Rubicon, a subunit of the Beclin 1-PI3-kinase complex and its homologue, PLEKHM1, negatively regulate endocytic pathway through the interaction with Rab7. Synchronous association with the Beclin 1–PI3-kinase complex and Rab7 is necessary for the function of Rubicon, but not PLEKHM1.
Abstract
The endocytic and autophagic pathways are involved in the membrane trafficking of exogenous and endogenous materials to lysosomes. However, the mechanisms that regulate these pathways are largely unknown. We previously reported that Rubicon, a Beclin 1–binding protein, negatively regulates both the autophagic and endocytic pathways by unidentified mechanisms. In this study, we performed database searches to identify potential Rubicon homologues that share the common C-terminal domain, termed the RH domain. One of them, PLEKHM1, the causative gene of osteopetrosis, also suppresses endocytic transport but not autophagosome maturation. Rubicon and PLEKHM1 specifically and directly interact with Rab7 via their RH domain, and this interaction is critical for their function. Furthermore, we show that Rubicon but not PLEKHM1 uniquely regulates membrane trafficking via simultaneously binding both Rab7 and PI3-kinase.
INTRODUCTION
Endocytosis involves the intracellular transport of extracellular and plasma membrane substrates to endosomes/lysosomes and is involved in many physiological processes (Mukherjee et al., 1997). Autophagy is also a process in which cytoplasmic constituents, including organelles, are transported within double-membraned autophagosomes to lysosomes for degradation (Xie and Klionsky, 2007; Yoshimori and Noda, 2008). Autophagy has divergent physiological roles in cancer, infection, immunity, and other processes (Mizushima et al., 2008). Many reports suggest there is a common element between the endosomal and autophagic pathways (Raiborg and Stenmark, 2009; Simonsen and Tooze, 2009), but these commonalities have not been fully elucidated.
Recently, two groups identified an interesting protein, Rubicon, that may link these two pathways (Matsunaga et al., 2009; Zhong et al., 2009). Rubicon was identified as a protein that binds to Beclin 1, a tumor suppressor protein and mammalian homologue of the yeast autophagy protein Atg6/Vps30 (Liang et al., 1999; Kihara et al., 2001a). Yeast Atg6/Vps30 facilitates two distinctive processes: autophagy and retrograde traffic from endosomes to the Golgi apparatus by two different phosphatidylinositol 3-kinase (PI3-kinase) complexes: class I (Atg14, Atg6, Vps34, Vps15) and class II (Atg38, Atg6, Vps34, Vps15) (Kihara et al., 2001b). Mammalian Beclin 1 has a similar, albeit distinct, complex organization. One complex consists of Beclin 1, hAtg14L/Atg14/Barkor, hVps34, and p150/hVps15 and acts in autophagosome formation (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Another complex contains Beclin 1, UVRAG, hVps34, and p150/hVps15 and functions in the endocytic pathway and autophagosome–lysosome fusion (Liang et al., 2006; Itakura et al., 2008; Liang et al., 2008). A third Beclin 1 complex includes Rubicon and the UVRAG complex (Matsunaga et al., 2009). Knockdown of Rubicon enhances endocytic transport and autophagosome–lysosome fusion (Matsunaga et al., 2009; Zhong et al., 2009). In addition, autophagy is induced even in the absence of nutrient starvation (Matsunaga et al., 2009; Zhong et al., 2009). Thus, the third complex negatively regulates both the endocytic and autophagic pathways. A Rubicon homologue has not been identified in yeast, suggesting that this potential complex regulatory mechanism is specific to higher eukaryotes. However, it is unclear how Rubicon regulates membrane traffic.
To further determine the function of Rubicon, we searched mammalian gene databases for Rubicon homologues. As a result, we identified PLEKHM1, which also contains an RH domain and is reportedly a causative gene for osteopetrosis due to mutations, but the mechanisms remain unknown (Van Wesenbeeck et al., 2007). Rubicon and PLEKHM1 share a common C-terminal domain, which we termed the RH (Rubicon homologous) domain. Here, we show that the RH domains of Rubicon and PLEKHM1 specifically interact with Rab7, a Rab GTPase that localizes to the late endosome/lysosome. These interactions are necessary for their functions in the endocytic pathway. In addition to binding to Rab7, Rubicon, but not PLEKHM1, also associates with the Beclin 1–PI3-kinase complex. Synchronous associations of Rubicon with the Beclin 1-PI3-kinase complex and Rab7 are necessary to regulate the endocytic and autophagic pathways. These findings provide new insights into the mechanisms that regulate membrane trafficking toward lysosomes.
MATERIALS AND METHODS
Cells and Transfections
HeLa, MCF7, human embryonic kidney (HEK)293A, and A549 cells were cultured in DMEM D6546 (Sigma, St. Louis, MO) containing 10% fetal bovine serum supplemented with 4 mM L-glutamine in 5% CO2 at 37°C. Cells were transiently transfected using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Plasmid Construction
The plasmids encoding monomeric red fluorescent protein (mStrawberry) and mCherry were generously provided by Dr. Roger Y. Tsien (Shaner et al., 2004). The cDNAs encoding Rab5 WT, Rab5 Q79L, Rab5 S34N, Rab7 WT, Rab7 Q67L, and Rab7 T22N derived from Madin-Darby canine kidney (MDCK) cells were gifts from Dr. K. Umebayashi. Human Rab9 cDNA was reverse transcribed and subcloned into the polymeric enhanced green fluorescent protein (pEGFP)-C1 vector (Clontech, Palo Alto, CA). pEGFP-LC3 (Kabeya et al., 2000) and mouse Rubicon (mRubicon) cDNA (Matsunaga et al., 2009) were previously described. The human Rubicon (hRubicon) was reverse transcribed using RNA extracted from A549 cells. For yeast two-hybrid analysis, the pGAD-C1 vector, pGBD-C1 vector and PJ69–4A strain were kingly provided by Dr. P. James (James et al., 1996). cDNA encoding human PLEKHM1 (hPLEKHM1), also known as KIAA0356, was kindly provided by the Kazusa DNA Research Institute (Kisarazu, Japan). The PLEKHM1 (FL_CGHL) and Rubicon (FL_CGHL) mutants were generated by site-direct mutagenesis.
