Abstract
In 1992, Raymond et al. published a compilation of the 41 yeast vacuolar protein sorting (vps) mutant groups and described a large class of mutants (class E vps mutants) that accumulated an exaggerated prevacuolar endosome-like compartment. Further analysis revealed that this “class E compartment” contained soluble vacuolar hydrolases, vacuolar membrane proteins, and Golgi membrane proteins unable to recycle back to the Golgi complex, yet these class E vps mutants had what seemed to be normal vacuoles. The 13 class E VPS genes were later shown to encode the proteins that make up the complexes required for formation of intralumenal vesicles in late endosomal compartments called multivesicular bodies, and for the sorting of ubiquitinated cargo proteins into these internal vesicles for eventual delivery to the vacuole or lysosome.
INTRODUCTION
By the early 1990s, it had become clear that the yeast Saccharomyces cerevisiae was an excellent model system for understanding the molecular mechanisms of Golgi and endosomal sorting of proteins destined for the lysosome-like vacuole. It had also become clear that there were dozens and dozens of proteins involved in this complex sorting pathway and that analysis of the vacuolar protein sorting (VPS) genes and Vps proteins would reveal novel insight into the sorting machinery. We were still trying to grasp the sheer number of VPS genes and to put some order to the large collection of vps mutants. To this end, Christopher Raymond, a talented graduate student in the lab, set himself upon the enormous task of analyzing all the vps mutants by protein immunolocalization using fluorescence microscopy. He had a hunch that there were many secrets yet to be revealed based on the localization patterns of vacuolar proteins in yeast mutants with vacuolar protein localization defects. After extensive analysis of the vps mutants, his efforts paid off handsomely.
Leading up to our 1992 article, many articles had been published describing yeast S. cerevisiae mutants that either secreted vacuolar proteins (vpl and vpt mutants) (Bankaitis et al., 1986; Rothman and Stevens, 1986; Robinson et al., 1988; Rothman et al., 1989) or had reduced vacuolar protease activity (pep mutants) (Hemmings et al., 1981; Jones, 1984). Large-scale complementation analysis revealed at least 40 genes that came to be known as VPS, for vacuolar protein sorting, because all of these yeast mutants were defective for the sorting of proteins to the vacuole (Rothman et al., 1989). Based on light and electron microscopic analyses of vacuole morphology, Scott Emr's lab divided the 33 vpt gene groups into three distinct classes, class A, B, and C (Banta et al., 1988). Class A mutants had normal vacuole morphology (1–3 large vacuoles), class B mutants exhibited a fragmented vacuole morphology, and class C mutants exhibited extreme defects in vacuole morphology and biogenesis. In fact, class C vps mutants contained only small vesicular remnants of a vacuole. The phenotypic classification of the very large collection of vps mutants by Emr and colleagues was very important to the field because it grouped the genes encoding proteins likely to have related or common functions and thus facilitated the molecular and functional analysis of the genes that followed.
Our highly cited article in Molecular Biology of the Cell (Raymond et al., 1992) built on these previous publications. Christopher Raymond had been perfecting his immunofluorescence microscopy technique with yeast, and having read all the previous papers on vps mutants, he proceeded to immunolocalize various vacuolar proteins in a subset of these mutants. He quickly noticed that there were many more than three classes of mutants and convinced himself that a large-scale analysis of the entire vps mutant collection was in order.
In concert with the immunolocalization, Christopher Raymond together with Isabelle Howald-Stevenson and Carol Vater carried out thorough genetic and phenotypic analyses of the vps mutants. This analysis identified mutants in five additional complementation groups and revealed previously undetected overlaps in mutant complementation groups, yielding a final tally of 41 distinct vps mutant groups. But most important to the analysis was that we immunolocalized a late-Golgi membrane reporter protein (A-ALP; the C-terminal Golgi retention region of DPAP A/Ste13 fused to the membrane and lumenal domains of alkaline phosphatase) and three vacuolar proteins (alkaline phosphatase, ALP/Pho8; Vma2, subunit of the vacuole membrane V-ATPase; and carboxypeptidase Y, CPY) in multiple representatives of each of the 41 vps mutant groups. This approach was important because it allowed us to see details of vacuole morphology in these mutants that were not obvious by techniques used previously. Based on this expanded phenotypic analysis the 41 vps mutant groups were now divided into six classes, classes A through F, with classes A, B, and C as defined previously by Emr and colleagues. Class D vps mutants contained a single large vacuole in mother cells, but the mutants were largely defective for segregation of the vacuole from the mother cells to budding daughter cells, and the mutants exhibited a partial vacuole acidification defect due to the loss of the peripheral ATP-catalytic portion of the V-ATPase from the membrane. Class F vps mutants contained a large central vacuole surrounded by a number of “class B-like” fragmented vacuole structures.
