The TatABC subunits of the twin-arginine translocation machinery allow transport of folded proteins by an unknown mechanism. Here we show that the entire surfaces of folded Tat substrates contact TatB via both of its predicted helices. Our data suggest that TatB forms an oligomeric binding site that transiently accommodates folded Tat precursors.
Abstract
Twin-arginine-containing signal sequences mediate the transmembrane transport of folded proteins. The cognate twin-arginine translocation (Tat) machinery of Escherichia coli consists of the membrane proteins TatA, TatB, and TatC. Whereas Tat signal peptides are recognized by TatB and TatC, little is known about molecular contacts of the mature, folded part of Tat precursor proteins. We have placed a photo-cross-linker into Tat substrates at sites predicted to be either surface-exposed or hidden in the core of the folded proteins. On targeting of these variants to the Tat machinery of membrane vesicles, all surface-exposed sites were found in close proximity to TatB. Correspondingly, incorporation of the cross-linker into TatB revealed multiple precursor-binding sites in the predicted transmembrane and amphipathic helices of TatB. Large adducts indicative of TatB oligomers contacting one precursor molecule were also obtained. Cross-linking of Tat substrates to TatB required an intact twin-arginine signal peptide and disappeared upon transmembrane translocation. Our collective data are consistent with TatB forming an oligomeric binding site that transiently accommodates folded Tat precursors.
INTRODUCTION
The twin-arginine translocation (Tat) system transports proteins in a fully folded conformation across membranes of bacteria, archaea, and plant chloroplasts (recently reviewed in Panahandeh et al., 2009). Proteins are targeted to the Tat pathway by N-terminal signal sequences harboring the conserved sequence motif S-R-R-x-F-L-K (Berks, 1996). The transport is energized solely by the transmembrane H+-motive force (Mould et al., 1991; Cline et al., 1992; Bageshwar and Musser, 2007). In Gram-negative bacteria and chloroplasts, the Tat system comprises the three essential membrane proteins TatA, TatB, and TatC. Some bacteria also express a paralogue of TatA named TatE. TatD, which is expressed from the same operon as TatA, TatB, and TatC, is a soluble protein (Wexler et al., 2000). Recent studies suggest that it could be involved in the degradation of malfolded FeS-containing Tat substrates (Matos et al., 2009). Although a quality control system rejecting malfolded Tat substrates is well documented (Sanders et al., 2001; DeLisa et al., 2003; Matos et al., 2008; Panahandeh et al., 2008), folding is not a prerequisite for being transported via the Tat pathway (Roffey and Theg, 1996; Hynds et al., 1998; Cline and McCaffery, 2007; Richter et al., 2007).
TatC that is predicted to span the membrane six times provides the primary recognition site for Tat signal peptides (Cline and Mori, 2001; Alami et al., 2003; Gerard and Cline, 2006; Marrichi et al., 2008). The precise signal peptide-binding site of TatC has not yet been identified, but it seems that the entire N-terminal half of TatC is involved in precursor recognition (Holzapfel et al., 2007), with the first cytosolic loop of TatC being of particular importance (Kreutzenbeck et al., 2007; Strauch and Georgiou, 2007).
TatA, much like TatB, consists of a single transmembrane domain and one amphipathic helix most likely located on the cytoplasmic leaflet of the membrane. Differently sized homooligomeric complexes of TatA were described after various in vitro and in vivo analyses (Barrett et al., 2005; Gohlke et al., 2005; McDevitt et al., 2006; Greene et al., 2007). Assembly of these TatA complexes, presumably originating from tetrameric units, requires the presence of precursor and TatBC (Mori and Cline, 2002; Dabney-Smith et al., 2006; Leake et al., 2008; Ridder et al., 2009). Homooligomeric complexes of TatA are consistent with the idea that TatA forms a size-fitting protein-conducting channel. However, supporting data on translocation intermediates stalling near TatA are scarce (Panahandeh et al., 2008). On the contrary, one Tat substrate was in fact found arrested in a membrane environment completely devoid of any Tat components (Cline and McCaffery, 2007). In combination with the finding that exposure of hydrophobic side chains blocked Tat-dependent transport (Richter et al., 2007), these data seem to support a “membrane-weakening” function of TatA by which it would allow transmembrane passage next to membrane lipids (Brüser and Sanders, 2003). Even more controversial views are that TatA might undergo topology inversion (Gouffi et al., 2004; Chan et al., 2007) or even function as a soluble protein (Schreiber et al., 2006; Berthelmann et al., 2008; Frielingsdorf et al., 2008).
Despite 25% sequence homology, TatB cannot functionally be replaced by TatA (Sargent et al., 1999). Discrete mutations at the extreme N terminus of TatA, however, render it bifunctional now being able to suppress a TatB deficiency (Blaudeck et al., 2005; Barrett et al., 2007). This is the natural situation in Gram-positive bacteria that only possess TatAC-systems (Jongbloed et al., 2006). In E. coli, however, TatB is necessary for the transport of all tested natural Tat precursors. TatB forms 1:1 complexes with TatC (Bolhuis et al., 2001) that associate into higher oligomeric assemblies (McDevitt et al., 2005; Oates et al., 2005; Orriss et al., 2007). Together with TatC, TatB is thus part of a receptor complex for Tat precursors directly interacting with Tat signal peptides (Alami et al., 2003; Kreutzenbeck et al., 2007; Panahandeh et al., 2008). Several reports suggest that before recognition by TatBC, a Tat precursor might first bind to membrane lipids (Hou et al., 2006; Shanmugham et al., 2006; Bageshwar et al., 2009).
While previous studies on the interaction between Tat precursors and the subunits of the Tat machinery were largely restricted to the area of the signal sequence, we wanted to analyze which contacts the folded mature domains of Tat precursors would enter upon membrane targeting. Unexpectedly we find that the major binding site for folded substrates is formed by TatB.
MATERIALS AND METHODS
Plasmids
The precursors of SufI and TorA-PhoA and their KK-mutants were expressed from plasmids pKSMSufI-RR, pKSMSufI-KK (Alami et al., 2003) and pET28aTorA-PhoA, pET28aTorA-PhoAKK (Panahandeh et al., 2008), respectively. These plasmids also served as templates for the introduction of TAG stop codons at different amino acid positions of the mature domains of SufI and PhoA, and p8737 (Alami et al., 2002) for the insertion of stop codons into tatB. PCR-based site-specific mutations were introduced using the QuickChange Site-directed Mutagenesis kit system (Stratagene). The individual primer pairs used to introduce the amber stop codons are given in the Supplemental Material. Plasmid pKSM717SufI, S26L, A27L, Q30L carrying the indicated amino acid exchanges was constructed using pKSMSufI-RR as template and the primers listed under Supplemental Material.
In Vitro Reactions
S-135 cell extracts were prepared either from E. coli strain SL119 (Lesley et al., 1991) or Top10 (Invitrogen) transformed with plasmid pSup-BpaRS-6TRN(D286R) (Ryu and Schultz, 2006) to express amber mutants of Tat precursors. Cells were grown overnight in the presence of 10 μg/ml tetracycline (SL119) and 35 μg/ml chloramphenicol (Top10, pSup-BpaRS-6TRN(D286R), respectively, and were used to inoculate fresh medium without antibiotics at a 1:100 ratio. Cells were grown and S-135 cell extracts were prepared as described (Moser et al., 2007).
Coupled transcription/translation of pSufI and pTorA-PhoA from plasmid DNAs was performed as described (Moser et al., 2007). In case of pTorA-PhoA, oxidizing conditions were provided by the addition of 5 mM oxidized glutathione (GSSG) at the start of synthesis. Incorporation of Bpa was performed as described (Panahandeh et al., 2008). For cross-linking and transport experiments, membrane vesicles were added 10 min after starting the synthesis reaction and incubated for 25 min at 37°C. To assay translocation of precursors into INVs, samples were treated with proteinase K according to ref. (Moser et al., 2007). Before UV-induced cross-linking (Panahandeh et al., 2008), reactions were stopped with 0.4 mM puromycin (final concentration) for 5 min at 37°C.