Antibodies and Reagents
Rabbit polyclonal anti-PLEKHM1 antibodies were raised against a recombinant protein encoding the C-terminal 300 amino acids, which were subcloned into the KpnI-XbaI sites of the pCold II DNA vector (TAKARA Bio, Tokyo, Japan). The plasmids were transformed into BL2 competent cells. The expressed protein was eluted from an SDS polyacrylamide gel, and the purified recombinant protein was used to immunize rabbits. The rabbit antisera against PLEKHM1 were affinity-purified using the same recombinant protein.
The following additional antibodies were used: rabbit polyclonal anti-FLAG (Sigma), mouse monoclonal anti-FLAG M2 (Sigma), rabbit polyclonal anti-Myc (MBL, Nagoya, Japan), rabbit polyclonal anti-GFP (MBL), rabbit polyclonal anti-RFP (MBL), rabbit polyclonal anti-LC3 (PM036, MBL), mouse monoclonal anti-EEA1 (BD Transduction Laboratories, Lexington, KY), mouse monoclonal anti-LAMP-1 (H4A3; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-α-tubulin (B5–1-2; Sigma), and sheep polyclonal anti-EGFR (Fitzgerald Industries, Concord, MA) antibodies. Rabbit polyclonal antibodies against Beclin 1, UVRAG, hVps34, p150 (hVps15), and Rubicon were previously described (Kihara et al., 2001a; Matsunaga et al., 2009).
Preparation of Recombinant PLEKHM1 and Rubicon Proteins
cDNA encoding PLEKHM1 (FL) or Rubicon (FL) was subcloned into pCold TF DNA (TAKARA Bio). Recombinant proteins were expressed in BL21 cells and purified with Ni-NTA (Qiagen) according to manufacturer's instructions.
In vitro Glutathione S-transferase Pulldown Assay
In vitro glutathione S-transferase (GST) pulldown assays were performed as previously described (Christoforidis and Zerial, 2000) with some modifications. Briefly, to prepare the inactive and active forms of GST-Rab7 by nucleotide exchange, buffer 1 (50 mM Tris-HCl, pH 8.0, 2.5 mM DTT, 25 mM EDTA, and 12.5 mM MgCl2) containing 20 μM GDP or GTPγS and buffer 2 (20 mM Tris-Cl, pH 8.0, 1 mM DTT, 1 mM EDTA, 5 mM MgCl2, and 0.6% CHAPS) containing GST-Rab7 were mixed and incubated for 10 min at 30°C. Subsequently, MgCl2 at a final concentration of 10 mM was added to this mixture to stabilize the nucleotides. GDP- or GTPγS-loaded GST-Rab7 was incubated with glutathione Sepharose 4B (GE Heathcare Bioscience, Piscataway, NJ) for 1 h at 4°C and washed three times with buffer 3 (20 mM Tris-Cl, pH 8.0, 1 mM DTT, 1 mM EDTA, 5 mM MgCl2, and 0.3% CHAPS) and once with buffer 4 (20 mM Tris-Cl, pH 8.0, 1 mM DTT, 1 mM EDTA, and 5 mM MgCl2). Next, recombinant PLEKHM1 or Rubicon was added to the GST-Rab7 beads and incubated for 2 h at 4°C. Then, the beads were washed three times with buffer 4 and eluted with SDS-sample buffer. The samples were subjected to SDS-PAGE followed by immunoblotting.
Immunoblot Analysis
Cells were washed with ice-cold PBS, scraped, collected by centrifuging at 4°C, and then lysed in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail; Roche, Mannheim, Germany) for 10 min on ice. The lysates were centrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were collected. The samples were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 3% skim milk in 0.1% Tween-20/TBS and then incubated with the appropriate primary antibodies. When either the anti-PLEKHM1 antibody or anti-Rubicon antibody was used, the membranes were incubated in Can Get Signal Solution I (Toyobo, Tokyo, Japan) overnight at 4°C. Immunoreactive bands were detected using horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) and luminol solution (ECL plus; GE Healthcare Bioscience).
Adenoviral Overexpression System
3xFLAG-hPLEKHM1 was subcloned into the pENTR-1A plasmid (Invitrogen). The cDNA insert was subcloned into the pAd/CMV/V5-DEST vector (Invitrogen) using the Gateway system with LR clonase (Invitrogen). Recombinant adenovirus was prepared with the ViraPower Adenovirus Expression System (Invitrogen) according to the manufacturer's instructions. Adenovirus infections were performed as follows. One day before infection, ∼2 × 105 cells were seeded into six-well plates and incubated overnight at 37°C in a CO2 incubator. The medium was then replaced with 2 ml of culture medium containing recombinant adenoviruses. After a 12-h incubation, the medium was replaced with 2 ml of culture medium. At 48 h after infection, the cells were used for experiments.
