We have identified 7SK RNA to be enriched in nuclear speckles. Knock-down of 7SK results in the mislocalization of nuclear speckle constituents, and the transcriptional up-regulation of a reporter gene locus. 7SK RNA transiently associates with the locus upon transcriptional down-regulation correlating with the displacement of pTEF-b.
Abstract
Noncoding RNAs play important roles in various aspects of gene regulation. We have identified 7SK RNA to be enriched in nuclear speckles or interchromatin granule clusters (IGCs), a subnuclear domain enriched in pre-mRNA processing factors. 7SK RNA, in association with HEXIM 1 and 2, is involved in the inhibition of transcriptional elongation by RNA polymerase II. Inhibition occurs via sequestration of the active P-TEFb kinase complex (CDK 9 and Cyclin T1/T2a/b or K) that is involved in phosphorylating the C-terminal domain of RNA polymerase II. Our results demonstrate that knock-down of 7SK RNA, by specific antisense oligonucleotides, results in the mislocalization of nuclear speckle constituents in a transcription-dependent manner, and the transcriptional up-regulation of a RNA polymerase II transcribed reporter gene locus. Furthermore, 7SK RNA transiently associates with a stably integrated reporter gene locus upon transcriptional down-regulation and its presence correlates with the efficient displacement of P-TEFb constituents from the locus. Our results suggest that 7SK RNA plays a role in modulating the available level of P-TEFb upon transcriptional down-regulation by sequestering its constituents in nuclear speckles.
INTRODUCTION
The eukaryotic cell nucleus is a membrane-bounded organelle where many critical cellular functions are carried out within macromolecular protein complexes (reviewed in Prasanth and Spector, 2005). To efficiently carry out these functions, the mammalian cell nucleus is compartmentalized into specific subnuclear domains or nuclear bodies (reviewed in Spector, 2006; Matera et al., 2009). Nuclear bodies are broadly characterized by their macromolecular composition, number, size, and associated functions. The majority of nuclear bodies show little (PML bodies and Cajal bodies) to no mobility in the nucleus (Muratani et al., 2002; Platani et al., 2002). However, the proteins in these domains generally show high levels of exchange between the domains and the surrounding interchromatin space (reviewed in Misteli, 2001; Misteli, 2008). In addition to their protein constituents, some of these subnuclear domains also harbor RNA molecules (reviewed in Prasanth and Spector, 2007; Wilusz et al., 2009).
Recent large-scale genomic sequencing analyses have revealed that human and mouse bear very similar number of protein coding genes (∼23,000), surprisingly this number is not very different from Caenorhabditis elegans (∼20,000) or Drosophila (∼14,000) (www.ensembl.org). This suggests that the complexity associated with higher organisms is not solely related to the number of protein coding genes that they harbor (reviewed in Mattick, 2001; Frith et al., 2005; Mattick and Makunin, 2006; Kapranov et al., 2007; Pheasant and Mattick, 2007; Prasanth and Spector, 2007; Wilusz et al., 2009). Posttranscriptional modifications such as alternative pre-mRNA splicing and RNA editing, as well as posttranslational modifications of proteins, including phosphorylation, sumoylation, and ubiquitylation, can increase the proteome size, but this is unlikely to fully account for the complexity associated with higher eukaryotes (reviewed in Prasanth and Spector, 2007). Interestingly, transcriptome analysis has revealed that in human cells, a large fraction of the genome is transcribed into RNA, whereas the protein coding part comprises <5% of the genome (reviewed in Mattick and Makunin, 2006; Willingham and Gingeras, 2006; Kapranov et al., 2007; Prasanth and Spector, 2007; Wilusz et al., 2009). Sequence annotation of these transcripts has revealed that most if not all of these RNAs do not encode for proteins and are therefore classified as noncoding RNA (ncRNAs) or transcripts of unknown functions (TUFs) (reviewed in Mattick and Makunin, 2006; Willingham and Gingeras, 2006; Kapranov et al., 2007; Prasanth and Spector, 2007; Wilusz et al., 2009). Some of the ncRNAs are expressed in all cell types and are involved in executing essential housekeeping functions whereas a large proportion of them show tissue or cell-type specific expression and are suggested to be regulatory in nature (reviewed in Prasanth and Spector, 2007; Wilusz et al., 2009). Altered expression of some of these regulatory ncRNA transcripts has been directly implicated in various diseases including cancer and neurological disorders (reviewed in Costa, 2005; Szymanski et al., 2005; Calin and Croce, 2006; Costa, 2007; Prasanth and Spector, 2007; Taft et al., 2009). It has been proposed that the large number of regulatory RNAs present in higher eukaryotes contributes to the complex network that governs cellular functions (reviewed in Mattick, 2001, 2004a; Mattick, 2005; Mattick and Makunin, 2006).
Interestingly, a subset of ncRNAs [examples include snRNAs, snoRNAs, scaRNAs, MEN epsilon/beta (NEAT1), Air, Gomafu, TERRA] is retained in the nucleus and localizes to specific nuclear bodies (reviewed in Prasanth and Spector, 2007; Wilusz et al., 2009). It is hypothesized that many yet to be identified ncRNAs may also be involved in modulating vital gene regulatory pathways or act as structural components of some of the subnuclear domains (Prasanth et al., 2005; Pang et al., 2007; Prasanth and Spector, 2007; Amaral et al., 2008; Dinger et al., 2008; Mercer et al., 2008; Dinger et al., 2009; Mattick, 2009; Mercer et al., 2009; Pang et al., 2009). For example, the recently identified nuclear-retained ncRNAs; MEN epsilon/beta or NEAT1 play a crucial role in establishing and maintaining paraspeckle integrity in mammalian cells (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009).