To analyze translocation of membrane-bound precursor, pSufI was synthesized in the presence of Tat+-INV. INV were reisolated by centrifugation at 100,000 g for 30 min at 4°C and resuspended in buffer to give the same ionic conditions as before with either no further additives or an ATP-regenerating system (Moser et al., 2007). Incubation at 37°C for 30 min was followed by PK treatment and UV irradiation.
To analyze membrane-binding independently of translocation, pSufI was synthesized in the absence of INVs for 30 min. Reactions were stopped by incubating samples for 5 min with puromycin. To deplete any residual ATP, hexokinase and glucose were added at final concentrations of 0.01 u/ml and 12 mM respectively, and samples were incubated for 10 min at 37°C. To then allow membrane-binding of pSufI, samples were incubated with INV for 15 min at 37°C in the presence of 0.1 mM CCCP before PK treatment and UV irradiation.
Immunoprecipitations were performed as described (Alami et al., 2003) with some modifications. After the final washing step, antigen-antibody complexes were released from Protein A-Sepharose by shaking samples in SDS-PAGE loading buffer (Moser et al., 2007) for 10 min at 37°C at 1400 rpm. After centrifugation for 1 min at 16000 × g, the supernatant was transferred to a new reaction tube and incubated for 5 min at 95°C when immunoprecipitating TatA and TatB and for 10 min at 56°C when immunoprecipitating TatC.
Preparation of Membrane Vesicles
Tat+-INV were prepared as described previously (Moser et al., 2007) from E. coli strain BL21(DE3)pLysS transformed with plasmid p8737 (Alami et al., 2002). Tat genes were induced at OD600 = 0.5 with 1 mM isopropyl thio-ß-d-galactopyranoside (IPTG).
Tat+-INV containing TatB amber mutants were prepared from E. coli strain BL21(DE3) transformed with plasmids pSup-BpaRS-6TRN(D286R) and p8737 carrying the individual tatB mutations. Tat genes were induced at OD600 = 0.5 with 1 mM IPTG. At the same time 1 mM Bpa was added from a 1M stock solution made in 1 M NaOH. Preparation continued as described (Moser et al., 2007), avoiding extensive exposure to day light.
Miscellaneous
Polyclonal antibodies against TatA, TatB, and TatC were raised in rabbits as described (Alami et al., 2002; Alami et al., 2003). Immunoblotting was performed using the Lumi-LightPlus Western Blotting Kit (Roche) following the manufacturer's instructions. SDS-PAGE was performed as described previously (Moser et al., 2007). Transport efficiencies were calculated using ImageQuant 5.2 (GE Healthcare).
RESULTS
Site-Specific Introduction of a Photo-Cross-Linker into the Mature Domains of Tat Precursor Proteins
To probe for signal sequence-independent contact sites between a Tat precursor protein and subunits of the Tat translocation apparatus, we introduced the UV-sensitive p-benzoyl-derivative of phenylalanine (Bpa) into the mature part of SufI. SufI (FtsP) is a cofactor-free natural Tat substrate of E. coli (Tarry et al., 2009a) and has extensively been used for in vivo as well as in vitro analyses of the Tat export pathway in E. coli.
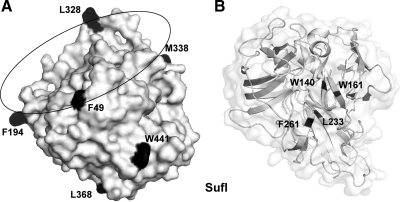
To incorporate the photo-cross-linker Bpa into SufI, amber (TAG) stop codons were introduced at selected sites of the sufI DNA. Based on the crystal structure of SufI (Tarry et al., 2009a), we chose the six surface-exposed sites illustrated in Figure 1A. Four of these positions (F49, F194, L328, and M338) are all located on the outer circumference on one side of SufI, whereas positions L368 and W441 lie at the opposite pole of the globular protein. As controls we selected the four positions, W140, W161, L233, and F261, for which the crystal structure of SufI predicts a central location thus hiding Bpa in the core of the molecule (Figure 1B).
Figure 1.
Surface-exposed and internal amino acid residues of SufI that were exchanged by the photo-cross-linker benzoyl-phenylalanine. (A) Surface-exposed residues of SufI. Structural data were taken from Tarry et al. (2009a). (B) Internal residues W140, W161, L233, and F261 of SufI.
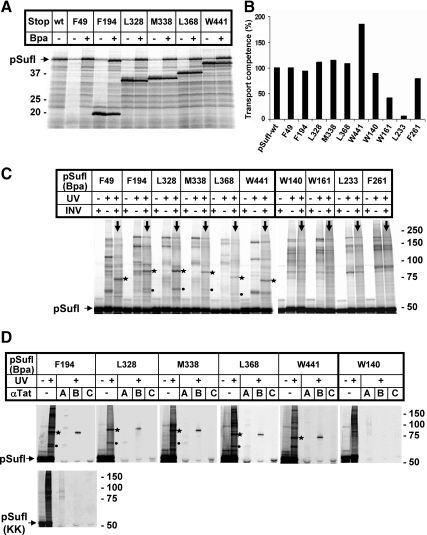
The SufI amber mutants were expressed in a cell-free transcription/translation system equipped with an amber-specific suppressor tRNA and a cognate tRNA-synthetase (Ryu and Schultz, 2006). Because this tRNA-synthetase charges Bpa to the tRNAsup, amber codons are suppressed if the in vitro translation is performed in the presence of Bpa. This is illustrated in Figure 2A displaying radiolabeled translation products by SDS-PAGE and phosphorimaging. In the absence of Bpa (−Bpa), prominent truncates of SufI were obtained due to premature termination at the individual amber stop codons introduced. The only fragment that was too short to be retained on the gel was that of the F49 mutant. In addition to the termination products, small amounts of full-size SufI precursor (pSufI) were obtained even in the absence of added Bpa, suggesting some read-through of the amber stop codons under these in vitro conditions. In the presence of added Bpa, however, the amounts of full-size pSufI had considerably increased for all amber mutants, indicating suppression of the stop codons by incorporation of Bpa (the same results were obtained for the internal sufI mutants, not shown).
Figure 2.
On membrane-targeting, the surface of pSufI is found predominantly in the vicinity of TatB. (A) Codons of the sufI DNA that specify the indicated amino acids were mutated to amber stop codons. The resulting DNAs were transcribed and translated in vitro by an E. coli cell-free extract prepared from a strain that expresses an amber suppressor tRNA and the cognate Bpa-specific tRNA-synthetase. Radiolabeled translation products were separated by SDS-PAGE and are visualized by Phosphorimaging. The electrophoretic mobilities of molecular mass standards are indicated on the left. In contrast to the approx. 50-kDa wild-type (wt) precursor of SufI (pSuf), smaller translation products were predominantly expressed from the amber mutant DNAs when Bpa was missing. These smaller translation products match the molecular masses expected for premature termination at the individual amber codons. The shortest fragment of 48 amino acids was not retained by the polyacrylamide gel used here. In addition to each premature termination product, some full-size pSufI was synthesized in the absence of Bpa, obviously resulting from read-through of the mutant mRNAs. Addition of Bpa led to a clear increase in full-size pSufI of each variant, indicating suppression of the amber codons by the incorporation of the cross-linker Bpa. (B) Wild-type pSufI and its indicated Bpa variants were synthesized in vitro in the presence of inside-out inner membrane vesicles, and the transport efficiency of each mutant was analyzed as described in Materials and Methods. Transport efficiency of wt-pSufI was set 100%. (C) After synthesis of each indicated Bpa variant of pSufI in the presence or absence of membrane vesicles (INV), samples were irradiated with UV light or mock-incubated before SDS-PAGE and Phosphorimaging. UV irradiation led to numerous radiolabeled bands larger in size than pSufI, only few of which were specifically obtained in the presence of INV (lanes marked with downward pointing arrows). The most prominent of those photo-adducts are labeled with asterisks and dots. Of notice, no membrane-specific adducts were obtained for SufI variants W140, W161, L233, F261 carrying Bpa in the interior of the folded structure. (D) The indicated Bpa variants of pSufI were synthesized in vitro in the presence of INVs. Samples irradiated with UV light were either directly prepared for SDS-PAGE or only after coimmunoprecipitation with antibodies directed against TatA, TatB, and TatC (αTat). Asterisk-labeled adducts are recognized by anti-TatB antibodies and dot-labeled ones by anti-TatA antibodies. No Tat-specific adduct was observed for the internal W140 variant of pSufI, nor were any specific cross-links obtained with the nonfunctional KK-precursor of SufI.