Adenovirus Small Hairpin RNA Expression System
hPLEKHM1 or hRubicon was knocked down using an adenovirus vector-based shRNA. The RNAi oligonucleotide sequences for hRubicon and GL2 luciferase (negative control) were previously described (Matsunaga et al., 2009). The target sequences for hPLEKHM1 were chosen using the siDirect algorithm (http://design.rnai.jp) and are located at base pairs 909–931 (5′-CCGGTCTCTGCAAGAGGTATTGT-3′). Oligonucleotides pairs were designed from these target sequences based on the Block-iT U6 RNAi Entry Vector Kit (Invitrogen). These pairs were annealed, ligated into the pENTR/U6 vector (Invitrogen), and transferred into the pAd/BLOCL-iT DEST vector (Invitrogen) through an LR recombination reaction (Invitrogen). The recombinant adenovirus was generated using ViraPower Adenoviral technology (Invitrogen). For knockdown analysis, subconfluent A549 cells in 35-mm dishes were infected with adenovirus and transferred to 60-mm dishes after 12 h. After an additional 24 h, the cells were reinfected with the virus. The medium was changed after an additional 12 h. At 5 d after infection, the cells were subjected to experiments.
Yeast Two-Hybrid Analysis
The yeast two-hybrid protocol was previously described (James et al., 1996). The yeast strain PJ69–4A was transformed with pGBD-Rab and pGAD-hPLEKHM1 or pGAD-hRubicon. Transformants were grown on a yeast nitrogen base that was supplemented with or without histidine and contained 2.5 mM 3-amino-1,2,4-triazole (3-AT) at 30°C for 3–4 d.
Fluorescence and Immunofluorescence Microscopy
Cells were cultured on coverslips and then fixed with 3% paraformaldehyde in PBS for 20 min. For immunofluorescence microscopy, the cells were permeabilized with 50 μg/ml digitonin in PBS for 10 min at room temperature. The permeabilized cells were incubated in blocking solution (0.1% gelatin in PBS) and then stained with primary and secondary antibodies diluted in blocking solution. The coverslips were mounted in Slowfade Gold Reagent (Invitrogen), and images were obtained with an Olympus FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan).
Immunoprecipitation
Cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [wt/vol] glycerol, 100 mM NaF, 10 mM EGTA, 1 mM Na3VO4, 1% Triton X-100, 5 μM ZnCl2, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail; Roche). The cell lysates were clarified by centrifuging at 15,000 rpm for 10 min at 4°C. The supernatants were immunoprecipitated with FLAG M2 agarose beads (Invitrogen) or the indicated primary antibody and protein G-Sepharose 4FF (GE Healthcare Bioscience). Immunocomplexes were washed five times with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Triton X-100) and then subjected to immunoblot analysis.
EGF Transport to Labeled Lysosomes
A549 cells were incubated with 0.1 mg/ml dextran-conjugated tetramethylrhodamine (70,000 MW; Invitrogen) in DMEM containing 10% FBS for 16–18 h. The cells were washed twice with PBS and infected with adenovirus in fresh medium for 6–9 h to overexpress the indicated proteins. After infection, the cells were washed twice with PBS and incubated in fresh medium. At 30–36 h after infection, the cells were treated with serum-free DMEM for 2 h at 37°C and then incubated in uptake medium (DMEM, 2% BSA, and 20 mM HEPES, pH 7.5) containing 2 μg/ml Alexa 488-EGF (Invitrogen). After incubating for 15 min at 37°C, the cells were washed twice with PBS to remove unbound EGF and transferred to a 37°C incubator. After an additional incubation for 45 min, the cells were fixed and examined by confocal microscopy.
Epidermal Growth Factor Receptor Degradation
A549 cells were cultured in growth medium to ∼80% confluency, washed with PBS, and incubated in serum-free medium for 2 h. Epidermal growth factor receptor (EGFR) endocytosis was stimulated by adding 100 ng/ml EGF (Invitrogen) to the serum-free medium. At 15, 60, 120, and 180 min after EGF stimulation, the cells were lysed with lysis buffer and subjected to immunoblot analysis.
Statistical Analysis
Statistical analyses were performed using a two-tailed unpaired t test. p values < 0.05 were considered statistically significant.
RESULTS
Rubicon and PLEKHM1 Directly Interact with Rab7 via the RH Domain
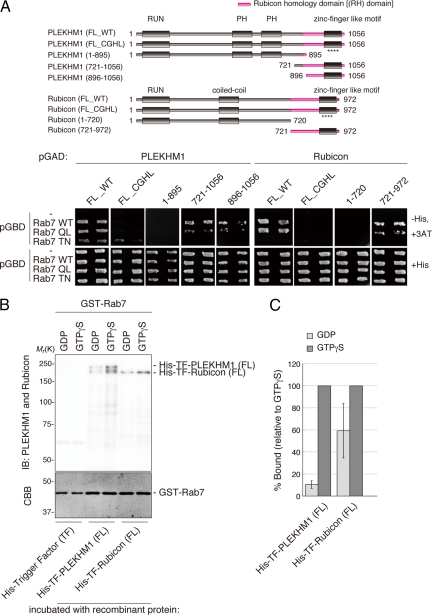
Rubicon functions in the late stage of the endocytic pathway and in the maturation step of autophagy (Matsunaga et al., 2009; Zhong et al., 2009). Because Rab7 is involved in both processes (Feng et al., 1995; Gutierrez et al., 2004; Jager et al., 2004; Stenmark, 2009), we examined whether Rubicon associates with Rab7. As shown in Supplementary Figure S1A, a yeast two-hybrid analysis revealed that Rubicon binds to Rab7, but not Rab5 or Rab9. In addition, a series of deletion mutants showed that Rubicon binds to Rab7 via the C-terminal RH domain (Figure 1A). Another RH domain–containing protein, PLEKHM1, also interacted with Rab7. Furthermore, PLEKHM1 specifically interacted with Rab7 through its RH domain (Supplementary Figure S1A; Figure 1A). Thus, the RH domain is a novel Rab7-binding motif that is not similar to other known Rab7-binding motifs.