Of particular interest in the present study is the population of unidentified nuclear RNA/s that are enriched in nuclear speckles or interchromatin granule clusters (IGCs) (Carter et al., 1991; Huang et al., 1994; Lamond and Spector, 2003; Hall et al., 2006; Hutchinson et al., 2007). Nuclear speckles are thought to be storage/modification/assembly centers for factors involved in pre-mRNA processing and factors are recruited from speckles to active transcription sites in a phosphorylation-dependent manner (reviewed in Misteli and Spector, 1997; Lamond and Spector, 2003; Hall et al., 2006). MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2 (Hutchinson et al., 2007), is a long (∼6.7 kb) ncRNA (Wilusz et al., 2008) that is misregulated in many human cancers (Ji et al., 2003; Lin et al., 2006) and was previously shown to be specifically retained in nuclear speckles (Hutchinson et al., 2007). However, knock-down of MALAT1 did not result in reorganization of nuclear speckles indicating that MALAT1 is not a structural RNA (Hutchinson et al., 2007; Clemson et al., 2009; Sasaki et al., 2009).
Here, we demonstrate by RNA-FISH that 7SK RNA, a 331-nt long, highly conserved, small nuclear RNA, is enriched in nuclear speckles where it colocalizes with pre-mRNA splicing factors. Knock-down of 7SK RNA in human cells resulted in the disorganization of various splicing factors from IGCs in a transcription-dependent manner. Using a stably integrated in vivo reporter cell line system we show that knock-down of 7SK RNA results in increased expression of the reporter gene as well as efficient recruitment of various pre-mRNA processing factors to the transcriptionally active gene locus. Furthermore, 7SK RNA transiently associated with the reporter gene locus upon transcriptional down-regulation, and its presence correlated with the displacement of P-TEFb constituents from the locus. Based upon these data we propose that 7SK RNA negatively regulates RNA pol II transcription by facilitating the displacement of P-TEFb components from the site of transcription, and sequestering them in nuclear speckles, thereby contributing to the regulation between gene expression and silencing.
MATERIALS AND METHODS
Cell Culture and Drug Treatment
HeLa and U2OS cells were grown in DMEM containing high glucose (Invitrogen, Carlsbad, CA) supplemented with penicillin-streptomycin and 10% fetal bovine serum (Hyclone Laboratories, Logan, UT). In addition, U2OS 2-6-3 tet-OFF and U2OS 2-6-3-TOLC cells were cultured in DMEM + 10% Tet system-approved FBS (BD Biosciences, Clontech, USA) in the presence of hygromycin (100 μg/ml) and G418 (600 μg/ml).
To inhibit RNA polymerase II–mediated transcription, HeLa cells were treated with α-amanitin (50 μg/ml) for 5 h at 37°C in the CO2 incubator. To induce transcription from the artificial locus in U2OS 2-6-3-TOLC cells, cells were treated with Doxycycline (DOX; 1 μg/ml) for various time points. Similarly, DOX was added to inhibit transcription from the locus in U2OS 2-6-3 tet-OFF cells.
Plasmid Constructs and DNA Transfection
Human 7SK promoter and coding region (Murphy et al., 1987) was PCR amplified and was cloned into pEYFP-N1 vector (BD Biosciences Clontech, Palo Alto, CA). The EYFP coding region was removed from the pEYFP-N1 vector during cloning. Next, a 6× repeat of the MS2 binding stem loop was subcloned in between the 7SK promoter and coding region. MS2-BP-YFP (Janicki et al., 2004) with the human pol II promoter was subcloned into pVitro-Tn5; pVitro-MS2-BP-YFP (Invitrogen, San Diego, CA). Finally, the 7SK-6xMS2 stemloop region, with native promoter, was subcloned into pVitro-MS2-YFP-NLS; pVitro-MS2-BP-YFP-7SK-6xMS2.
Plasmid DNAs (pVitro-MS2-BP-YFP-7SK-6xMS2, YFP-SF2/ASF, MS2-BP-YFP, LacI-YFP, LacI-CFP, YFP-SC35, U2AF65-CFP, U1-70K-CFP (2 μg/experiment) were individually transfected into cells either by electroporation or by lipid-based transfection (Lipofectamine-2000; Invitrogen, Carlsbad, CA). After electroporation, cells were seeded onto acid-washed coverslips and processed for immunofluorescence localization or RNA fluorescence in situ hybridization 24 h posttransfection.
Immunofluorescence
Cells were rinsed briefly in phosphate-buffered saline (PBS) and then fixed for 15 min in 1.7% formaldehyde in PBS (pH 7.4). Cells were permeabilized in PBS containing 0.5% Triton X-100 and 1% goat serum for 7–10 min on ice, washed in PBS +NGS, and were incubated with primary antibodies (anti-SC35 [1:200] (Fu and Maniatis, 1990), anti-SF2/ASF [1:200; gift from Dr. Adrian Krainer, CSHL, USA], anti-B” [1:50] (Habets et al., 1985), anti-CDK9 [1:100] and anti-Cyclin-T1 [1:100] (Santa Cruz Biotechnology, Santa Cruz, CA), anti-RNA pol II [H14; 1:20]) (Bregman et al., 1995) for 1 h at room temperature. Cells were rinsed in PBS containing 1% goat serum and then secondary anti–species-specific antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) were added for 30 min at room temperature: goat anti-rabbit (GAR) IgG1-Texas Red (1:1000), GAM IgG Texas Red (1:1000), goat anti-rabbit (GAR) IgG1-FITC (1:1000). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Cells were mounted in medium containing 90% glycerol, 10% PBS pH 8.0 plus 1 mg/ml paraphenylenediamine. Cells were examined using an Axioplan 2i fluorescence microscope (Carl Zeiss, Thornwood, NY) equipped with Chroma filters (Chroma Technology, Brattleboro, VT). OpenLab software (Improvision, Boston, MA) was used to collect digital images from an ORCA cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ). Images were also acquired using a DeltaVision deconvolution microscope system and softWoRx 2.50 software (Applied Precision, Issaquah, WA).