By far the major discovery reported in this article, and the reason we believe it has garnered so many citations, was the identification of the fifth class of vps mutant groups, the class E vps mutants. Many of these mutants were initially placed into class A (Banta et al., 1988) because they had generally normal looking vacuoles by light and electron microscopy. However, whereas the vacuole membranes contained the integral membrane protein ALP, the classic marker of the vacuole membrane, the class E vps mutants stood out from the class A members because the V-ATPase antibodies labeled a very prominent organelle (the class E compartment) distinct from the ALP-staining vacuoles (Raymond et al., 1992). We scratched our heads long and hard trying to understand how the V-ATPase might be trapped in some “prevacuolar” structure, yet ALP was transported to the vacuole so efficiently. Because all the mutant alleles in a given class E vps gene resulted in the same phenotype and there were so many class E vps mutant groups, we became convinced that class E vps mutants were indeed blocked for transport of most vacuolar proteins at some prevacuolar, endosomal organelle and that somehow ALP transport was unaffected.
We found that soluble vacuolar proteases were also localized to the class E compartment, suggesting that some vacuolar proteins (V-ATPase, CPY, proteinase A, and proteinase B) in transit through this organelle became trapped and could not progress to the vacuole. We were probably most surprised and delighted to discover that the late-Golgi membrane reporter protein A-ALP also accumulated in this class E compartment. In the early 1990s, our lab was also investigating the retention of late-Golgi membrane proteins (such as DPAP A/Ste13), and one model being considered was retention by recycling between the Golgi and endosomal compartments. Based on this very early analysis of the class E vps mutants, we quickly recognized that it was likely that the late-Golgi membrane proteins that we were studying were indeed transported to the endosome and then cycled back to the late-Golgi compartment under the control of some as yet unidentified Vps proteins.
Christopher Raymond's double-labeling immunofluorescence studies revealed that A-ALP, CPY, and the V-ATPase all colocalized to these same class E structures. In addition, in our neighbor lab at the University of Oregon, George Sprague and his colleagues had just identified an allele of the class E vps mutant vps2 in a screen for receptor endocytosis mutants, and these vps2/ren1 mutants accumulated the a-factor receptor Ste3 in the same class E compartment (Davis et al., 1993). Together, these data revealed that recycling Golgi membrane proteins, endocytosed plasma membrane proteins, and newly synthesized vacuolar proteins all passed through a common prevacuolar endosomal-like compartment and that loss of function of any of the 13 class E VPS genes resulted in the accumulation of all these proteins in the class E compartment.
These 13 genes have since been shown to be required for the formation of intralumenal vesicles (ILVs) at the multivesicular body (MVB) and for the sorting of cargo into these ILVs. Temperature-sensitive alleles of the class E VPS27 gene were found to rapidly accumulate endocytosed proteins (Ste3) and Golgi membrane proteins (the CPY sorting receptor Vps10) in the class E compartment upon shift to the restrictive temperature, and upon shift back to the permissive temperature Ste3 was transported to the vacuole and Vps10 was transported back to the Golgi puncta (Piper et al., 1995). A year later, the Emr lab published a detailed report on the class E vps mutant vps28 that also displayed this perivacuolar compartment that accumulated cargo and showed a dramatically altered multilamellar morphology by electron microscopy (Rieder et al., 1996). These two reports together with the Raymond et al. (1992) paper defined the class E compartment.