Figure 1.
PLEKHM1 and Rubicon preferentially bind directly to the GTP-bound form of Rab7. (A) Deletion analysis of PLEKHM1 and Rubicon. Deletion series of PLEKHM1 and Rubicon in which the indicated deletions in the Rubicon homology (RH) domains and CGHL mutagenesis are shown with magenta and asterisks (****), respectively. The interactions between the PLEKHM1 or Rubicon deletion mutants and Rab7 were examined by a yeast two-hybrid assay. In all cases, two independent colonies were tested. (B and C) The nucleotide dependency was examined by an in vitro GST pulldown assay as described in Materials and Methods. (C) Recombinant PLEKHM1 and Rubicon proteins were detected with anti-PLEKHM1 and anti-Rubicon antibodies. GST-Rab7 was visualized by staining with Coomassie Brilliant Blue (CBB). (D) The relative binding amounts were quantified and are shown as percentages.
The RH domain contains a zinc finger-like motif composed of nine cysteine residues and a histidine residue that is conserved among all five human RH domain–containing proteins (Supplementary Figure S2). We constructed a CGHL mutant in which three cysteine residues and a histidine residue in the putative core of the zinc finger-like motif were replaced with glycine and leucine residues, respectively. The CGHL mutants of PLEKHM1 and Rubicon failed to interact with Rab7 (Figure 1A).
Rubicon only interacted with the wild-type and GTP-bound forms (QL) (Supplementary Figure S1A; Figure 1A). However, PLEKHM1 bound to wild-type and both the GTP-bound form (QL) and GDP-bound form (TN) of Rab7 in the yeast two-hybrid assay (Supplementary Figure S1A; Figure 1A) and a coimmunoprecipitation analysis in mammalian cells (Supplementary Figure S1B). To clarify the nucleotide dependency of this interaction, we performed an in vitro binding assay with immobilized recombinant GST-Rab7 loaded with GTPγS or GDP analogs. As a result, recombinant PLEKHM1 strongly interacted with the GTPγS-loaded GST-Rab7, but only minimally interacted with the GDP-bound form (Figure 1, B and C). Recombinant proteins for Rubicon also preferentially interacted with the GTP-bound form (Figure 1, B and C). Together, these data demonstrated that PLEKHM1 and Rubicon directly interact with the GTP-bound form of Rab7.
PLEKHM1 Negatively Regulates Endocytic Transport But Not Autophagic Transport
Next, we examined if PLEKHM1, like Rubicon, functions in the endocytic and autophagic pathways. Some mutations in PLEKHM1 are known to cause osteopetrosis (Van Wesenbeeck et al., 2007), suggesting that PLEKHM1 plays a role in osteoclast function. However, the mechanisms underlying this causative link have not been elucidated. Moreover, although PLEKHM1 is ubiquitously expressed, its role in other cell types is also unknown. The ability of PLEKHM1 to bind to Rab7, as shown in this study, and to colocalize with late endosomal/lysosomal markers, as shown previously (Van Wesenbeeck et al., 2007), suggest that PLEKHM1 functions in the endocytic pathway.
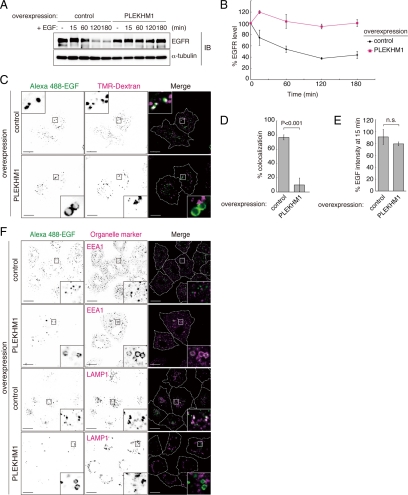
To determine the role of PLEKHM1 in the endocytic pathway, we first examined the effects of PLEKHM1 overexpression on the degradation of EGFR. EGFR degradation was significantly delayed in PLEKHM1-overexpressing cells compared with the controls (Figure 2, A and B). Next, we examined the integrity of the endocytic pathway. Internalized Alexa 488-EGF colocalized with lysosomes that were stained with tetramethylrhodamine (TMR)-dextran in the perinuclear region of control cells, but EGF minimally colocalized with TMR-dextran in PLEKHM1-overexpressing cells (Figure 2, C and D). The overall incorporation of Alexa 488-EGF into cells was not significantly affected (Figure 2E). In PLEKHM1-overexpressing cells, internalized EGF accumulated in EEA1-positive compartments but not in LAMP1-positive compartments (Figure 2F). Lysotracker staining showed that the endosomes in PLEKHM1-overexpressing cells were acidic, which suggests that the effects of overexpression on the transport of cargo proteins were not due to increased endosomal/lysosomal pH (Supplementary Figure S3A). Taken together, we concluded that PLEKHM1 overexpression inhibits the transport of EGF from early endosomes to lysosomes.
Figure 2.