To test to what extent the mutations affected translocation competence, transport of the pSufI variants into inside-out inner membrane vesicles of E. coli (INV) was measured (for experimental details see below Figure 3A). When compared with wild-type pSufI, all of the mutants carrying Bpa on the surface exhibited unimpaired translocation efficiencies. In contrast, transport of two internal mutants (W161 and L233) was markedly reduced suggesting a major destabilization of SufI's tertiary structure by Bpa at those locations.
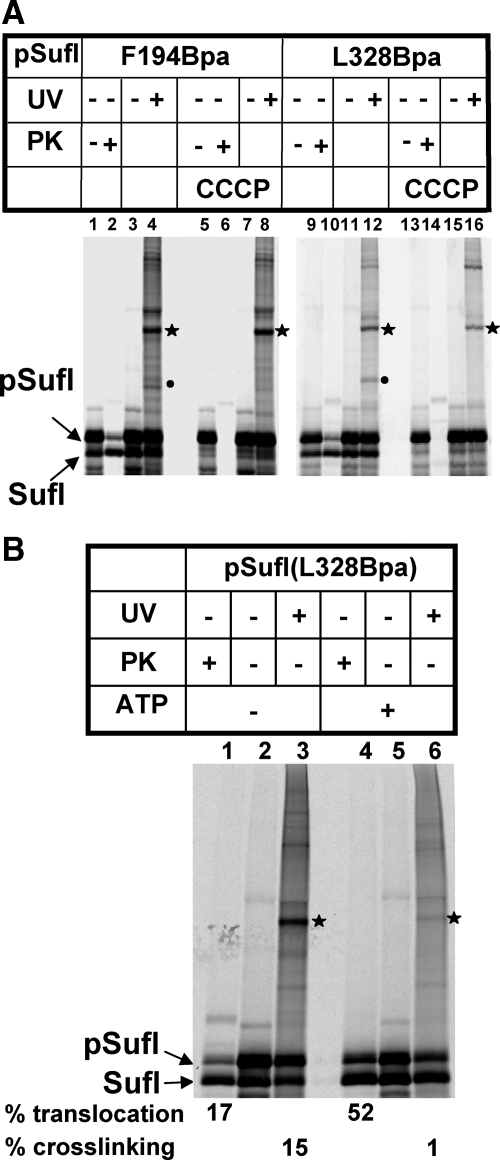
Figure 3.
A Tat precursor interacts transiently with TatB before its translocation. (A) The two indicated Bpa variants of pSufI were synthesized in the presence of INV, one-half of each reaction containing the uncoupler CCCP at 0.1 mM, the other half only the solvent DMSO. Samples were either digested with Proteinase K (PK) to visualize protease-resistant (i.e., translocated precursor and mature forms of SufI) or otherwise irradiated with UV light. The labeled photo-adducts are those identified in Figure 2. Whereas CCCP completely blocks translocation of pSufI into INVs and also abolishes cross-linking to TatA, the TatB adducts of pSufI were not affected by the uncoupler. (B) After synthesis of pSufI in the presence of INV, vesicles were collected by centrifugation, resuspended, and incubated in the absence or presence of an ATP regenerating system (cf. Materials and Methods). The ATP-dependent increase in translocation is paralleled by a loss of cross-linking to TatB.
The Predominant Interacting Partner of Folded SufI Is TatB
We then synthesized the ten sufI mutants in vitro in the presence of Bpa to allow for the incorporation of the photo-cross-linker and, after addition of membrane vesicles, for the interaction with the Tat apparatus. In the autoradiogram of Figure 2C, the results obtained for the individual pSufI variants are represented by three lanes each. Comparing from left to right each first and third lane (downward pointing arrows) reveals a UV-dependent appearance of multiple radiolabeled bands that are bigger than pSufI. The majority of these pSufI adducts were also obtained in the absence of INV (compare each second and third lane). However, for the surface-exposed Bpa variants F49 – W441 of pSufI, a small number of specific adducts were seen only after addition of INVs. The most prominent ones are those marked with an asterisk migrating around the 75-kDa marker position. Less prominent INV-specific adducts were smaller in mass and were seen clearly only for four of the Bpa variants of pSufI (dots).
To identify these INV-specific pSufI adducts, we performed immunoprecipitation with antibodies against TatA, TatB, and TatC. Figure 2D depicts the results for five of the surface-exposed Bpa variants of pSufI. The dot-labeled pSufI adducts were all recognized by anti-TatA antibodies. No contacts to TatC were detected. However, by far the most prominent cross-links were the asterisk-labeled ones that were precipitated with antibodies directed against TatB. The F49 variant not included in Figure 2D, had already previously been reported to cross-link to TatB (Alami et al., 2003). Thus all six cross-linker positions that cover a considerable surface area of the folded SufI (Figure 1A) were found in the preferential proximity of TatB. Cross-linking was strictly restricted to surface-exposed sites, because none of the four internal Bpa-variants yielded any INV-specific cross-link (Figure 2C, W140 – F261), nor did immunoprecipitation reveal any significant Tat-cross-reactive material (exemplified in Figure 2D for W140).
The five pSufI-TatB adducts displayed fairly different electrophoretic mobilities (asterisk-labeled bands in Figures 2, C and D). A possible explanation is different geometries of the branched adducts that are expected to form as a result of TatB cross-linking to distantly located contact sites on the surface of folded SufI. Position-specific cross-links varying in electrophoretic mobility have also been observed for Sec precursors proteins and the chaperone Trigger factor (Eisner et al., 2003).
To evaluate the functional relevance of the cross-linking of folded SufI to TatB, we constructed and tested variants of the SufI amber mutants that had the RR-motif replaced by the transport-deficient KK-pair. The results are shown in Figure 2D for SufI(F194Bpa) directly displaying the RR- and KK-variants on top of each other. The characteristic cross-links (dot and asterisk) of the RR-percursor were not obtained for the KK-mutant and no immunoreactive material could be recovered, except for some diffuse TatA-cross-reactive bands of higher molecular mass. These are contaminants recognized by our polyclonal antiserum, because they were obtained even when the cross-linker was hidden in the core of SufI (Figure 2D, right, W140Bpa). Thus the observed contacts between a considerable surface area of mature SufI and TatB require an intact Tat signal sequence to form and therefore are likely to reflect functional interactions of the in vitro synthesized pSufI with INVs. Because the contact sites were restricted to surface-exposed residues of pSufI, it is obvious that TatB interacted only with folded precursor molecules.
Folded pSufI Transiently Contacts TatB before Translocation
In the preceding experiments, interactions between pSufI and TatB/TatA were monitored while pSufI was synthesized in the presence of INVs (i.e., in conditions that would allow both binding and translocation into the vesicles). To assign the pSufI-TatB adducts to either of the two events, we performed the cross-linking experiment with the Bpa variants F194 and L328 of pSufI in the presence of carbonyl cyanide m-chlorophenyl-hydrazone (CCCP). CCCP dissipates the PMF (proton-motive force) of the vesicles and thereby abolishes Tat-dependent translocation into INVs but does not interfere with binding (Ma and Cline, 2000; Alami et al., 2002). Figure 3A depicts transport of pSufI into INV as indicated by the conversion of pSufI to SufI and the appearance of proteinase K (PK)-resistant pSufI and SufI species (lanes 1, 2 and 9, 10). After addition of the uncoupler CCCP, however, both signs of transport were missing for both pSufI variants tested (lanes 5, 6 and 13, 14). Whereas the cross-links to TatA (dot) also disappeared in the presence of CCCP, those to TatB (asterisk) were still visible (compare lanes 4–8 and 12–16). Consistent with a PMF-dependent recruitment of TatA to TatBC (Mori and Cline, 2002; Alami et al., 2003; Dabney-Smith et al., 2006), the pSufI-TatA adducts are therefore likely to represent later steps during translocation, whereas the PMF-insensitive TatB cross-links rather reflect a pretranslocational binding event. Because the size of the TatB adducts did not change upon addition of CCCP, which also abolishes signal sequence cleavage (compare lanes 1–5 and 9–13), they are likely adducts of the signal sequence-containing SufI precursor.