RNA-Fluorescence In Situ Hybridization (RNA-FISH)
To detect 7SK-RNA, cells were rinsed briefly in PBS, fixed in 2% formaldehyde in PBS (pH 7.4) for 15 min at RT. Cells were permeabilized in PBS containing 0.5% Triton X-100 and 5 mM VRC (New England Biolabs, Beverly, Massachusetts) on ice for 5–10 min. Cells were washed in PBS 3 times 10 min each and rinsed once in 2× SSC before hybridization. Hybridization was carried out using a Texas Red conjugated 24-mer DNA oligodeoxynucleotide probe (5′-cagatgagccgaatcaaccctggc-3′) complementary to 7SK RNA in a moist chamber at 42°C for 12–16 h as described earlier (Prasanth et al., 2005). Total poly(A+) RNA was visualized by RNA-FISH using a fluorescently-labeled oligo dT probe (Huang et al., 1994). For co-RNA-FISH and immunolocalization studies, after RNA-FISH, cells were again fixed for 5 min in 2% formaldehyde, and IF and imaging were performed as described earlier (Prasanth et al., 2003; Prasanth et al., 2005).
Antisense-Oligonucleotide Treatment
Phosphorothioate-modified DNA antisense oligodeoxynucleotides targeting 7SK RNA (AS99: CCTTGAGAGCTTGTTTGGAGG; AS94: CTAGCCAGCCAGATCAGCCG) were used to deplete 7SK RNA as described previously (Prasanth et al., 2005). Antisense DNA oligonucleotides (100 nM) were administered to HeLa and U2OS cells using Lipofectamine 2000 or Lipofectamine RNAiMax reagent as per the manufacturer's instructions (Invitrogen, USA). Cells were treated with antisense oligonucleotides (2 times at an interval of 24 h), were fixed post-24 h after the second antisense treatment and were processed for immunofluorescence localization or RNA fluorescence in situ hybridization.
RESULTS
7SK RNA Localizes to Nuclear Speckles
By using an RNA-FISH based approach in human and mouse cell lines, we have localized 7SK RNA in a fine punctate nuclear distribution, excluding the nucleoli, in mouse NIH-3T3 cells, human HeLa (Figure 1Aa), and U2OS cells. In addition, it was also enriched in 15–30 subnuclear domains, reminiscent of nuclear speckles or IGCs (Figure 1Aa). Colocalization studies of 7SK RNA in HeLa cells expressing the SR family splicing factor EYFP-SF2/ASF (Bubulya et al., 2004) confirmed the localization of 7SK RNA to nuclear speckles (Figure 1Aa–Ad). Furthermore, RNA-FISH in cells that were pre-extracted with detergent to remove most of the soluble homogenous pool of 7SK RNA showed a more pronounced distribution of 7SK RNA in nuclear speckles (data not shown). Pretreatment of cells with RNAse A (data not shown) as well as knock-down of 7SK RNA using multiple modified antisense oligonucleotides abolished the 7SK RNA-FISH signal in the nuclear speckles (Figure 2B & Supplemental Figure 1).
Figure 1.
7SK RNA is enriched in nuclear speckles. (Aa–Ad) Deconvolved images of RNA-FISH performed on HeLa cells using a 7SK oligonucleotide probe reveals enrichment of 7SK RNA (red, Aa) in nuclear speckles and colocalizes (Ac) with a known nuclear speckle marker, SF2/ASF (green, Ab). (Ba–Bd) Inhibition of RNA polymerase II by α-amanitin treatment shows 7SK RNA (red, Ba) enrichment in rounded up nuclear speckles (Bc). (Ca–Cc) Transient expression of an MS2 stem-loop tagged 7SK RNA (green, Ca) and MS2-BP-YFP in live HeLa cells shows colocalization of tagged 7SK RNA with SC35-RFP (red, Cb) in nuclear speckles (merge, Cc). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue, Ad, Bd). The scale bar represents 10 μm.
Figure 2.
Knock-down of 7SK RNA results in the disorganization of nuclear speckle components. (A) Antisense oligonucleotides designed against 7SK RNA were used to knock-down 7SK in HeLa cells. 90% knock-down was achieved as assessed by Q-PCR. Control oligonucleotide had no effect. 7SK RNA levels were normalized to β-actin mRNA and are presented relative to RNA levels in scrambled oligonucleotide transfected cells. The data are shown as mean and SD values of three measurements per data point. (B) YFP-SF2/ASF (green, Ba) and 7SK RNA FISH (red, Bb) show prominent colocalization in speckles in HeLa cells treated with control antisense oligonucleotides (control oligo). Antisense knock-down of 7SK results in the appearance of large intranuclear foci of SF2/ASF (green, Bc, see arrowheads) and its disorganization from speckles. (C) YFP-SF2/ASF (green, Ca) and SC35 immunofluorescence (red, Cb) show prominent colocalization in speckles (merge, Cc) in HeLa cells treated with scrambled antisense oligonucleotides (control oligo). Antisense knock-down of 7SK results in the appearance of large intranuclear foci of SF2/ASF (green, Cd) and its disorganization from speckles. The cell that shows SF2/ASF foci also shows appearance of small rounded up SC35 foci (see arrows, red, Ce). Interestingly, both SF2/ASF and SC35 foci do not colocalize (merge, Cf). (D) YFP-SF2/ASF (green, Da) and immunolocalization of B” U2snRNP (red, Db) show prominent colocalization at the speckles in HeLa cells treated with control antisense oligonucleotides (control oligo). Cells lacking 7SK RNA demonstrate intranuclear foci of SF2/ASF (green, Dc). However, B” U2 snRNP shows a more homogenous distribution but continues to localize in Cajal bodies (red, Dd, see arrow). (E) RNA FISH using oligo dT probes shows poly(A+) RNA enrichment in speckles of control oligonucleotide treated cells (red, Eb). In contrast, following 7SK knock-down poly (A+) RNA appears as a more homogenous nuclear pool (red, Ed). Scale bar represents 5 μm.
Nuclear speckles contain various pre-mRNA processing factors and have been suggested to play a role in modulating transcription and pre-mRNA splicing (reviewed in Lamond and Spector, 2003; Hall et al., 2006). Treatment of cells with RNA pol II inhibitors was previously shown to result in the rounding up of nuclear speckles (Spector et al., 1983; Huang et al., 1994). Incubation of YFP-SF2/ASF expressing HeLa cells with the RNA pol II inhibitor α-amanitin clearly showed the enrichment of 7SK RNA in the rounded up nuclear speckles (Figure 1B, a–d). It has previously been demonstrated that only a fraction of the SR protein SF2/ASF associates with rounded up nuclear speckles in transcriptionally inhibited cells, whereas the rest of the pool localizes around the nucleoli (Figures 1Bb and 3Ce) (Bubulya et al., 2004). However, the distribution of 7SK RNA in α-amanitin–treated cells shows that it does not localize around nucleoli and its distribution in RNA pol II–inhibited cells is very similar to other known non-SR splicing factors (Figure 3, Cb and Cf) (Bubulya et al., 2004).