The effects of PLEKHM1 overexpression on the endocytic pathway. (A and B) (A) EGFR degradation was impaired by overexpressing PLEKHM1. A representative anti-EGFR immunoblot of EGF-stimulated A549 cells. The same membrane was immunoblotted with an anti-α-tubulin antibody as a loading control. (B) The immunodetectable levels of EGFR (relative to α-tubulin) remained constant in PKLEMH1 cells relative to the controls (mean ± SEM). (C) EGF transport was disrupted by overexpressing PLEKHM1. EGF transport to lysosomes was evaluated based on the colocalization of EGF and dextran. (D) The percentages of colocalization are shown as merged EGF/total EGF (mean ± SD, n > 20). Bars, 20 μm. (E) The effects of PLEKHM1 overexpression on EGF uptake. The EGF intensities 15 min after adding EGF are shown as percentages (mean ± SD, n > 20). (F) Accumulation of EGF on early endosomes in PLEKHM1-overexpressing cells. Cells were stained with an anti-EEA1 antibody, an early endosomal marker, and an anti-LAMP1 antibody, a late endosomal/lysosomal marker. Bars, 20 μm.
Next, we examined the effects of PLEKHM1 knockdown on endocytic transport. Two PLEKHM1-specific shRNAs strongly decreased PLEKHM1 protein expression in A549 cells (Figure 3A). We found that the total EGFR levels were already decreased in PLEKHM1 knockdown cells before EGF treatment (Figure 3, A and B). The steady-state EGFR levels were significantly decreased to 40–50% of the control (Figure 3B), and this decrease was largely due to lysosomal degradation because lysosomal protease inhibitors blocked this decrease (Figure 3, C and D). In addition, we kinetically measured the rate at which Alexa 488-EGF colocalized with LAMP1 in PLEKHM1 knockdown cells. The colocalization rates were increased compared with the control cells at each time point (Figure 3E; Supplementary Figure S4), supporting the idea that PLEKHM1 knockdown accelerates transport to lysosomes. Thus, we concluded that PLEKHM1 depletion constitutively enhances EGFR degradation. Both the overexpression and knockdown experiments are consistent with the idea that PLEKHM1 negatively regulates the endocytic trafficking of EGFR.
Figure 3.
The effects of PLEKHM1 knockdown on the endocytic pathway. (A and B) The effects of PLEKHM1 knockdown on the EGFR levels. (A) Whole cell extracts from knockdown cells were analyzed by immunoblotting using the indicated antibodies. The experiments were performed in triplicate and a representative gel is shown. (B) Each protein level was normalized to the α-tubulin level and expressed as a percentage relative to control. Data are shown as the mean ± SEM. (C and D) PLEKHM1 knockdown enhances the degradation of EGFR. (C) Knockdown cells were repeatedly treated with lysosomal inhibitors (1 mg/ml leupeptin and 5 μg/ml E-64d) for 50 h and subjected to immunoblotting. (D) The EGFR levels were normalized to the α-tubulin levels and are shown as a percentage relative to the control. (E) The effects of knockdown on the efficiency of EGF transport to LAMP1-positive endosomes. Knocked down cells were stained with an anti-LAMP1 antibody at each time point. The percentages of colocalization are shown as merged EGF/total EGF (mean ± SD, n > 20). Colocalization images are shown in Supplementary Figure S4.
In addition to the endocytic pathway, Rubicon negatively regulates the fusion between autophagosomes and lysosomes, and overexpressing Rubicon compromises this step (Matsunaga et al., 2009; Supplementary Figure S3B). In contrast, GFP-LC3 colocalized to LAMP1-labeled lysosomes in PLEKHM1-overexpressing cells, suggesting that PLEKHM1 does not affect fusion (Supplementary Figure S3B). To further verify this finding, we examined the effects on PLEKHM1 knockdown on autophagy flux. First, we performed a tfLC3 (GFP-mRFP-LC3) assay in which the ratio of the number of red dots to that of green/red dots represents the efficiency of autophagosome–lysosome fusion in tfLC3-expressing cells. There were no differences in the ratio between the control and PLEKHM1 knockdown cells (Supplementary Figure S5, A and B). Next, we monitored LC3 flux by immunoblotting. As shown in Supplementary Figure S5, C and D, the increase in the amount of LC3 after treating with protease inhibitors was slightly larger in PLEKHM1 knockdown cells than in control cells, but this difference was significantly smaller compared with Rubicon knockdown cells. Taken together, we conclude that PLEKHM1 is not directly involved in autophagy.
Function of PLEKHM1 and Rubicon in Endosomal Transport Depends on Their Abilities to Bind to the GTP-bound Form of Rab7
Given that both Rubicon and PLEKHM1 bind to Rab7 and play a role in endosomal transport, we next examined the effects expressing Rab7 dominant-active or -negative mutants on the endosomal localization of Rubicon and PLEKHM1. On expression of the dominant-active Rab7 QL mutant, mStrawberry-PLEKHM1 was increasingly recruited to endosomes with Rab7 QL, as shown previously (Van Wesenbeeck et al., 2007; Figure 4A). On the other hand, expression of the dominant-negative Rab7 TN mutant inhibited the localization of PLEKHM1 to endosomal membranes (Figure 4A). Similarly, we also found that the recruitment of mStrawberry-Rubicon to endosomal membranes was enhanced by Rab7 QL, but inhibited by the TN mutant (Figure 4A). Neither the dominant-negative Rab5 SN mutant nor the Rab9 SN mutant affected the localization of mStrawberry-Rubicon (Supplementary Figure S6). Furthermore, the non–Rab7-binding CGHL mutants of PLEKHM1 and Rubicon did not localize to endosomal membranes (Figure 4B). These data show that interactions with the GTP-bound form of Rab7 are required for their localization to endosomes, which is consistent with the in vitro binding assay.
Figure 4.