One of the more serendipitous aspects of this study (Raymond et al., 1992) was in the choice of the two vacuolar membrane proteins to immunolocalize in the vps mutants, ALP and the V-ATPase. Christopher Raymond chose these proteins partly because we had generated antibodies to these proteins that allowed clear visualization by indirect immunofluorescence, and our lab had developed a strong interest in the biogenesis of the multisubunit V-ATPase. Whereas the V-ATPase accumulated in the class E compartment, ALP was very efficiently transported to the vacuole. We considered two models to explain these observations (see figure 9 from Raymond et al., 1992); the first hypothesized that ALP was transported on a pathway distinct from that used by the V-ATPase and soluble vacuolar proteins such as CPY, proteinase A, and proteinase B, with this “CPY pathway” intersecting the endocytic pathway at the class E compartment. An alternative model suggested that all of these proteins followed the same pathway to the vacuole, but for unknown reasons only ALP was transported efficiently through the class E compartment in class E vps mutants. Therefore, the Raymond et al., 1992 article laid the groundwork for contemporaneous articles some 5 years later from our lab as well as the labs of Scott Emr, Gregory Payne, and Sandra Lemmon demonstrating that the ALP and CPY pathways were in fact distinct (Cowles et al., 1997; Piper et al., 1997; Stepp et al., 1997) and that the CPY pathway merged with the endocytic pathway before the vacuole.
Figure 9.
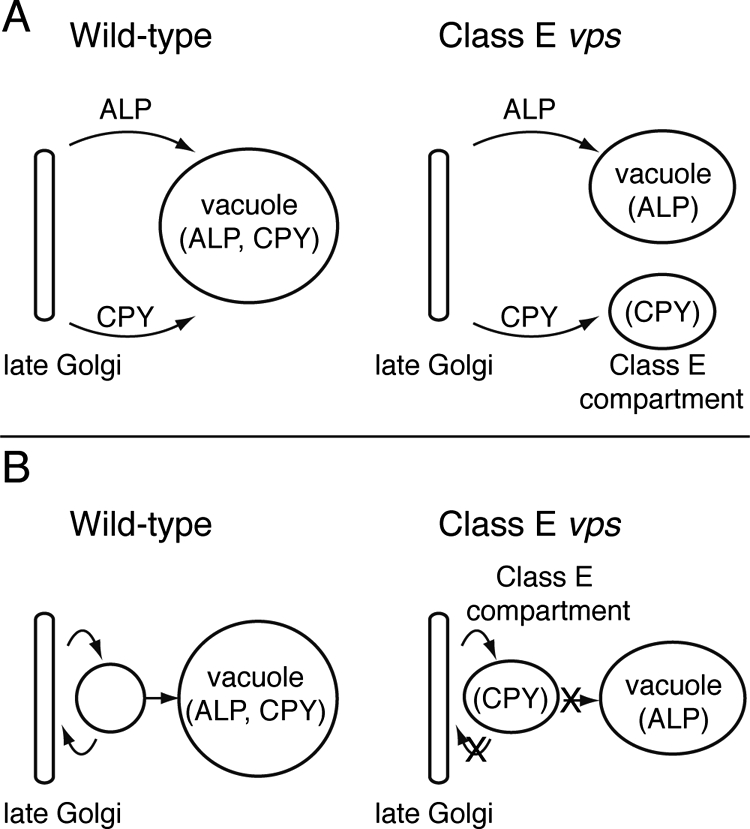
Two models for the formation of the new compartment in class E vps mutants. (A) Proteins bound for the vacuole in wild-type cells are normally packaged into at least two separate classes of transport vesicles that subsequently fuse with the vacuole. In class E vps mutants, these pathways fail to merge. In addition, late-Golgi proteins such as A-ALP enter the pathway defined by CPY as a consequence of the class E vps mutation. (B) Proteins bound for the vacuole in wild-type cells normally pass through a prevacuolar endosome-like compartment. Late-Golgi proteins are hypothesized to cycle between the late-Golgi compartment and the prevacuole. In class E vps mutants, most protein traffic (an exception being ALP) from the putative endosome-like organelle either to the vacuole or back to the late Golgi is restricted, leading to morphological exaggeration of the organelle. The steady-state resident proteins for each compartment are shown in parentheses.