To examine whether the multiple contacts, observed between folded pSufI and TatB, reflected a functional intermediary stage of the Tat export process, we synthesized pSufI(L328Bpa) in the presence of INV to allow for binding and transport and then recovered the vesicles with the bound precursor by ultracentrifugation. When the resuspended INVs were mock incubated in buffer solution followed by treatment with PK, ∼17% of the INV-associated SufI species turned out to be protease-protected (Figure 3B, lanes 1 and 2). Most of this PK-protected SufI must have originated from translocation into the vesicle lumen during the initial period of synthesis, because no ATP was added for reenergization to the isolated vesicles. UV irradiation of these INVs revealed that ∼15% of the vesicle-associated pSufI was present in the 75-kDa TatB adduct (lane 3). Incubation of the recovered INVs in the presence of an ATP-regenerating system, however, changed the situation completely (lanes 4–6). Now virtually all of the SufI-TatB adducts, which could be recovered from the mock-treated INV, had vanished (lane 6), and concomitantly the amount of SufI that was translocated into the PK-protected environment of the INVs had substantially increased (52%; lane 4). These results therefore clearly indicate that the cross-links observed between the surface of a folded Tat precursor and TatB represent a temporary binding event that precedes Tat-dependent translocation.
Site-Specific Introduction of a Photo-Cross-Linker into TatB
The results presented so far would be consistent with TatB functioning as a pretranslocational attachment site for a folded Tat precursor. If so, the question arises where on TatB the precursor-binding sites would be located. To address this experimentally, we introduced the cross-linker Bpa now into TatB rather than into the Tat precursor protein. Codons specifying a total of nine amino acids were mutated to amber stop codons. The selected amino acids (Figure 4A) are located throughout the essential parts of TatB as known from truncation analysis (Lee et al., 2002). Mutagenizing PCR was performed on tatABCD templates contained in a high expression plasmid. These constructs were transformed into otherwise wild-type E. coli cells together with plasmid pSup-BpaRS-6TRN(D286R) encoding the amber suppressor tRNABpa and the cognate tRNA synthetase. When grown in the presence of Bpa, transformants expressed full-size TatB variants (not shown) indicating successful suppression of the stop codons and incorporation of Bpa into TatB. All membrane vesicles isolated from the tatB mutant strains contained TatB levels comparable to that of the control strain transformed with the wild-type tatABCD plasmid and were active in pSufI translocation (see below and Supplemental Figure).
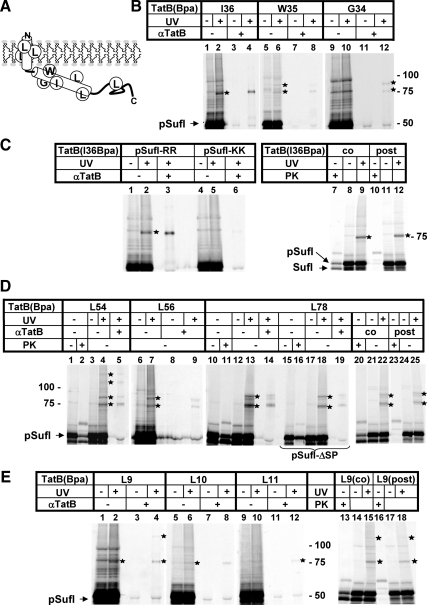
Figure 4.
Incorporation of the cross-linker Bpa into TatB reveals multiple contacts between an extended part of the molecule and precursor proteins. (A) Model of TatB depicting its predicted transmembrane and amphipathic helices and the approximate positions of Bpa. (B) In vitro synthesis of radiolabeled pSufI in the presence of INVs containing the indicated Bpa variants of TatB. UV-irradiation yielded cross-links of varying intensities (asterisks) that were immunoprecipitated by anti-TatB antibodies. Note the different intensities by which the three positions on a helix turn of TatB cross-link to pSufI. (C) In vitro synthesis of radiolabeled pSufI in the presence of INVs containing the I36Bpa variant of TatB. Cross-linking was completely abolished if the signal sequence of pSufI carried a KK-mutation of the consensus RR-motif (lanes 1–6). Cross-linking was unimpaired (lanes 7–12), if INVs were added posttranslationally after depleting all energy sources to prevent translocation into INV (cf. Materials and Methods). Protease-protected SufI species shown in lane 7 demonstrate the translocation activity of the TatB(I36Bpa) variant and the lack of PK protection in lane 10 confirms the successful depletion of energy. (D) In vitro synthesis of radiolabeled pSufI in the presence of INVs containing the indicated Bpa variants of TatB. Note the adducts to TatB (L54Bpa) larger than 100 kDa that likely contain more than one TatB monomer. The origin of the TatB-adduct running at 90 kDa is not clear. This and the 75-kDa adduct were also obtained with a noncleavable mutant of pSufI (pSufI-ΔSP) indicating that all cross-links were between TatB and the precursor of SufI (lanes 15–19). Cross-linking of TatB to pSufI occurred independently of translocation (lanes 20–25). (E) In vitro synthesis of radiolabeled pSufI in the presence of INVs containing the indicated Bpa variants of TatB reveals a position-dependent cross-linking of the transmembrane helix of TatB to pSufI (lanes 1–12). Note the high molecular mass adduct to TatB(L9Bpa) indicative of a TatB oligomer. Cross-linking of TatB to pSufI occurred independently of translocation (lanes 13–18).
Membrane-Targeting of a Tat Precursor Is Characterized by Multiple Contacts with the Cytosolic Domains of TatB
The tatB mutant vesicles were then used in the same type of in vitro assays shown in Figures 2 and 3, except that the cross-linker was now present in the vesicles rather than in the radiolabeled pSufI. Figure 4B shows that UV-irradiation of Bpa placed in the three consecutive positions I36, W35, G34 of the amphipathic helix of TatB yielded radiolabeled (and hence pSufI-containing) adducts, which were recognized by antibodies against TatB (asterisks). The intensity of cross-linking dropped from position I36 over W35 to G34, the adducts of which could only be detected by immunoprecipitation (lane 12). These findings suggest that the predicted amphipathic helix of TatB is in fact in a helical conformation when contacting the substrate. Because Bpa is virtually a spacer-less cross-linker, the weak adducts obtained when it was moved around the perimeter of the amphipathic helix most likely reflect some rotational mobility of the helix. Except for position I36, the other Bpa locations yielded double TatB adducts (see below).
To discriminate between mere molecular proximity and functional binding, we tested cross-linking of the TatB (I36Bpa) variant also to a KK-mutant precursor of SufI. As shown in Figure 4C (lanes 1–6), no cross-linking of TatB to pSufI carrying the inactive Tat signal sequence was observed, suggesting that the interaction with TatB required a previous proof of Tat compatibility. Moreover, interaction of the amphipathic helix of TatB with pSufI did not simply result from a stalling of precursor at the TatB(I36Bpa) variant, as could have been expected if the Ile-to-Bpa exchange had impaired the ability of TatB to functionally interplay with the Tat machinery. As shown by protease protection, the TatB(I36Bpa)-INVs were, however, active in translocation of pSufI (Figure 4C, lane 7 and Supplemental Figure).
To determine more closely at which step of the Tat export pathway the amphipathic helix of TatB came into contact to pSufI, we performed synthesis of pSufI first in the absence of INVs, stopped translation with puromycin, and then depleted samples of any residual ATP by the addition of hexokinase and glucose before adding membranes together with the uncoupler CCCP. As expected, no translocation of pSufI occurred under these completely energy-deprived conditions (Figure 4C, compare lanes 7 and 10). Nevertheless the 75-kDa adduct between pSufI and the TatB of the deenergized INVs was formed (compare lanes 9 and 12), indicating that it must result from the pretranslocational binding of pSufI to the vesicles.