Figure 3.
Displacement of splicing factors from nuclear speckles upon 7SK depletion is transcription dependent. (A) Immunofluorescence localization of CDK9 (green, Aa) and the hyperphosphorylated form of RNA pol II (H14 ab, red, Ab) in control oligonucleotide treated U2OS cell shows a punctate nuclear localization excluding the nucleoli (arrow). 7SK antisense olgonucleotide-treated cell shows a reorganization of the distribution of CDK9 to nucleoli (green, Ac), whereas the RNA pol II localization remains unaltered (red, Ad). (B) Immunoblot analysis using antibodies against P-TEFb components and splicing factors in HeLa cells shows no change in their levels in control versus 7SK AS treated cells. (C) YFP-SF2/ASF (green, Ca) and B” (red, Cb) localization in control oligonucleotide treated HeLa cells followed by α-amanitin incubation shows that YFP-SF2/ASF is enriched around the nucleoli whereas B” protein localizes to the rounded up speckles (merge, Cc). α-amanitin incubated 7SK antisense oligonucleotide treated HeLa cells also shows a similar distribution of YFP-SF2/ASF and B” (Ce–Ch) indicating the disorganization of splicing factors from nuclear speckles observed in 7SK depleted cells is transcription dependent. DNA is counterstained with DAPI (blue, Cd, Ch). The bar indicates 10 μm.
We have also tagged the human 7SK RNA with bacteriophage MS2 stem-loop repeats (7SK-MS2) and transiently expressed this construct with an MS2 binding protein-YFP (MS2-BP-YFP) fusion protein in human cells. The cells that expressed MS2-BP-YFP alone showed homogenous nuclear staining of YFP, including in the nucleoli (data not shown). However, cells that coexpressed both 7SK-MS2 and MS2-BP-YFP showed a fine punctate nuclear distribution of YFP. In addition, prominent YFP signal was also enriched in nuclear speckles as confirmed by coimmunolocalization with an IGC marker protein SC35 (Figure 1, Ca–Cc). From these results, it is clear that a significant fraction of the nuclear retained 7SK RNA is localized to nuclear speckles.
7SK RNA Modulates the Distribution of Nuclear Speckle Proteins
The involvement of nuclear retained 7SK RNA in transcriptional regulation has been well documented (Blencowe, 2002; Peterlin and Price, 2006; Barrandon et al., 2008). In the nucleus, 7SK facilitates the interaction of active P-TEFb with HEXIM1 and/or 2. HEXIM proteins inactivate the kinase activity of P-TEFb, which in turn inhibits RNA pol II transcription elongation (Peterlin and Price, 2006; Barrandon et al., 2008). Previous studies have reported the presence of components of the P-TEFb complex (CDK9, Cyclin T1 and HEXIM1) in nuclear speckles (Herrmann and Mancini, 2001; Pendergrast et al., 2002; Haaland et al., 2005). However, we were interested in examining whether 7SK RNA that localizes to nuclear speckles is part of the inactive P-TEFb kinase complex and/or whether it has an independent role in the organization of nuclear speckle constituents. To address the role of 7SK in the subnuclear localization of pre-mRNA splicing factors, the distribution of various nuclear speckle components were examined in human cells after depletion of 7SK RNA. More than 90% knock-down of 7SK RNA was achieved by using modified antisense DNA oligonucleotides (Figure 2, A and B). Two independent antisense oligonucleotides were used to target 7SK RNA and both showed similar results (data not shown). Cells were analyzed for 7SK knock-down and for immunolocalization of various nuclear speckle marker proteins 48 h post antisense oligonucleotide treatment. In control oligonucleotide (scrambled oligo) treated cells, YFP-SF2/ASF distributed in nuclear speckles and colocalized with 7SK RNA (Figure 2B, a–b). In cells that were depleted of 7SK RNA, YFP-SF2/ASF showed a weak homogenous localization. Intriguingly, these cells also showed 2–8 very prominently labeled round YFP-SF2/ASF foci (Figure 2, Bc, Cd, Dc, and Ec) (number of nuclei counted = 1000). The appearance of the ‘SF2/ASF foci’ in 7SK-depleted cells served as a good marker to differentiate cells showing efficient knock-down of 7SK RNA. Similar distribution was also observed with endogenous SF2/ASF, as observed by immunostaining using SF2/ASF antibodies (Supplemental Figure 1). Interestingly, several of the other nuclear speckle-associated splicing factors (SC35, Figure 2C; U2AF65, Supplemental Figure 2, A and C; U1-70K, Supplemental Figure 2, B and D) also formed prominent nuclear foci in 7SK RNA depleted cells. Interestingly, none of these nuclear foci colocalize with the SF2/ASF foci indicating that they form independent foci and do not represent remnants of nuclear speckles (Figure 2C, d–f). The prominent nuclear foci observed in the 7SK depleted cells did not correspond to rounded up nuclear speckles observed during transcription inhibition, as these were fewer in number and also did not contain all the other nuclear speckle proteins nor poly(A+) RNAs (Figures 2C, d–f, 2D, c–d, and 2E, c–d). In addition, the SF2/ASF foci that appeared after 7SK RNA depletion did not colocalize with any of the known subnuclear domains including PML bodies and Cajal bodies (data not shown). The nonSR family splicing factor B”, which is part of the U2-snRNP complex, also showed a change in its distribution in the 7SK-depleted cells. In control oligonucleotide-treated cells, B” localized in nuclear speckles and also in Cajal bodies (Figure 2Db). In the 7SK-depleted cells, B” localization in the nuclear speckles was less apparent, as compared with control oligonucleotide-treated cells, and showed a more homogenous distribution (Figure 2Dd). However, the B” staining in the Cajal bodies remained unaltered (see the arrow in Figure 2Dd). Together, these results show that 7SK depletion changes the distribution of pre-mRNA splicing factors between nuclear speckles and the nucleoplasm, but not from other subnuclear structures (i.e., Cajal bodies). Interestingly, the poly(A+)RNA pool that is localized in nuclear speckles also showed a more homogenous distribution with a few foci after 7SK RNA depletion (Figure 2E, a–d) suggesting that the presence of 7SK RNA is critical for appropriately maintaining the speckle constituents in human cells. Importantly, immunoblot analysis from total cell extracts revealed that the levels of various speckle components including, B” U2snRNP, SF2/ASF, CDK9, and Cyclin T1 in control versus 7SK depleted samples were comparable (Figure 3B). This suggests that 7SK depletion alters the nuclear distribution of nuclear speckle proteins without modulating their total cellular levels.