The localization of PLEKHM1 and Rubicon is dependent on the Rab7 interaction. (A) The effects of coexpressing Rab7 mutants on the localization of mStrawberry-PLEKHM1 and mStrawberry-Rubicon. HeLa cells were transiently cotransfected with the indicated plasmids and then examined by confocal microscopy. Bars, 20 μm. (B) Mutants that cannot interact with the GTP-bound form of Rab7, such as PLEKHM1 (FL_CGHL), PLEKHM1 (1–895), and Rubicon (FL_CGHL), localize to the cytoplasm. HeLa cells were transfected with each GFP-tagged mutant. Bars, 20 μm.
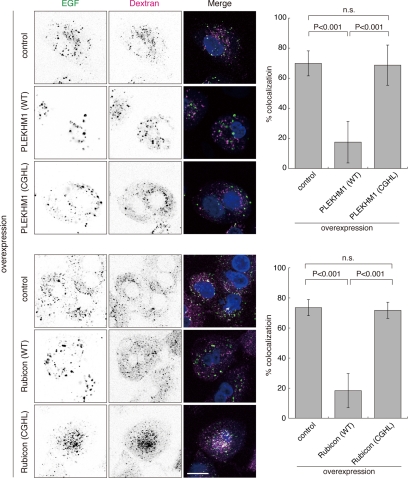
To explore whether binding to Rab7 through the RH domain is important for the function of these proteins, we tested the effects of overexpressing the CGHL mutants on EGF transport to lysosomes. Whereas PLEKHM1 or Rubicon overexpression inhibited the transport of EGF to TMR-labeled lysosomes (Figures 2, C and D, and 5), overexpression of the CGHL mutants did not affect EGF transport (Figure 5). Overexpression of PLEKHM1 (CGHL) also did not affect EGFR degradation (Supplementary Figure S7A). Moreover, although wild-type overexpression caused the endosomes to become abnormally enlarged, overexpression of the CGHL mutant did not affect endosomal morphology (Supplementary Figure S3C).
Figure 5.
The interactions with Rab7 are important for the regulation of the endocytic pathway. The effects of overexpressing the non–Rab7-binding CGHL mutant on endocytic transport were evaluated by assessing the transport of EGF to lysosomes as shown in Figure 2, C and D. The percentages of colocalization are shown as merged EGF/total EGF (mean ± SD, n > 30). Bars, 20 μm.
In contrast to wild-type Rubicon, overexpressing the Rubicon CGHL mutant did not affect autophagy induction and autophagosome maturation (Supplementary Figure S7, B–E), indicating that Rab7 binding is required for Rubicon to function in autophagy as well as the endocytic pathway. These findings raise the possibility that PLEKHM1 and Rubicon act as GTP-activating proteins for Rab7. However, our attempts to monitor the GAP activity of the RH domain toward Rab7 in vitro have been unsuccessful to date (Supplementary Figure S8).
Simultaneous Binding to the Beclin 1-hVps34 Complex and Rab7 Is Critical for Rubicon Function
Although Rubicon was originally identified as a Beclin 1–binding protein (Matsunaga et al., 2009; Zhong et al., 2009), we found that PLEKHM1 does not bind to Beclin 1 (Supplementary Figure S9). Therefore, we next examined the relationship between the ability of Rubicon to bind to Beclin 1 and to Rab7. First, we determined that neither the Rab7 dominant-negative nor -active mutant affects the interaction between Rubicon and Beclin 1 (Figure 6A). Second, we showed that the Rubicon (CGHL) mutants could also bind to Beclin 1 (Figure 6A). These data demonstrate that the interaction with Rab7 is not required for the Rubicon–Beclin 1 association. Third, we examined whether Rab7 interacts with Rubicon in the Beclin 1 complex or only with free Rubicon. Pulldown assays with FLAG-Rab7 successfully coprecipitated endogenous Rubicon together with Beclin 1, p150 (hVps15) and hVps34 (Figure 6, B and C). Fourth, Rubicon knockdown conspicuously disturbed the coimmunoprecipitation of Rab7 with Beclin 1, p150 (hVps15), and hVps34 (Figure 6, B and C). Taken together, these results suggest that Rubicon forms a complex that includes both the Beclin 1 complex and Rab7. Previous reports have shown that hVps34 associates with Rab7 via p150 (hVps15) (Stein et al., 2003; Cao et al., 2007). Our data suggest that the ability of Rab7 to bind to hVps34 and p150 (hVps15) is also mediated by Rubicon.
Figure 6.
Rubicon mediates simultaneous binding between the Beclin 1–PI3-kinase complex and Rab7. (A) The interaction between Beclin 1 and Rubicon is Rab7-independent. Lysates from HEK293A cells expressing a Rab7 mutant, Myc-Beclin 1 and FLAG-Rubicon (wild-type) or FLAG-Rubicon (CGHL) mutant were immunoprecipitated with anti-FLAG M2 beads. (B and C) Rubicon mediates simultaneous binding between the endogenous Beclin 1–PI3-kinase complex and Rab7. (B) Lysates from knocked down cells that were transiently transfected with the vector control or FLAG-Rab7 were immunoprecipitated with anti-FLAG M2 beads and subjected to immunoblotting with the indicated antibodies. (C) The immunoprecipitated protein levels in B were quantified.