Bpa was also incorporated into residues L54 and L56 at the end of the predicted amphipilic helix of TatB and further distally at position L78 (cf. Figure 4A). Like the I36 – G34 mutants the more distal TatB variants yielded ∼75-kDa adducts to pSufI that could be immunoprecipitated with antibodies against TatB (Figure 4D, lanes 4 and 5; 7 and 9; 13 and 14). In addition, all three positions yielded bigger TatB adducts (asterisks). One of them had an approximate mass of 90 kDa, similar to what was found for TatB(W35Bpa) and TatB(G34Bpa) (cf. Figure 4B, lanes 6 and 12). TatB(L54Bpa) gave rise to even two additional adducts of ∼110 kDa and 130 kDa (Figure 4D, lane 4). To rule out that any of these multiple adducts resulted from cross-linking of TatB to the processed form of SufI, INVs containing the TatB(L78Bpa) variant were also probed with a noncleavable mutant precursor of SufI (pSufI-ΔSP). As shown in Figure 4D (lanes 15 – 19), the mutant precursor although virtually not being processed (lanes 15 and 16) displays the same cross-linking pattern as its wild-type counterpart (lanes 13, 14 and 18, 19).
Thus all these prominent cross-links are adducts to the precursor of SufI. The 90-kDa adduct seems to be too small to reflect two TatB monomers contacting one pSufI molecule and in theory could originate from two distinct species of pSufI resulting in differently branched adducts. In contrast, the 110-kDa and 130-kDa adducts of the TatB(L54Bpa) variant (lanes 4, 5) by size are likely to represent complexes of two to three TatB monomers with a single precursor. These cross-links therefore seem to indicate an oligomeric structure of TatB, to which pSufI had been targeted. Similar to the more proximal Bpa position I36 in the amphipathic helix of TatB, the TatB variants depicted in Figure 4D were active in translocation (lanes 2, 11 and 20 and Supplemental Figure) and interacted with pSufI at a stage preceding the actual translocation step, because they were not affected by de-energizing the INVs (lanes 23–25). These results indicate that during functional membrane-targeting of pSufI, a stretch of at least 42 amino acids of the cytosolic domain of TatB comes in close proximity to the Tat precursor.
The Transmembrane Helix of TatB Is Also in Contact with a Bound Tat Precursor
An amino acid exchange in the N-terminal part of the transmembrane domain of TatB was found to suppress defective Tat signal sequences (Kreutzenbeck et al., 2007), suggesting functional interactions of the membrane domain of TatB and Tat substrate proteins. We therefore also investigated TatB variants that contained the cross-linker in the transmembrane helix of TatB (Figure 4E). Bpa at position 9 gave a clear cross-link to pSufI (lane 2, asterisk). Cross-links became weaker when Bpa was moved around the helix turn (lanes 6, 10) indicating that the face of the transmembrane helix encompassing residue Leu 9 is closest to the Tat substrate. Interestingly, also TatB(L9Bpa) yielded an additional adduct bigger than 100 kDa (lanes 4, 15, 18) again indicative of more than one TatB monomer contacting one radioactively labeled pSufI. As summarized in lanes 13–18, the TatB(L9Bpa) variant exhibited translocation activity (cf. also the Supplemental Figure), but formed cross-links with pSufI, even if translocation was completely blocked (lanes 16–18). These results would be consistent with both an interaction of the transmembrane domain of TatB with the signal sequence of SufI or with the folded SufI moiety, in which case the transmembrane helices of TatB would be part of a binding platform for folded Tat precursors.
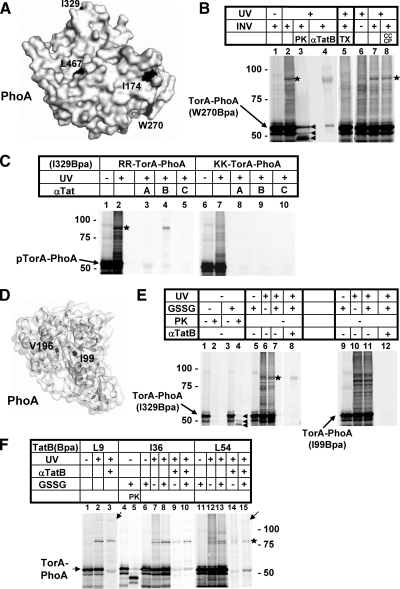
TorA-PhoA Displays the Same TatB Binding as pSufI
To see whether these findings could be confirmed for a second Tat precursor, we also analyzed TorA-PhoA, which is a model Tat precursor consisting of the Tat signal sequence of E. coli TorA (TMAO-reductase) and the mature part of PhoA (alkaline phosphatase) (DeLisa et al., 2003). Four surface-exposed (Figure 5A, I174, W270, I329, and L467) and two internal residues (Figure 5D, I99 and V196) of the PhoA moiety were chosen for replacement by amber stop codons and subsequent incorporation of the photo-cross-linker Bpa. As exemplified for one of the surface located Bpa variants (Figure 5B, W270), UV-irradiation in the presence of INVs again yielded one prominent adduct (lanes 2, 7, 8, asterisk) that could be immunoprecipitated with anti-TatB antibodies (lane 4). It was absent from reactions that did not contain vesicles (compare lanes 6 and 7). Different from the findings with pSufI, no visible cross-linking of the Bpa mutants of TorA-PhoA to TatA was obtained (exemplified for the surface-exposed Bpa variant I329 in Figure 5C, lane 3). Like pSufI, folded TorA-PhoA was found close to TatB, only if it carried an intact RR-motif in its signal sequence (Figure 5C, compare lanes 4 and 9).
Figure 5.
TorAPhoA shows the same interaction with TatB as pSufI. (A) Surface-exposed residues I174, W270, I329, and L467 of PhoA used for the incorporation of the cross-linker Bpa. Structural data were taken from Wang et al., (2005). (B) The 55 kDa Tat precursor protein TorA-PhoA carrying Bpa at the surface position W270 (arrow) was synthesized in vitro with INVs added as indicated. When irradiated with UV-light, a major photo-cross-link (asterisk) was obtained that was recognized by anti-TatB antibodies (αTatB) and was absent from reactions devoid of membranes (lane 6). It was completely digested by Proteinase K (PK) (lane 3) in contrast to the PK-resistant translocated TorA-PhoA species (arrowheads) and therefore not membrane-protected. The three translocated species of TorA-PhoA from top to bottom are precursor, mature form and a translocation-arrested species (Panahandeh et al., 2008). Solubilization of the INVs' Tat machinery by 1% Triton X-100 prevented formation of the TatB adduct of TorA-PhoA (lane 5), whereas dissipation of the PMF by CCCP did not (lane 8). (C) TorA-PhoA (arrow) carrying Bpa at the surface position I329 was synthesized in vitro in the presence of INVs, irradiated with UV-light, and immunoprecipitated with antisera against TatA, TatB, and TatC (lanes 1–5). The only Tat-specific photo-adduct obtained was the one recognized by anti-TatB antibodies (asterisk). Cross-linking was completely abolished if the signal sequence of TorA-PhoA carried a KK-mutation of the consensus RR-motif (lanes 6–10). (D) Internal residues I99 and V196 of PhoA used for the incorporation of Bpa. (E) Comparison between the surface-exposed Bpa variant I329 of TorA-PhoA and the internal variant I99. Lanes 1–4 illustrate that transport of TorA-PhoA into INV proceeded only when TorA-PhoA was synthesized under oxidizing conditions, which were achieved by the addition of oxidized glutathione (GSSG). Transport is indicated in lane 4 by the appearance of the PK-resistant TorA-PhoA species specified in B (arrowheads). In contrast to the internal Bpa variant I99, the surface-exposed variant I329 formed adducts with TatB (asterisk) both under reducing and oxidizing conditions. (F) Cross-linking of the transmembrane and amphipathic helix of TatB to TorA-PhoA. The two TatB variants L9Bpa and L54Bpa again yielded higher molecular mass adducts (arrows). Cross-linking was observed both under reducing and oxidizing conditions.