Components of the inactive P-TEFb complex, CDK9, Cyclin-T1 and HEXIM1 have previously been reported to localize to nuclear speckles (Herrmann and Mancini, 2001; Pendergrast et al., 2002; Haaland et al., 2005). Because 7SK RNA is a part of the inactive P-TEFb complex, we were next interested to address whether 7SK RNA plays any role in the recruitment of these proteins to nuclear speckles. In cells depleted of 7SK RNA, Cdk9 redistributed to nucleoli whereas in control oligonucleotide-treated cells, it showed more punctate nuclear distribution excluding nucleoli (Figure 3A).
Next, we examined whether the changes in distribution of nuclear speckle components in 7SK RNA depleted cells were sensitive to the overall transcriptional status of the cells. To test this, we treated HeLa cells with or without 7SK RNA with α-amanitin to inhibit RNA pol II transcription and compared the distribution of nuclear speckle proteins. Intriguingly, no significant difference in SF2/ASF and B” distribution was observed between 7SK RNA-depleted cells and control oligonucleotide-treated cells (Figure 3C). From these data we conclude that the changes in localization of nuclear speckle proteins we observed in 7SK RNA-depleted cells is sensitive to ongoing transcription.
Knock-Down of 7SK RNA Increases the Transcriptional Output of a Stably Integrated Reporter Locus
7SK RNA is known to play a crucial role in the transcriptional regulation of HIV viral RNAs (reviewed in Peterlin and Price, 2006). We were interested to monitor whether nuclear levels of 7SK RNA modulate cellular transcription. To test this, we took advantage of a stably integrated in vivo reporter cell line system where levels of gene expression at a single genetic locus can be monitored in real-time (Janicki et al., 2004; Shav-Tal et al., 2004). Tet-ON and LacI-mCherry plasmids were stably integrated into the original U2OS 2-6-3 cell line (U2OS 2-6-3-TOLC; Figure 4A). U2OS 2-6-3-TOLC cells were transiently transfected with MS2-BP-YFP, and its recruitment was monitored at the locus during gene activation following doxycycline (DOX) treatment, in the presence or absence of 7SK RNA. In cells that were treated with control oligonucleotides, MS2-BP-YFP was recruited to the transcriptionally active locus within 15 min post DOX addition (Figure 4Bc). Interestingly, in cells that were depleted of 7SK RNA, DOX treatment (15 min) resulted in an increased accumulation of MS2-BP-YFP at the locus (Figure 4Bd). This suggested that 7SK depletion increased the transcriptional activity of the reporter gene. To confirm this further, we analyzed the levels of reporter RNA by RT-PCR (Figure 4C). The primer pairs for the PCR were designed to differentiate reporter mRNA from pre-mRNA based on the amplicon size. An increased level of amplification of reporter mRNA was observed from total RNA of cells that were treated with 7SK antisense oligonucleotides compared with control oligonucleotides (Figure 4C). Finally, a concomitant increase in the levels of reporter Cyan fluorescent protein (CFP) was also observed in cells that were depleted of 7SK RNA (Figure 4D). From these results, it is clear that upon depletion of 7SK RNA, the reporter gene locus is hyperactivated and the resulting transcripts are processed and translated efficiently into protein.
Figure 4.
7SK depletion up-regulates reporter gene expression using the pTet-ON system. (A) Schematic representation of the reporter construct used for the analysis. The arrows in the figure indicate the positions of the primer used for the RT-PCR analysis. The figure is adapted and modified from (Janicki et al., 2004). (B) In control cells MS2-YFP localization shows an accumulation of the reporter RNA at the site of transcription 15 min post gene activation (green, Bc, arrowhead). In the absence of 7SK RNA, higher levels of reporter RNA accumulate at the transcription site (green, Bd, arrow). Scale bar represents 5 μm. (C) RT-PCR analysis of RNA samples from cells isolated 15 min and 30 min post DOX treatment shows up-regulation of the reporter gene in cells that are depleted of 7SK RNA (D) Immunoblot analysis using an anti-GFP antibody demonstrates the up-regulation of reporter CFP protein in 7SK knock-down cells corroborating the increased transcriptional activation of the reporter gene in the absence of 7SK RNA. α-tubulin was used as a loading control.