Next, we examined whether the ability of Rubicon to bind to the Beclin 1 complex is also indispensable for Rubicon function. The Beclin 1–binding region of Rubicon maps to a. a. residues 393–521, which contain a coiled-coil motif that is sufficient to associate with endogenous Beclin 1 (Figure 7). When the sequence corresponding to this Beclin 1–associating domain (BA domain) was overexpressed in cells, endogenous Rubicon did not coimmunoprecipitate with Beclin 1, probably due to competition (Figure 7). Moreover, overexpression of the BA domain impaired the coimmunoprecipitation of Beclin 1 with Rab7, although it did not affect the ability of Rubicon to bind to Rab7 (Figure 8, A and B). Therefore, Rab7 appears to be the only available binding partner for endogenous Rubicon in cells that overexpress the BA domain. In these cells, EGFR degradation and autophagy are significantly accelerated (Figure 8, C and D; Supplementary Figure S10), similar to previously reported Rubicon knockdown phenotypes (Matsunaga et al., 2009). These findings strongly suggest that even if Rab7 is bound, loss of Beclin 1 association causes defects in Rubicon function. Taken together, we conclude that Rubicon functions by binding to Beclin 1 and Rab7 simultaneously.
Figure 7.
The Beclin 1–associating (BA) domain has a dominant-negative effect on Beclin 1 binding. Schematic view of the mouse Rubicon (mRubicon) structure and its deletion mutants. The mRubicon deletion mutants were subjected to immunoprecipitation analyses in HEK293A cells to map the Beclin 1–binding region. An anti-RFP antibody was used to detect mStrawberry (mSt) and mCherry. Lysates from HEK293A cells expressing each plasmid were immunoprecipitated with an anti-Beclin 1 antibody and immunoblotted with an anti-Rubicon antibody. An asterisk indicates a nonspecific band.
Figure 8.
The ability of Rubicon to interact with both Rab7 and Beclin 1 is important to regulate the endocytic pathway. (A and B) The effects of overexpressing the BA domain (393–521) on the simultaneous interactions among Beclin 1, Rubicon, and Rab7. (A) Coimmunoprecipitation of endogenous Beclin 1 with Rab7 was disrupted in HEK293A cells overexpressing the BA domain, whereas Rubicon coimmunoprecipitated with Rab7 in these cells. An asterisk indicates a nonspecific band. (B) Immunoprecipitated protein levels in A were quantified. (C and D) The effects of overexpressing the BA domain on EGFR degradation. (C) EGFR levels were monitored in A549 cells overexpressing the BA domain as shown in Figure 2A. (D) Immunodetectable levels of EGFR (relative to α-tubulin) remained constant in cells overexpressing the BA domain relative to the controls (mean ± SEM). An anti-RFP antibody was used to detect mCherry-mRubicon (393–521). (E) PLEKHM1 and Rubicon negatively regulate the endocytic pathway through their interactions with Rab7. Their interactions with Rab7 are also required for their localization to endosomal membranes. Rubicon mediates the simultaneous interaction between the Beclin 1–PI3-kinase complex and Rab7, which is an important step in the regulation of the endocytic pathway and autophagosome maturation.
DISCUSSION
In this study, we described a novel RH protein domain that associates directly with Rab7. Rubicon and PLEKHM1 negatively regulate endosomal transport by binding to Rab7 via their RH domains (Figure 8E). Furthermore, this study provides novel insight into the function of PLEKHM1. These two RH domain–containing proteins also have several significant differences. Rubicon must bind to the Beclin 1–PI3-kinase complex in addition to Rab7 for its function, whereas PLEKHM1 only requires Rab7 binding. Rubicon, unlike PLEKHM1, is involved in autophagosome maturation. Both of these proteins localize to endosomes but do not show complete colocalization. Therefore, these two RH domain proteins seem to function through different mechanisms (Figure 8E).
PLEKHM1 bound not only to wild-type Rab7 and the Rab7QL mutant but also to the Rab7TN mutant in a yeast two-hybrid assay and immunoprecipitation analyses in mammalian cells, suggesting that PLEKHM1 may bind to both the GTP-bound and GDP-bound forms of Rab7. However, the TN mutant is predicted to have reduced affinity for both GDP and GTP and may behave as a nucleotide-free Rab7 depending on the assay conditions. Hence, in order to determine the precise nucleotide dependence, we performed an in vitro GST pulldown assay and found that recombinant PLEKHM1 strongly interacted with GTPγS-loaded Rab7 but only minimally interacted with GDP-loaded Rab7. The data convincingly confirm that PLEKHM1 preferentially binds to the GTP-bound form of Rab7, corroborating our hypothesis that PLEKHM1, like Rubicon, is a Rab7 effector.
The structure of the RH domain appears to be important for the recognition of Rab7. Point mutations at cysteine residues in the RH domain abrogate the interaction with Rab7, similar to the relationship between Rab3a and its effector Rabphilin3a (Stahl et al., 1996). The Rab3a-binding site in Rabphilin3a has an architecture that is similar to the RH domain in Rubicon, including a long-α-helix-zinc finger–short-α-helix, in which both the long and short α-helices interact with Rab3a (Ostermeier and Brunger, 1999). A mutation in the zinc finger of Rabphilin3a disrupts this configuration and abolishes Rab3a binding (Stahl et al., 1996). Likewise, the RH domain structure may be important for binding with Rab7.
How do Rubicon and PLEKHM1 negatively regulate EGF and its receptor transport to lysosomes? It was recently determined that Rab7 catalyzes retrograde trafficking from the endosomes to the Golgi apparatus by interacting with a retromer complex (Rojas et al., 2008; Seaman et al., 2009). Likewise, Rab7 may positively act in the retrieval pathway of EGFR to the plasma membrane. When Rubicon and PLEKHM1 are knocked down, retrieval trafficking to the plasma membrane may be impaired, leading to enhanced trafficking to the lysosomes. Alternatively, Rubicon and PLEKHM1 may compete with other Rab7 effectors that positively function in transport to the lysosomes. The adenovirus protein RIDα, which functionally mimics Rab7 and can directly bind to the Rab7 effectors RILP and ORP1L, enhances the lysosomal degradation of EGFR even at a resting state in the absence of EGF activation (Shah et al., 2007). Rab7 functions in many cell types, including osteoclasts (Zhao et al., 2001). Given its ubiquitous expression pattern, PLEKHM1 also likely functions as a universal effector of Rab7.