Previous studies had shown that TorA-PhoA, which harbors four cysteines within its PhoA moiety, can only be transported by the Tat machinery if oxidizing conditions allow formation of two disulfide bonds (DeLisa et al., 2003; Panahandeh et al., 2008). This is also shown in Figure 5E, in which protease-protected species sequestered by the INVs (lane 4, arrowheads) were only obtained when the TorA-PhoA variants were synthesized in the presence of oxidized glutathione (GSSG, compare lanes 2 and 4). Cross-links to TatB were, however, obtained under both reducing and oxidizing conditions (lanes 6 and 7). Neither in the reduced nor in the oxidized conformation did internal Bpa variants of TorA-PhoA cross-link to TatB (demonstrated for the I99 mutant, lanes 10–11) indicating again that TatB contacts only the surface of folded Tat precursors.
We also tested whether the adducts between TatB and vesicle-bound TorA-PhoA was accessible to externally added PK. PK, while sparing the TorA-PhoA species that were translocated into the membrane vesicles, completely digested the adduct between TatB and TorA-PhoA (Figure 5B, compare lanes 2 and 3). This is consistent with TatB forming an attachment site exposed on the membrane surface. On the other hand, disrupting the vesicles by Triton X-100 before irradiation abolished interaction with TatB (Figure 5B, compare lanes 5 and 7), indicating that a membrane-embedded Tat translocase was required for these precursor-TatB adducts to form. As with pSufI, TatB adducts of TorA-PhoA were insensitive to CCCP (Figure 5B, compare lanes 7 and 8).
TatB variants carrying the cross-linker either within the transmembrane or the amphipathic helix were examined for interaction also with TorA-PhoA. All three positions yielded cross-links with radioactively labeled precursor (Figure 5F, asterisk) that could be immunoprecipitated with anti-TatB antiserum (lanes 3, 9, 10, 14, 15). Lack of disulfide bond formation by omission of GSSG did not affect interaction of TorA-PhoA with TatB (cf. lanes 7–10; 14, 15). Again, Bpa incorporated at the positions L9 and L54 of TatB yielded additional high-molecular-mass adducts (lanes 3 and 15, arrows), which could correspond in size at least to a TatB dimer plus one precursor molecule. Collectively, the results summarized in Figure 5 confirm that membrane targeting of folded Tat precursors leads to multiple contacts with the transmembrane and the amphipathic helices of TatB.
DISCUSSION
Previous studies revealed that the signal sequence of Tat precursor proteins is primarily recognized by TatC. TatB was found as a constituent of a TatBC receptor complex but its function remained poorly characterized. We now show that upon functionally targeting the Tat translocase, folded Tat precursors become surrounded by TatB. Juxtaposition of a folded Tat substrate and TatB requires an active RR-motif but is independent of the PMF at the membrane. TatB proximity is lost upon transmembrane transport of the precursor indicating that it represents an intermediary binding step of a Tat precursor before its translocation. Interaction with TatB occurs at multiple contact sites along the predicted transmembrane and amphipathic helices of TatB. Collectively these data suggest that TatB assembles into an oligomeric binding site that is able to transiently accommodate large parts of a folded Tat precursor.
Because no TatB interactions with sites hidden in the folded Tat precursors were detected, the observed interactions of TatB occurred only with folded precursors. Therefore, TatB adducts obtained with TorA-PhoA under reducing conditions suggest that, although the two disulfide bonds of PhoA could not form (Panahandeh et al., 2008), the precursor was probably not completely unfolded. This conclusion was recently also drawn from results obtained with truncated variants of TorA-PhoA (Maurer et al., 2009). Thus it seems likely that the lack of two disulfide bonds in TorA-PhoA either leads to only a local unfolding or to a generally relaxed conformation with an enlarged surface area. This would still allow reduced TorA-PhoA to bind to TatB, provided that TorA-PhoA contained an intact RR-motif for targeting the Tat machinery. TatB-precursor interactions of the type described here would then precede the clearance of translocation-incompetent substrates.
Among the cross-linker positions tested, L9 and L54 of TatB yielded additional adducts with both pSufI and TorA-PhoA that were bigger than 1:1 precursor-TatB complexes (Figures 4, D and E and 5F). Because in these experiments the cross-linker was not incorporated into the precursor molecules and otherwise we never observed any substantial formation of SDS-resistant precursor dimers under our in vitro conditions, the larger complexes that TatB(L9Bpa) and TatB(L54Bpa) formed are likely homooligomeric TatB assemblies with one precursor molecule each. By size, these adducts could contain two and three TatB monomers. Using cysteine cross-linking, dimerization of TatB via its transmembrane and amphipathic helix has in fact recently been demonstrated to occur (Lee et al., 2006).
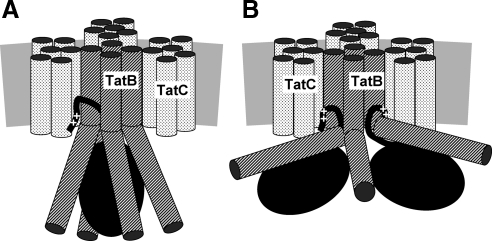
Our combined findings would be compatible with models depicted in Figure 6. A folded precursor is shown that has been targeted to TatBC via its twin-arginine signal peptide. The exact binding site(s) for the Tat signal sequence on TatC and TatB are not known and the ones depicted in Figure 6 are therefore speculative. The models are further based on the assumption that the TatBC receptor consists of four TatBC protomers (for clarity not all stoichiometric TatC subunits are shown). A tetrameric TatBC assembly would be in line with previous cysteine-based cross-linking studies (Lee et al., 2006), but our results could also be explained by TatBC assemblies of higher order, such as heptamers, that were recently deduced from single particle electron microscopy of isolated TatBC complexes (Tarry et al., 2009b). Positioning of TatC on the periphery of the complex is consistent with a previous cysteine cross-linking analysis of TatC (Punginelli et al., 2007).
Figure 6.
Models of possible TatB-precursor interactions. (A) The gray bar represents the lipid bilayer. Each TatC molecule is depicted by six stippled transmembrane helices. Four TatB monomers are shown (diagonally hatched cylinders) with their transmembrane helices contacting one TatC protomer each (the front TatC subunit has been omitted for clarity) and with their cytosolic amphipathic helices encapsulating a folded Tat precursor (black ellipse). The twin-arginine signal sequence (++) is represented by a black line. Its contacts to TatB and TatC are speculative. The tetrameric nature of the TatBC complex is based on previous findings (Lee et al., 2006), but the model would easily accommodate higher order oligomeric structures. (B) Two Tat precursor molecules simultaneously binding to TatB as suggested by recent data (Tarry et al., 2009b; Ma and Cline, 2010).
Using site-directed photo-cross-linking, TatB contact sites were found distributed over the major part of the precursors' surfaces. These adducts generally appeared as major distinct bands on SDS-PAGE representing substantial fractions of the in vitro synthesized Tat precursors (cf. Figure 3B). This fact actually rules out that the multiple TatB contact sites detected on the surfaces of Tat precursors each result from a different orientation of a highly mobile Tat precursor with respect to a single TatB monomer. Such a situation would hardly lead to discrete and prominent adducts but would rather result in a background noise of cross-links only. We therefore conclude that the multiple contact sites identified both on the substrates and on TatB rather reflect a static binding of Tat precursors to stable oligomeric TatB assemblies. These could be realized by several TatB monomers forming a cage-like structure around the folded precursor as depicted in Figure 6A.
Recently, Ma and Cline provided experimental evidence that the homologous Tat system of plant chloroplasts is able to accommodate and collectively translocate dimers or even tetramers of Tat precursors (Ma and Cline, 2010). The model depicted in Figure 6B combines such a situation with the contacts between precursor molecules and TatB that our cross-linking approach revealed. Such a scenario might require a larger TatB assembly such as the heptameric arrays of TatBC inferred from single particle analysis of isolated TatBC complexes (Tarry et al., 2009b).