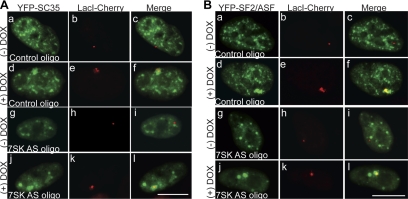
Nuclear speckles play critical roles in modulating RNA pol II–mediated transcription and pre-mRNA processing (reviewed in Lamond and Spector, 2003), and disruption of nuclear speckles in cells results in aberrant transcription and pre-mRNA splicing (Sacco-Bubulya and Spector, 2002). Depletion of 7SK RNA resulted in the mislocalization of various nuclear speckle-associated pre-mRNA splicing factors. Therefore, we were interested to study whether 7SK RNA depletion influences the recruitment of splicing factors to a transcription site. We have analyzed the recruitment of YFP-tagged SF2/ASF and SC35 to the DOX induced transcription locus in U2OS 2-6-3-TOLC cells in the presence or absence of 7SK RNA. In control oligonuclotide-treated cells, both SC35 (Figure 5A, a–c) and SF2/ASF (Figure 5B, a–c) localized to nuclear speckles and did not associate with the lacI-mCherry-tagged transcriptionally inactive locus (Figure 5, Ac and Bc). After addition of DOX to the medium (180 mins), both SC35 and SF2/ASF localized to the transcriptionally active decondensed gene locus (Figures 5A, d–f and 5B, d–f). Both proteins continue to show nuclear speckle association in DOX treated cells (Figures 5, Ad and Bd). In 7SK AS oligonucleotide-treated cells both SC35 and SF2/ASF formed the prominent round nuclear foci (Figure 5A, g and j and 5B, g and j). Interestingly, DOX-treated 7SK RNA–depleted cells continue to show strong association of both SC35 and SF2/ASF to the transcriptionally active locus (Figure 5A, j–l and 5B, j–l). These results imply that 7SK RNA does not influence the recruitment of SR-splicing factors to the transcription site.
Figure 5.
The recruitment of SR-family splicing factors to a transcriptionally active locus remains unaffected in cells that are depleted of 7SK RNA. In U2OS 2-6-3-TOLC cells, YFP-SC35 (A) and YFP-SF2/ASF (B) are recruited to the transcriptionally active locus ([+] DOX, Ad–f; Aj–l; Bd–f; Bj–l) in the presence (YFP-SC35, Ad–f; YFP-SF2/ASF, Bd–f) or absence (YFP-SC35, Aj–l; YFP-SF2/ASF, Bj–l) of 7SK RNA. Note that the 7SK depleted cells show prominent round YFP-SC35 (Ag–l) and YFP-SF2/ASF (Bg–l) subnuclear foci. The scale bar represents 10 μm.
pTEF-b Protein Constituents, but Not 7SK RNA, Are Efficiently Recruited to the Transcribing Reporter Locus
It is known that P-TEFb phosphorylates serine 2 within the carboxy-terminal domain (CTD) of RNA pol II, a posttranslational modification that is essential for efficient transcriptional elongation (Peterlin and Price, 2006; Zhou and Yik, 2006). We analyzed the recruitment of P-TEFb components to the reporter gene locus during transcriptional activation. When the locus was inactive, neither CDK9 (Figure 6A, a–c) nor 7SK RNA (Figure 6Ba-c) was present at the locus. After addition of DOX (240 min), CDK9 was efficiently recruited to the actively transcribing locus (Figure 6C, a–c and 6E, a–d). Similar results were also observed with cyclin T1 (data not shown). However, we did not observe 7SK RNA at the transcriptionally active locus (Figure 6D, a–c and 6F, a–d). These results indicate that only CDK9 and CyclinT1, and not 7SK RNA, are recruited to the gene locus during active transcription.
Figure 6.
P-TEFb, but not 7SK, is present at sites of active transcription of a stably integrated reporter locus. (A and B) Immunolocalization to CDK9 (red, Aa) and 7SK RNA-FISH (red, Ba) in U2OS 2-6-3 tet-ON cells. These constituents (Ac, Bc) are not visualized at the inactive locus (LacI-YFP, green, Ab and Bb). (C and D) 240 min after Dox addition and induction of transcription CDK9 (red, Ca), but not 7SK RNA (red, Da), is concentrated at the active locus (green, Cb and Db). While CDK9 (Cc) shows direct overlap with decondensed chromatin, 7SK RNA (Dc) is absent from the region. (E and F) CDK9 (red, Ea) but not 7SK RNA (red, Fa) is enriched at the actively transcribing reporter gene locus (green, Eb and Fb) as observed by MS2-YFP bound to nascent RNA (Ec and Fc). DNA is counterstained with DAPI (blue, Ed, Fd). Scale bar represents 10 μm.
7SK RNA Transiently Associates with the Gene Locus Upon Transcriptional Down-Regulation and Plays a Role in the Displacement of P-TEFb from the Locus
7SK RNA is involved in the cessation of transcription of cellular genes by facilitating the interaction of HEXIMs with the CDK9/Cyclin T kinase complex (Peterlin and Price, 2006; Zhou and Yik, 2006; Barrandon et al., 2008). Because 7SK RNA was absent from the transcriptionally active locus, we were interested in analyzing whether 7SK RNA associates with the gene locus during the inactivation process. We used a U2OS 2-6-3 Tet-OFF cell line for this experiment, where the reporter gene is active in the absence of DOX and upon addition of DOX, it becomes inactivated. Within 5–10 min post DOX addition, transcription at the locus ceased with a concomitant loss of detectable levels of CDK9 (Figures 7Db, 7Eb, and 8 and Supplemental Figure 3). Dual RNA-FISH and immunolocalization analysis revealed the presence of CDK9 in cells where the locus was transcriptionally active (see arrowhead in Figure 7Ab). However, there was no 7SK RNA associated with the active locus (Figure 7Ac). Surprisingly, within 2 min 30 s post-DOX addition, 7SK RNA was enriched at the locus, which also had CDK9 associated with it (arrowhead in Figure 7B, a–d). 7SK RNA association with the locus was prominent 5 min post DOX addition (arrowheads in Figure 7C, a–d and see inset in Figure 7C, b–d). However, within 7.5–10 min post-addition of DOX, 7SK RNA was no longer associated with the locus (Figure 7, Dc and Ec). Interestingly, CDK9 also dissociated from the locus concomitant with the disappearance of 7SK RNA (Figure 7, Db and Eb). These data indicate that 7SK RNA is transiently enriched at the reporter gene locus during its transcriptional down-regulation. Interestingly, during gene inactivation, the dissociation of 7SK RNA from the locus coincided with the loss of CDK9 suggesting that this transient association of 7SK RNA is likely crucial for the efficient displacement of P-TEFb constituents from the gene locus.
Figure 7.