Previously, it was shown that Rab7 associates with hVps34 and p150 (hVps15) (Stein et al., 2003; Cao et al., 2007). In this study, we demonstrated that Rubicon mediates this interaction. Notably, the function of Rubicon in the endocytic and autophagic pathways requires binding to both Rab7 and the Beclin 1–PI3-kinase complex. This unique feature may allow Rubicon to control these two membrane trafficking pathways, although its exact function remains unclear. One possibility is that synchronized suppression of these two positive regulators during membrane trafficking guarantees that transport can be stopped at a certain step. Rubicon may also facilitate the recycling of receptors to the plasma membrane through Rab7 and increments of local PI3P (or anchoring PI3-kinase to endosomes by binding to Rab7), causing a secondary reduction in the transport to lysosomes (Matsunaga et al., 2009). This scenario is analogous to earlier findings that retromer-mediated retrograde transport from endosomes to the Golgi apparatus is PI3P-dependent (Burda et al., 2002). Furthermore, recycling of the cation-independent mannose 6-phosphate receptor, EGFR, and transferrin receptor is mediated by PI3P-binding protein, SNX1, SNX4, and SNX16 (Kurten et al., 1996; Carlton et al., 2004; Choi et al., 2004; Traer et al., 2007). A previous report demonstrated that UVRAG, a component of the Beclin 1 complex, binds to the HOPS complex that has GEF activity toward Rab7 (Liang et al., 2008). Therefore, Rubicon may terminate Rab7-dependent processes by modulating UVRAG in the Beclin 1 complex, and this hypothesis is supported by findings that the phenotype of the endocytic pathway in Rubicon knockdown cells is the exact opposite of the UVRAG knockdown phenotype (Liang et al., 2008; Matsunaga et al., 2009).
Together, our present study provides new insights into the regulation of the endocytic/autophagic pathway. We not only identified new Rab7 effectors but also discovered a direct connection between two key players in membrane traffic, a Rab protein and PI3-kinase. Rab7 and PI3-kinase function at specific stages in a concerted manner through Rubicon.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Roger Y. Tsien (University of California, San Diego, CA) for kindly providing the mStrawberry and mCherry cDNAs, Dr. Kyohei Umebayashi (University of Osaka, Japan) for kindly providing the Rab7 cDNA, Dr. P. James (University of Wisconsin, Madison) for kindly providing the pGAD-C1 vector, pGBD-C1 vector and PJ69–4A strain, and all members of the Yoshimori laboratory for helpful discussions. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology (T.Y. and T.N.) and the Takeda Science Foundation (T.Y.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0495) on October 13, 2010.
REFERENCES
- Burda P., Padilla S. M., Sarkar S., Emr S. D. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Cao C., Laporte J., Backer J. M., Wandinger-Ness A., Stein M. P. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 2007;8:1052–1067. doi: 10.1111/j.1600-0854.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B. J., Oorschot V.M.J., Rutherford A., Mellor H., Klumperman J., McMahon H. T., Cullen P. J. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- Choi J. H., Hong W. P., Kim M. J., Kim J. H., Ryu S. H., Suh P. G. Sorting nexin 16 regulates EGF receptor trafficking by phosphatidylinositol-3-phosphate interaction with the Phox domain. J. Cell Sci. 2004;117:4209–4218. doi: 10.1242/jcs.01233. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- Feng Y., Press B., Wandingerness A. Rab-7—-an important regulator of late endocytic membrane traffic. J. Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M. G., Munafo D. B., Beron W., Colombo M. I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with ;Mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E. L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001a;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001b;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurten R. C., Cadena D. L., Gill G. N. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–689. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C., et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Matsunaga K., et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Ghosh R. N., Maxfield F. R. Endocytosis Physiol. Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- Ostermeier C., Brunger A. T. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Rojas R., van Vlijmen T., Mardones G. A., Prabhu Y., Rojas A. L., Mohammed S., Heck A. J., Raposo G., van der Sluijs P., Bonifacino J. S. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J., Harbour M. E., Tattersall D., Read E., Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. H., Cianciola N. L., Mills J. L., Sonnichsen F. D., Carlin C. Adenovirus RID alpha regulates endosome maturation by mimicking GTP-Rab7. J. Cell Biol. 2007;179:965–980. doi: 10.1083/jcb.200702187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Tooze S. A. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl B., Chou J. H., Li C., Sudhof T. C., Jahn R. Rab3 reversibly recruits rabphilin to synaptic vesicles by a mechanism analogous to raf recruitment by ras. EMBO J. 1996;15:1799–1809. [PMC free article] [PubMed] [Google Scholar]
- Stein M. P., Feng Y., Cooper K. L., Welford A. M., Wandinger-Ness A. Human VPS34 and p150 are rab7 interacting partners. Traffic. 2003;4:754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sun Q. M., Fan W. L., Chen K. L., Ding X. J., Chen S., Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer C. J., Rutherford A. C., Palmer K. J., Wassmer T., Oakley J., Attar N., Carlton J. G., Kremerskothen J., Stephens D. J., Cullen P. J. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 2007;9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- Van Wesenbeeck L., et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J. Clin Invest. 2007;117:919–930. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. P., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 2008;20:401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Zhao H., Laitala-Leinonen T., Parikka V., Vaananen H. K. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J. Biol. Chem. 2001;276:39295–39302. doi: 10.1074/jbc.M010999200. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Wang Q. J., Li X. T., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. Y. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.