In principle, the observed cross-links between Tat precursors and TatB could also be explained by an even more speculative conformation, in which the amphipathic helices of TatB cover the folded substrate only after having flipped across the bilayer. We, however, regard such a scenario less likely (i) because in contrast to the topology-related TatA protein (Gouffi et al., 2004; Chan et al., 2007) there is no experimental evidence for such a movement of the amphipathic helices of TatB, and (ii) if they did so, it would be at high energy expenditure, which would be at odds with the observation that the precursor-TatB adducts formed even after dissipation of the PMF. Incorporated into all proposed models is our finding that cross-linking from the amphipathic helix of TatB was position-dependent thus indicating that this precursor-interacting part of TatB in fact has a helical conformation. Second, the calculated length of the predicted amphipathic helix of TatB would be sufficient to cover the entire surface at least of one folded SufI molecule.
The model of Figure 6A could also illustrate how at a later step the signal sequence is threaded into the TatBC complex to stay in contact with TatC (Gerard and Cline, 2006) and reach the far end of the TatB transmembrane helices as suggested by tatB mutations of residue Glu 8 suppressing defective signal sequences (Kreutzenbeck et al., 2007). How the actual transmembrane transport of the folded substrate domain could proceed from there, for example by recruiting TatA protomers, remains to be solved. Nevertheless, a situation as depicted in Figure 6A would be fully compatible with precursor molecules found attached to the periphery of isolated TatBC complexes (Tarry et al., 2009b). Even the changed dimensions of the membrane-embedded TatBC complex observed upon precursor binding (Tarry et al., 2009b) could be attributed to a movement of the TatB amphipathic helices from the surface of the complex to a cage-like structure forming around a folded and targeted Tat precursor.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Peter Schultz and Eric Tippman, The Scripps Research Institute, La Jolla, for plasmid pSup-BpaRS-6TRN(D286R) and Dr. Enguo Fan for critically reading the manuscript. This work was supported by Grant LHSG-CT-2004-005257 of the European Union, the collaborative research center 746 and the research unit 929 of the German Research Council, and the Excellence Initiative of the German Federal and State Governments (GSC-4, Spemann Graduate School).
Abbreviations used:
- Bpa
p-benzoyl-phenylalanine
- CCCP
carbonyl cyanide m-chlorophenyl-hydrazone
- GSSG
oxidized glutathione
- INV
inside-out inner membrane vesicles
- PK
proteinase K
- PMF
H+-motive force
- Tat
twin-arginine translocation
- TorA-PhoA
fusion between the Tat signal sequence of TorA (TMAO reductase) and PhoA (alkaline phosphatase).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0585) on October 6, 2010.
REFERENCES
- Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell. 2003;12:937–946. doi: 10.1016/s1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Alami M., Trescher D., Wu L. F., Müller M. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 2002;277:20499–20503. doi: 10.1074/jbc.M201711200. [DOI] [PubMed] [Google Scholar]
- Bageshwar U. K., Musser S. M. Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J. Cell Biol. 2007;179:87–99. doi: 10.1083/jcb.200702082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bageshwar U. K., Whitaker N., Liang F. C., Musser S. M. Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol. Microbiol. 2009;74:209–226. doi: 10.1111/j.1365-2958.2009.06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C. M., Freudl R., Robinson C. Twin arginine translocation (Tat)-dependent export in the apparent absence of TatABC or TatA complexes using modified Escherichia coli TatA subunits that substitute for TatB. J. Biol. Chem. 2007;282:36206–36213. doi: 10.1074/jbc.M704127200. [DOI] [PubMed] [Google Scholar]
- Barrett C. M., Mangels D., Robinson C. Mutations in subunits of the Escherichia coli twin-arginine translocase block function via differing effects on translocation activity or Tat complex structure. J. Mol. Biol. 2005;347:453–463. doi: 10.1016/j.jmb.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Berks B. C. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- Berthelmann F., Mehner D., Richter S., Lindenstrauss U., Lunsdorf H., Hause G., Brüser T. Recombinant expression of tatABC and tatAC results in the formation of interacting cytoplasmic TatA tubes in Escherichia coli. J. Biol. Chem. 2008;283:25281–25289. doi: 10.1074/jbc.M707757200. [DOI] [PubMed] [Google Scholar]
- Blaudeck N., Kreutzenbeck P., Müller M., Sprenger G. A., Freudl R. Isolation and characterization of bifunctional Escherichia coli TatA mutant proteins that allow efficient Tat-dependent protein translocation in the absence of TatB. J. Biol. Chem. 2005;280:3426–3432. doi: 10.1074/jbc.M411210200. [DOI] [PubMed] [Google Scholar]
- Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- Brüser T., Sanders C. An alternative model of the twin arginine translocation system. Microbiol. Res. 2003;158:7–17. doi: 10.1078/0944-5013-00176. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Zlomislic M. R., Tieleman D. P., Turner R. J. The TatA subunit of Escherichia coli twin-arginine translocase has an N-in topology. Biochemistry. 2007;46:7396–7404. doi: 10.1021/bi7005288. [DOI] [PubMed] [Google Scholar]
- Cline K., Ettinger W. F., Theg S. M. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J. Biol. Chem. 1992;267:2688–2696. [PubMed] [Google Scholar]
- Cline K., McCaffery M. Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 2007;26:3039–3049. doi: 10.1038/sj.emboj.7601759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Mori H. Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 2001;154:719–729. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney-Smith C., Mori H., Cline K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 2006;281:5476–5483. doi: 10.1074/jbc.M512453200. [DOI] [PubMed] [Google Scholar]
- DeLisa M. P., Tullman D., Georgiou G. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci USA. 2003;100:6115–6120. doi: 10.1073/pnas.0937838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner G., Koch H. G., Beck K., Brunner J., Müller M. Ligand crowding at a nascent signal sequence. J. Cell Biol. 2003;163:35–44. doi: 10.1083/jcb.200306069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frielingsdorf S., Jakob M., Klösgen R. B. A stromal pool of TatA promotes Tat-dependent protein transport across the thylakoid membrane. J. Biol. Chem. 2008;283:33838–33845. doi: 10.1074/jbc.M806334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard F., Cline K. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 2006;281:6130–6135. doi: 10.1074/jbc.M512733200. [DOI] [PubMed] [Google Scholar]
- Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., Berks B. C. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouffi K., Gerard F., Santini C. L., Wu L. F. Dual topology of the Escherichia coli TatA protein. J. Biol. Chem. 2004;279:11608–11615. doi: 10.1074/jbc.M313187200. [DOI] [PubMed] [Google Scholar]
- Greene N. P., Porcelli I., Buchanan G., Hicks M. G., Schermann S. M., Palmer T., Berks B. C. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J. Biol. Chem. 2007;282:23937–23945. doi: 10.1074/jbc.M702972200. [DOI] [PubMed] [Google Scholar]
- Holzapfel E., Eisner G., Alami M., Barrett C. M., Buchanan G., Lüke I., Betton J. M., Robinson C., Palmer T., Moser M., Müller M. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry. 2007;46:2892–2898. doi: 10.1021/bi062205b. [DOI] [PubMed] [Google Scholar]
- Hou B., Frielingsdorf S., Klösgen R. B. Unassisted membrane insertion as the initial step in DeltapH/Tat-dependent protein transport. J Mol Biol. 2006;355:957–967. doi: 10.1016/j.jmb.2005.11.029. [DOI] [PubMed] [Google Scholar]
- Hynds P. J., Robinson D., Robinson C. The sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J. Biol. Chem. 1998;273:34868–34874. doi: 10.1074/jbc.273.52.34868. [DOI] [PubMed] [Google Scholar]