7SK transiently accumulates at the reporter gene locus during cessation of transcription. Immunolocalization of CDK9 (green; Ab) and 7SK RNA FISH (red; Ac) in representative U2OS 2-6-3 tet-OFF cells demonstrates that at the marked locus (LacI-CFP, blue; Aa) CDK9, but not 7SK RNA, is present during active transcription (Aa–Ad). At 2 min 30s post Dox addition to turn the locus “OFF” CDK9 (Bb) is still detectable at the locus (Ba) and a slight enrichment in 7SK RNA (Bc) that colocalizes with CDK9 (Bd) is now apparent. Five minutes post-Dox perfusion 7SK RNA (Cc) is now highly enriched at the locus (Ca) and colocalizes strongly with CDK9 (Cd). At 7 min 30 s post-Dox perfusion, the marked chromatin locus (Da) shows decrease in both CDK9 (Db) and 7SK RNA (Dc) signals, with only partial colocalization (Dd). By 10 min post-Dox perfusion neither CDK9 (Eb) nor 7SK RNA (Ec) is present at the marked locus (Ea). Scale bar represents 5 μm.
Figure 8.
7SK RNA depletion affects the displacement of Cdk9 from the reporter gene locus during transcriptional silencing. Immunolocalization of CDK9 (Ab; Aa–Ja) in U2OS 2-6-3 tet-OFF cells demonstrates that in the majority of control oligo treated cells (A–E), CDK9 is displaced within 10–15 min post DOX addition (Ca–c). However, in cells that are depleted of 7SK RNA (F–J) CDK9 continues to associate with the gene locus for a longer duration of time post-DOX addition (H–I). DNA is counterstained with DAPI (Ac–Jc). The bar represents 5 μm. (K) Histogram showing reduced displacement of CDK9 from the gene locus in cells that are transfected with ASO against 7SK RNA (n = 50). Scale bar represents 5 μm.
To understand the role of 7SK RNA in the displacement of Cdk9 from the locus during transcriptional inactivation, we examined the displacement kinetics of Cdk9 from the locus in control and 7SK-depleted cells during transcriptional inactivation. In a large fraction of control oligo-treated cells (>50%), Cdk9 was displaced from the locus 10 min post-DOX addition (Figures 8, C and K). However, the majority of 7SK-depleted cells, Cdk9 continued to associate with the locus even at 20 min post DOX addition (Figures 8, I and K). These results indicate that 7SK plays a critical role in the efficient release or displacement of CDK9 from the gene locus during transcriptional inactivation.
DISCUSSION
7SK is a nuclear-retained ncRNA that executes a housekeeping function in the cell by negatively regulating RNA pol II–mediated transcriptional elongation (Nguyen et al., 2001; Yang et al., 2001; Peterlin and Price, 2006). 7SK RNA is an abundant (∼2 × 105 copies per cell), 331-nucleotide-long RNA pol III–transcribed RNA that exhibits high sequence conservation among vertebrates (Zieve and Penman, 1976; Kruger and Benecke, 1987; Murphy et al., 1987; Wassarman and Steitz, 1991). Biochemical and cell biological studies have demonstrated that 7SK is a bonafide nuclear RNA (Zieve and Penman, 1976; Gurney and Eliceiri, 1980; Matera and Ward, 1993). However, the precise localization of 7SK RNA in the nucleus had not been clearly elucidated. A previous study reported “a slightly speckled nucleoplasmic localization” of 7SK RNA in human cells (Egloff et al., 2006). However, our RNA-FISH immunocytochemical analysis clearly demonstrated the presence of a fraction of 7SK RNA in nuclear speckles. Although speckles contain RNA, they are not sites of active transcription and are located in interchromatin regions with little to no DNA (reviewed in Lamond and Spector, 2003).
Previous studies have shown the presence of P-TEFb protein components (CDK9, CyclinT1 and HEXIM1) in nuclear speckles (Herrmann and Mancini, 2001; Pendergrast et al., 2002; Haaland et al., 2005). In HeLa cells, ∼50% of the P-TEFb is thought to be sequestered into the large inactive complex that also contains 7SK RNA, whereas the active P-TEFb complex is localized at sites of gene transcription (Yang et al., 2001; Michels et al., 2003; Yik et al., 2003; Byers et al., 2005; Yik et al., 2005; Michels and Bensaude, 2008). Because speckles do not correspond to active sites of transcription, the P-TEFb components present in speckles may provide a mechanism by which 7SK RNA can sequester the P-TEFb complex away from transcription sites. Our finding of an increased level of 7SK in nuclear speckles upon transcriptional inhibition and P-TEFb components (Herrmann and Mancini, 2001) further corroborates this possibility. In contrast, biochemical studies have revealed that in response to transcription inhibition, 7SK RNA and HEXIM proteins disassociate from the P-TEFb complex (Nguyen et al., 2001; Yang et al., 2001; Michels et al., 2003; Yik et al., 2003) and 7SK RNA shows increased association with different sets of proteins including hnRNPs (Barrandon et al., 2007; Hogg and Collins, 2007; Van Herreweghe et al., 2007). However, this may be due to a lack of nuclear speckles in the in vitro assay system. Our data suggest that 7SK RNA sequesters components of P-TEFb in nuclear speckles, and when required these factors are relocalized to the transcription sites for gene activation. This suggests yet another important function for nuclear speckles in regulating gene expression by modulating the levels of active P-TEFb complex in the nucleoplasm.
