolates of the A-lineage were distributed in the South region. The B-lineage was likely to exist in the Northeast and North central regions. The isolates belonging to the C-, E-, F-, G-, H- and I-lineages were distributed in the Northwest region. The G-, H- and I-lineages have narrow distributions and are apparently separated by mountain ridges (Figure 2; Area I), while the A-lineage was distributed in a wide range throughout a Southeastern basin (Figure 2; Area II). The isolates of the D-lineage were plotted on an eastern edge of the North central region. The J-lineage had a wide geographical distribution in Goiás.
Figure 2.
Distribution of vampire bat-transmitted RABV isolates in Goiás. Area I: distribution of G-, H- and I-lineages in mountain ridges. Area II: distribution of A-lineage in the basin. Symbols are same as those used in Fig. 1. Numbers inside the symbols indicate the number of isolates obtained in the same region. Small and open symbols indicated that only one isolate was obtained.
Discussion
Previous studies had elucidated that the distributions of vampire bat and transmitted RABV are delimited by mountain chains [5]. In the present study, it was reconfirmed that mountain chains divide the distribution patterns of each viral lineage. Furthermore, the isolates belonging to C-5 having a wide range in Goiás were divided into the A- and B-lineages, and were found to be distributed in the South and North regions on either side of a mountain chain. This finding supports Kobayashi's hypothesis that distribution of vampire bat-transmitted RABV is affected by mountain chains.
The same variants of vampire bat-transmitted RABV were spread widely in flat low lands (< 800 m), but at higher elevations (800-1600 m), they had a narrower distribution [9]. However, the G-, H- and I-lineages were found to be separated by mountain ridges in low areas (400-800 m) located in a southern undulating area of the Northwestern region (Figure 2; Area I). Furthermore, the A-lineage was located in an eastern basin of the South region (> 800 m; Figure 2; Area II). Considering that the higher lands showed an undulating landscape, the results suggest that the distribution patterns of vampire bat-transmitted RABV variants depend on such undulations. On the other hand, the distribution of common vampire bats in a valley is limited by the ridges that form the valley [10], thus supporting the notion that the distribution of RABV variants is affected by smaller topography than mountain chains.
Conclusions
The present study analyzed the epidemiology of vampire bat-transmitted RABV using a 619-nt region containing the partial glycoprotein gene and the G-L intergenic region, and indicated that the isolates can be further divided into several phylogenetic lineages with significant bootstrap values when compared to characterization based on the 203-nt partial N gene. Furthermore, the phylogenetic lineages were divided by both mountain chains and mountain ridges. In future studies, it will be important to analyze samples from different time points and to elucidate the dynamics of vampire bat-transmitted rabies in order to establish effective and sustainable control measures for preventing rabies circulation among vampire bats.
Methods
Samples
A total of 204 samples obtained from 192 cattle, 10 horses and 2 sheep in Goiás from October 2001 to August 2002 were employed, which had been confirmed as rabies positive through fluorescent antibody test and mouse inoculation test (Table 1). Viral RNA was extracted from the brain as described previously [11]. Lineages of the 164 cattle isolates, C-1, C-3, C-5, C-6, C-12, C-20, C-21 and C-22, were previously characterized based on a 203-nt sequence of the nucleoprotein gene [5], and are shown in Table 1.
Determination of nucleotide sequences
RT-PCR and direct sequencing with the HmG5-1302 (4619TGTTGAAGTTCACCTYCCMGATGT4642, positions relative to PV strain genome (Accession No. M13215) and RVLa-1 (5435ATRGGGTCATCATAAACCTC5416) primer pair were performed as described previously [11]. The target sequence includes the partial glycoprotein gene and G-L intergenic region. Nucleotide sequences were determined using the ATGC program version 4.02 (GENETYX Co., Tokyo, Japan).
Phylogenetioc analysis
Multiple nucleotide sequence alignments of the partial glycoprotein gene and G-L intergenic region were generated by the ClustalW package in MEGA ver. 4.0 [12]. A phylogenetic tree was constructed by the neighbor joining (NJ) method with bootstrap analysis (1000 psuedoreplicates) under the p-distance model. Phylogenetic clustering supported by a bootstrap value exceeding 70% was regarded as a reliable lineage [13]. Results were validated by the maximum likelihood method using PhyML [14]. In order to reconfirm the shape of the NJ tree, the ML tree was constructed under HKY substitution model justified by MODELTEST packaged in Hyphy program [15].