- Jongbloed J. D., van der Ploeg R., van Dijl J. M. Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol. 2006;14:2–4. doi: 10.1016/j.tim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Kreutzenbeck P., Kroger C., Lausberg F., Blaudeck N., Sprenger G. A., Freudl R. Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J. Biol. Chem. 2007;282:7903–7911. doi: 10.1074/jbc.M610126200. [DOI] [PubMed] [Google Scholar]
- Leake M. C., Greene N. P., Godun R. M., Granjon T., Buchanan G., Chen S., Berry R. M., Palmer T., Berks B. C. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. USA. 2008;105:15376–15381. doi: 10.1073/pnas.0806338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. A., Buchanan G., Stanley N. R., Berks B. C., Palmer T. Truncation analysis of TatA and TatB defines the minimal functional units required for protein translocation. J. Bacteriol. 2002;184:5871–5879. doi: 10.1128/JB.184.21.5871-5879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. A., Orriss G. L., Buchanan G., Greene N. P., Bond P. J., Punginelli C., Jack R. L., Sansom M. S., Berks B. C., Palmer T. Cysteine-scanning mutagenesis and disulfide mapping studies of the conserved domain of the twin-arginine translocase TatB component. J. Biol. Chem. 2006;281:34072–34085. doi: 10.1074/jbc.M607295200. [DOI] [PubMed] [Google Scholar]
- Lesley S. A., Brow M. A., Burgess R. R. Use of in vitro protein synthesis from polymerase chain reaction-generated templates to study interaction of Escherichia coli transcription factors with core RNA polymerase and for epitope mapping of monoclonal antibodies. J. Biol. Chem. 1991;266:2632–2638. [PubMed] [Google Scholar]
- Ma X., Cline K. Precursors bind to specific sites on thylakoid membranes prior to transport on the delta pH protein translocation system. J. Biol. Chem. 2000;275:10016–10022. doi: 10.1074/jbc.275.14.10016. [DOI] [PubMed] [Google Scholar]
- Ma X., Cline K. Multiple precursor proteins bind individual Tat receptor complexes and are collectively transported. EMBO J. 2010;29:1477–1488. doi: 10.1038/emboj.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrichi M., Camacho L., Russell D. G., DeLisa M. P. Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J. Biol. Chem. 2008;283:35223–35235. doi: 10.1074/jbc.M802660200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos C. F., Di Cola A., Robinson C. TatD is a central component of a Tat translocon-initiated quality control system for exported FeS proteins in Escherichia coli. EMBO Rep. 2009;10:474–479. doi: 10.1038/embor.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos C. F., Robinson C., Di Cola A. The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J. 2008;27:2055–2063. doi: 10.1038/emboj.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer C., Panahandeh S., Moser M., Müller M. Impairment of twin-arginine-dependent export by seemingly small alterations of substrate conformation. FEBS Lett. 2009;583:2849–2853. doi: 10.1016/j.febslet.2009.07.038. [DOI] [PubMed] [Google Scholar]
- McDevitt C. A., Buchanan G., Sargent F., Palmer T., Berks B. C. Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expressed at native levels. FEBS J. 2006;273:5656–5668. doi: 10.1111/j.1742-4658.2006.05554.x. [DOI] [PubMed] [Google Scholar]
- McDevitt C. A., Hicks M. G., Palmer T., Berks B. C. Characterisation of Tat protein transport complexes carrying inactivating mutations. Biochem. Biophys. Res. Commun. 2005;329:693–698. doi: 10.1016/j.bbrc.2005.02.038. [DOI] [PubMed] [Google Scholar]
- Mori H., Cline K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid [Delta]pH/Tat translocase. J. Cell Biol. 2002;157:205–210. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Panahandeh S., Holzapfel E., Müller M. In vitro analysis of the bacterial twin-arginine-dependent protein export. Methods Mol. Biol. 2007;390:63–80. doi: 10.1007/978-1-59745-466-7_5. [DOI] [PubMed] [Google Scholar]
- Mould R. M., Shackleton J. B., Robinson C. Transport of proteins into chloroplasts. Requirements for the efficient import of two lumenal oxygen-evolving complex proteins into isolated thylakoids. J. Biol. Chem. 1991;266:17286–17289. [PubMed] [Google Scholar]
- Oates J., Barrett C. M., Barnett J. P., Byrne K. G., Bolhuis A., Robinson C. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J. Mol. Biol. 2005;346:295–305. doi: 10.1016/j.jmb.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Orriss G. L., Tarry M. J., Ize B., Sargent F., Lea S. M., Palmer T., Berks B. C. TatBC, TatB, and TatC form structurally autonomous units within the twin arginine protein transport system of Escherichia coli. FEBS Lett. 2007;581:4091–4097. doi: 10.1016/j.febslet.2007.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahandeh S., Maurer C., Moser M., DeLisa M. P., Müller M. Following the Path of a Twin-arginine Precursor along the TatABC Translocase of Escherichia coli. J. Biol. Chem. 2008;283:33267–33275. doi: 10.1074/jbc.M804225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahandeh S., Holzapfel E., Müller M. The twin-arginine translocation pathway. In: Wooldridge K., editor. In Bacterial Secreted Proteins. Norfolk, UK: Caister Academic Press; 2009. pp. 23–43. [Google Scholar]
- Punginelli C., Maldonado B., Grahl S., Jack R., Alami M., Schroder J., Berks B. C., Palmer T. Cysteine scanning mutagenesis and topological mapping of the Escherichia coli twin-arginine translocase TatC component. J. Bacteriol. 2007;189:5482–5494. doi: 10.1128/JB.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Lindenstrauss U., Lucke C., Bayliss R., Brüser T. Functional Tat transport of unstructured, small, hydrophilic proteins. J. Biol. Chem. 2007;282:33257–33264. doi: 10.1074/jbc.M703303200. [DOI] [PubMed] [Google Scholar]
- Ridder A. N., de Jong E. J., Jongbloed J. D., Kuipers O. P. Subcellular localization of TatAd of Bacillus subtilis depends on the presence of TatCd or TatCy. J. Bacteriol. 2009;191:4410–4418. doi: 10.1128/JB.00215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffey R. A., Theg S. M. Analysis of the import of carboxyl-terminal truncations of the 23-kilodalton subunit of the oxygen-evolving complex suggests that its structure is an important determinant for thylakoid transport. Plant Physiol. 1996;111:1329–1338. doi: 10.1104/pp.111.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y., Schultz P. G. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- Sanders C., Wethkamp N., Lill H. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 2001;41:241–246. doi: 10.1046/j.1365-2958.2001.02514.x. [DOI] [PubMed] [Google Scholar]
- Sargent F., Stanley N. R., Berks B. C., Palmer T. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J. Biol. Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- Schreiber S., Stengel R., Westermann M., Volkmer-Engert R., Pop O. I., Muller J. P. Affinity of TatCd for TatAd elucidates its receptor function in the Bacillus subtilis twin arginine translocation (Tat) translocase system. J. Biol. Chem. 2006;281:19977–19984. doi: 10.1074/jbc.M513900200. [DOI] [PubMed] [Google Scholar]
- Shanmugham A., Wong Fong Sang H. W., Bollen Y. J., Lill H. Membrane binding of twin arginine preproteins as an early step in translocation. Biochemistry. 2006;45:2243–2249. doi: 10.1021/bi052188a. [DOI] [PubMed] [Google Scholar]
- Strauch E.M., Georgiou G. Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J. Mol. Biol. 2007;374:283–291. doi: 10.1016/j.jmb.2007.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarry M., Arends S. J., Roversi P., Piette E., Sargent F., Berks B. C., Weiss D. S., Lea S. M. The Escherichia coli cell division protein and model Tat substrate SufI (FtsP) localizes to the septal ring and has a multicopper oxidase-like structure. J. Mol. Biol. 2009a;386:504–519. doi: 10.1016/j.jmb.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarry M. J., Schäfer E., Chen S., Buchanan G., Greene N. P., Lea S. M., Palmer T., Saibil H. R., Berks B. C. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl. Acad. Sci. USA. 2009b;106:13284–13289. doi: 10.1073/pnas.0901566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Stieglitz K. A., Kantrowitz E. R. Metal specificity is correlated with two crucial active site residues in Escherichia coli alkaline phosphatase. Biochemistry. 2005;44:8378–8386. doi: 10.1021/bi050155p. [DOI] [PubMed] [Google Scholar]
- Wexler M., Sargent F., Jack R. L., Stanley N. R., Bogsch E. G., Robinson C., Berks B. C., Palmer T. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in Sec-independent protein export. J. Biol. Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.