Eighteen years later, it is clear that the ALP and CPY pathways are distinct (Bowers and Stevens, 2005) as suggested in part A of the figure. The 1997 studies from the Emr, Payne, and Lemmon labs further demonstrated that the adaptor protein-3 complex functioned in the cargo-selective transport of ALP from the Golgi complex directly to the vacuole (Cowles et al., 1997; Stepp et al., 1997).
When we assembled the Raymond et al., 1992 manuscript, we decided that Molecular Biology of the Cell was the perfect journal for what we believed was a groundbreaking but data-intensive manuscript. The editors of this brand-new journal professed the need in the field for a journal that reported important molecular and cellular research yet did not limit the data that could be presented to make the authors' points. This philosophy appealed to us and therefore we chose this bold new journal for our manuscript.
In the years following the discovery of the class E Vps group of proteins, they became the focus of studies designed to understand the mechanism of class E compartment formation and protein sorting through the CPY pathway. Another set of classic studies in the field came in 1997 and 1998 with the description of the ATPase Vps4, a class E Vps protein, and its role in endosomal protein sorting by Scott Emr and his colleagues (Babst et al., 1997, 1998). Later studies explored the connections between the class E Vps proteins and showed that they form four complexes termed endosomal sorting complex required for transport (ESCRT)-0, ESCRT-I, ESCRT-II, and ESCRT-III and that these complexes form oligomeric “sorting machines” on the endosomal membrane that are required for cargo sorting in yeast (Katzmann et al., 2001, 2003; Babst et al., 2002a,b; Bilodeau et al., 2002; Bowers et al., 2004) and are functionally conserved in mammals (Babst, 2005). Incidentally, the fact that the ESCRT complexes are composed of the class E Vps proteins is a good example of serendipity. Our identification of the class E grouping preceded the first report of ESCRT complexes (where the E stands for “endosomal”) by almost 10 years and the “E” in class E was merely to reflect one of the six classes of vps mutants (A–F) reported in 1992 and was not a prescient knowledge by us of the subsequent naming of the ESCRT complexes. Defining the interactions between the class E Vps proteins was the beginning of an explosion of activity in this field. Over the last decade, work from many labs has significantly advanced our understanding of the mechanism of cargo sorting and recognition at the MVB, the structure of the ESCRT complexes, the precise mode of ESCRT interactions, and the role of ubiquitin in cargo sorting.
Recently, several very important steps have been taken to understand the exact mechanism of ILV formation, membrane deformation, and vesicle scission by the ESCRT complexes. The formation and budding of ILVs into the MVB is unique in that the vesicles form and bud away from the cytoplasm with cargo included but ESCRTs excluded from the ILV. Several studies showed that ESCRT-III subunits form filaments both in vitro and when overexpressed in vivo and that these filaments can bend membranes (Ghazi-Tabatabai et al., 2008; Hanson et al., 2008; Lata et al., 2008; Saksena et al., 2009). Most impressively, in vitro assays have been developed very recently as tools to molecularly dissect the role of ESCRTs in ILV formation (Tran et al., 2009; Wollert et al., 2009; Sun et al., 2010; Wollert and Hurley, 2010). Together, these tools give the field an unprecedented look into the molecular mechanism of ILV formation and have led to new models to explain how the ESCRT complexes drive ILV formation in all eukaryotic cells.
Another very exciting aspect to this field that was unexpected by us at the time came from labs studying the life cycle of human immunodeficiency virus (HIV) type I and other retroviruses. These groups discovered that the HIV virus hijacks the ESCRT machinery to bud from infected cells as part of its normal life cycle (Garrus et al., 2001; Martin-Serrano et al., 2001). Wes Sundquist and his colleagues also defined the components and interactions of the human homologues of the yeast class E Vps proteins that make up the human ESCRT network (von Schwedler et al., 2003). Studying human homologues of the yeast ESCRT complexes has made significant contributions to our understanding of ESCRT function, the interactions between ESCRTs and viral proteins, and the consequences of these interactions for both host cells and viruses in vivo.