Geographical plotting
The 204 RABV isolates were plotted onto a geographical map described using the DIVA-GIS program [16] with GIS data from Instituto Brasileiro de Geografia e Estatística [17] and DIVA-GIS gData [18].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SH participated in the design of the study, performed the experimental procedures and the data analysis, and wrote the manuscript. TI, AABC, FHI and TS elaborated the study design, management, coordination, and drafting the manuscript. The authors have read and approved the final manuscript.
Contributor Information
Shinji Hirano, Email: s.hirano19821007@gmail.com.
Takuya Itou, Email: itou.takuya@nihon-u.ac.jp.
Adolorata AB Carvalho, Email: adbianco@fcav.unesp.br.
Fumio H Ito, Email: fumio@usp.br.
Takeo Sakai, Email: sakai.takeo@nihon-u.ac.jp.
Acknowledgements
This work was partly supported by the "Strategic Research Base Development" Program for Private Universities subsidized by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- Romijn PC, van der Heide R, Cattaneo CA, Silva RC, van der Poel WH. Study of lyssaviruses of bat origin as a source of rabies for other animal species in the State of Rio De Janeiro, Brazil. Am J Trop Med Hyg. 2003;69:81–86. [PubMed] [Google Scholar]
- Goncalves MA, Sa Neto RJ, Brazil TK. Outbreak of aggressions and transmission of rabies in human beings by vampire bats in northeastern Brazil. Rev Soc Bras Med Trop. 2002;35:461–464. doi: 10.1590/S0037-86822002000500006. [DOI] [PubMed] [Google Scholar]
- Mayen F. Haematophagous bats in Brazil, their role in rabies transmission, impact on public health, livestock industry and alternatives to an indiscriminate reduction of bat population. J Vet Med B Infect Dis Vet Public Health. 2003;50:469–472. doi: 10.1046/j.1439-0450.2003.00713.x. [DOI] [PubMed] [Google Scholar]
- Massad E, Coutinho FA, Burattini MN, Sallum PC, Lopez LF. A mixed ectoparasite--microparasite model for bat-transmitted rabies. Theor Popul Biol. 2001;60:265–279. doi: 10.1006/tpbi.2000.1494. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Sato G, Mochizuki N, Hirano S, Itou T, Carvalho AA, Albas A, Santos HP, Ito FH, Sakai T. Molecular and geographic analyses of vampire bat-transmitted cattle rabies in central Brazil. BMC Vet Res. 2008;4:44. doi: 10.1186/1746-6148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Sota C. Biology, ecology, and control of the vampire bat. Rev Infect Dis. 1988;10:S615–619. doi: 10.1093/clinids/10.supplement_4.s615. [DOI] [PubMed] [Google Scholar]
- Nel LH, Sabeta CT, von Teichman B, Jaftha JB, Rupprecht CE, Bingham J. Mongoose rabies in southern Africa: a re-evaluation based on molecular epidemiology. Virus Res. 2005;109:165–173. doi: 10.1016/j.virusres.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Ngoepe CE, Sabeta C, Nel L. The spread of canine rabies into Free State province of South Africa: A molecular epidemiological characterization. Virus Res. 2009;142:175–180. doi: 10.1016/j.virusres.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Ogawa A, Sato G, Sato T, Itou T, Samara SI, Carvalho AA, Nociti DP, Ito FH, Sakai T. Geographical distribution of vampire bat-related cattle rabies in Brazil. J Vet Med Sci. 2006;68:1097–1100. doi: 10.1292/jvms.68.1097. [DOI] [PubMed] [Google Scholar]
- Trajano E. Movements of Cave Bats in Southeastern Brazil, with Emphasis on the Population Ecology of the Common Vampire Bat, Desmodus rotundus (Chiroptera) Biotropica. 1996;28:121–129. doi: 10.2307/2388777. [DOI] [Google Scholar]
- Sato G, Itou T, Shoji Y, Miura Y, Mikami T, Ito M, Kurane I, Samara SI, Carvalho AA, Nociti DP, Ito FH, Sakai T. Genetic and phylogenetic analysis of glycoprotein of rabies virus isolated from several species in Brazil. J Vet Med Sci. 2004;66:747–753. doi: 10.1292/jvms.66.747. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Hills DM, Bull JJ. An empirical test of boostrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Guarino L, Cruz M, Rojas E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genet Res Newsl. 2002;127:15–19. [Google Scholar]
- Instituto Brasileiro de Geografia e Estatística (IBGE) http://www.ibge.gov.br/servidor_arquivos_geo
- DIVA-GIS gData. http://www.diva-gis.org/gData