In addition to the well-defined function of ESCRTs at endosomes, some of these proteins also have been implicated in such diverse processes as control of chromatin structure, cell cycle control, modification of RNA polymerase activity, and tumor suppression in mammalian cells (Slagsvold et al., 2006) as well as pH signaling in yeast (Bowers et al., 2004). Several groups have also demonstrated an intriguing role for ESCRTs in cytokinesis in mammalian cells (Carlton and Martin-Serrano, 2007; Morita et al., 2007) and Arabidopsis (Spitzer et al., 2006). Another incredible finding is that Vps4 and the ESCRTIII proteins are conserved in Archaebacteria where they interact and play a role in cell division (Lindas et al., 2008; Samson et al., 2008). Not surprisingly, the human homologues of the yeast class E Vps proteins are involved in many diseases, including cancer and neurodegeneration (Saksena and Emr, 2009; Stuffers et al., 2009). These findings all highlight the importance of the class E Vps proteins in multiple biological processes in multiple systems throughout evolution, and they go far beyond anything we had imagined when we first defined the class E compartment in yeast almost 20 years ago.
Interested readers are encouraged to consult some of the many excellent reviews that have recently been published on this topic (Bowers and Stevens, 2005; Piper and Katzmann, 2007; Saksena et al., 2007; Hurley, 2008; Carlton and Martin-Serrano, 2009; Raiborg and Stenmark, 2009; Hurley and Hanson, 2010). It has been enormously gratifying to watch what began as an ambitious effort by a young graduate student to characterize vps mutants evolve into such a large field with such important and timely issues for cell biology and human disease.
ACKNOWLEDGMENTS
We thank Christopher Raymond for hard work and insightful contributions that led to the discovery of the class E vps mutants. We also thank Nia Bryant, Elizabeth Conibear, and Kate Bowers for helpful comments related to this manuscript. This work was supported by National Institutes of Health grant GM-32448 (to T.H.S.).
REFERENCES
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T. K., Banta L. M., Emr S. D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V. A., Johnson L. M., Emr S. D. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta L. M., Robinson J. S., Klionsky D. J., Emr S. D. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., Piper R. C. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bowers K., Lottridge J., Helliwell S. B., Goldthwaite L. M., Luzio J. P., Stevens T. H. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem. Soc. Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- Cowles C. R., Odorizzi G., Payne G. S., Emr S. D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Davis N. G., Horecka J. L., Sprague G. F., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus J. E., et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S., Saksena S., Short J. M., Pobbati A. V., Veprintsev D. B., Crowther R. A., Emr S. D., Egelman E. H., Williams R. L. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings B. A., Zubenko G. S., Hasilik A., Jones E. W. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1981;78:435–439. doi: 10.1073/pnas.78.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Hanson P. I. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. The synthesis and function of proteases in Saccharomyces: genetic approaches. Annu. Rev. Genet. 1984;18:233–270. doi: 10.1146/annurev.ge.18.120184.001313. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Stefan C. J., Babst M., Emr S. D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas A. C., Karlsson E. A., Lindgren M. T., Ettema T. J., Bernander R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P. D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Bryant N. J., Stevens T. H. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J. Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Cooper A. A., Yang H., Stevens T. H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. E., Banta L. M., Kohrer K., McCaffery J. M., Emr S. D. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. H., Howald I., Stevens T. H. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. H., Stevens T. H. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Saksena S., Emr S. D. ESCRTs and human disease. Biochem. Soc. Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- Saksena S., Sun J., Chu T., Emr S. D. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R. Y., Obita T., Freund S. M., Williams R. L., Bell S. D. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Muller S., Hanisch F. G., Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Stepp J. D., Huang K., Lemmon S. K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers S., Brech A., Stenmark H. ESCRT proteins in physiology and disease. Exp. Cell Res. 2009;315:1619–1626. doi: 10.1016/j.yexcr.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Sun W., Vida T. A., Sirisaengtaksin N., Merrill S. A., Hanson P. I., Bean A. J. Cell-free reconstitution of multivesicular body formation and receptor sorting. Traffic. 2010;11:867–876. doi: 10.1111/j.1600-0854.2010.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran J. H., Chen C. J., Emr S., Schekman R. Cargo sorting into multivesicular bodies in vitro. Proc. Natl. Acad. Sci. USA. 2009;106:17395–17400. doi: 10.1073/pnas.0909473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U. K., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Wollert T., Hurley J. H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]