The changes in the distribution of nuclear speckle components in cells depleted of 7SK RNA indicates another potential role for 7SK RNA in organizing nuclear speckle constituents. RNA pol II transcription inhibition as well as pre-mRNA splicing inhibition is known to affect speckle morphology (reviewed in Lamond and Spector, 2003). The reporter cell line studies clearly indicate that 7SK knock-down does not inhibit transcription and constitutive splicing of pre-mRNAs at the reporter gene locus (Figure 4) and the pattern of distribution of splicing factors observed after 7SK RNA depletion is quite different from what is observed after inhibition of transcription or splicing (Spector et al., 1983; O'Keefe et al., 1994). It is also unlikely that 7SK RNA has a structural role in maintaining the integrity of nuclear speckles. It is known that hyper-phosphorylation of SR-splicing factors (examples include SF2/ASF and SC35) by SR protein kinases disrupt the nuclear speckles and such studies failed to observe any organized underlying nuclear skeleton stabilizing nuclear speckles supporting the conclusion that RNA(s) is not likely providing the structural framework for speckle organization (Sacco-Bubulya and Spector, 2002). We speculate that depletion of 7SK RNA in cells increases total RNA pol II transcription, which in turn affects the intracellular distribution of pre-mRNA processing factors by recruiting them from nuclear speckles to transcription sites. Previous studies have also indicated increased CDK9 activity and hyperactivation of reporter genes in cells that were depleted of 7SK RNA (Yang et al., 2001; Haaland et al., 2005). Disassociation of 7SK RNA from the P-TEFb complex by depletion of 7SK RNA leading to increased mRNA and protein production has also been shown in the case of cardiac muscle-cell hypertrophy (Sano et al., 2002). On the other hand, any alteration in nuclear speckle organization affects transcription and pre-mRNA processing from a reporter locus (Sacco-Bubulya and Spector, 2002). The changes in the level of RNA pol II–mediated transcription as well as the disorganization of speckle components, observed in the absence of 7SK RNA is likely interconnected. Recent studies have provided evidence of bidirectional coupling between transcriptional elongation and alternative splicing regulation (Lin et al., 2008; Barboric et al., 2009). At the cellular as well as organismal level, degradation of 7SK RNAs has been shown to result in changes in alternative splicing of pre-mRNAs (Barboric et al., 2009). It is speculated that upon 7SK-RNP degradation, the free pool of P-TEFb that is released from the inactive 7SK-RNP may promote alternative splicing via phosphorylating the RNA Pol II CTD, which in turn could facilitate enhanced binding of SR-family splicing factors to pre-mRNAs (Lin and Fu, 2007; Barboric et al., 2009; Long and Caceres, 2009; Zhong et al., 2009). A recent report has indicated a role for the SC35 splicing factor in transcription elongation, P-TEFb recruitment and RNA pol II CTD ser-2 phosphorylation in a gene-specific manner, further supporting the notion that splicing factors and transcription elongation components interact with each other to regulate gene expression (Lin et al., 2008).
7SK RNA has been shown to form an RNA-protein complex consisting of HEXIM1 and 2, LARP7 and BCDIN3 and P-TEFb (CDK9/Cyclin) to form an inactive P-TEFb complex (Michels et al., 2004; Byers et al., 2005; Peterlin and Price, 2006; He et al., 2008; Krueger et al., 2008; Markert et al., 2008; Michels and Bensaude, 2008; Barboric et al., 2009; Diribarne and Bensaude, 2009) The active P-TEFb is composed of CDK9 and one of the four c-types of Cyclins (Cyc-T1/T2a(b)/K) (Zhou and Yik, 2006). LARP7 and BCDIN3 are integral components of the 7SK-RNP complex and are essential for the stability of 7SK snRNP complex (He et al., 2008; Krueger et al., 2008; Barboric et al., 2009). The association of HEXIM1 and 2 with the P-TEFb complex through 7SK RNA inactivates the kinase activity of CDK9, thereby inhibiting the transcriptional elongation carried out by RNA pol II (Peterlin and Price, 2006; Barrandon et al., 2007; Michels and Bensaude, 2008). Two structurally different protein-binding elements situated in the 5′ and 3′ terminal hairpins of 7SK RNA facilitate complex formation of the inactive P-TEFb complex (Egloff et al., 2006). HEXIM proteins can inhibit the kinase activity of P-TEFb only in the presence of 7SK RNA, indicating that 7SK RNA may act as a molecular scaffold for the formation of the inactive P-TEFb complex (Yik et al., 2003; Michels et al., 2004; Yik et al., 2004; Barboric et al., 2005; Blazek et al., 2005; Byers et al., 2005; Yik et al., 2005). However, information regarding the spatial context in the nucleus where 7SK RNA interacts with HEXIM proteins and the CDK9/CyclinT1 complex to form the inactive complex has been lacking. Our data show an increased transient accumulation of 7SK RNA in the vicinity of a gene locus during its down-regulation, and this coincides with the loss of CDK9 from the locus. Transient overexpression of 7SK has been shown to inhibit transcription from a reporter plasmid indicating the relative levels of 7SK RNA in cells are crucial for proper gene expression (Egloff et al., 2006), and our data have shown that knock-down of 7SK results in increased transcription from a stably integrated reporter locus. Increased local concentration of 7SK RNA observed in the area of the gene locus during the cessation of transcription may set up a concentration gradient to facilitate the efficient transfer of CDK9 from the gene to 7SK RNA thereby down regulating gene expression. Changes in the displacement of Cdk9 from the gene locus of cells that were depleted of 7SK RNA further supports the notion that 7SK RNA plays a critical role in the efficient removal of P-TEFb components from transcription sites. It remains to be demonstrated whether HEXIM proteins and other components of the 7SK snRNP complex are already bound to the pool of 7SK RNA that is enriched at the gene locus during gene inactivation. Previous studies have shown that both HEXIM1 and P-TEFb interact at the 5′ and 3′ hairpins of 7SK RNA, respectively and that 7SK RNA can interact with HEXIM1 without the recruitment of P-TEFb (Egloff et al., 2006). Any alteration in 7SK RNA, which abolishes the binding of HEXIM1, also inhibits the recruitment of P-TEFb to 7SK RNA (Egloff et al., 2006). Similarly, in vitro reconstitution studies have revealed that the binding of HEXIM1-7SK RNA is required for the efficient recruitment of P-TEFb to the 7SK RNA containing complex (Michels et al., 2004). These results suggest that 7SK RNA first interacts with HEXIM proteins along with LARP7 and BCDIN3 and as a complex interacts with active P-TEFb. From our data, it is tempting to speculate that 7SK RNA, most probably as a complex with HEXIM proteins is recruited to a gene during its down-regulation, facilitating the efficient removal of P-TEFb components and their recruitment to nuclear speckles for storage/modification. Thus, nuclear speckles in turn may be involved in regulating gene expression by modulating the levels of the active P-TEFb complex in cells. Together, our findings have provided new insight into the mechanism of action of 7SK RNA and nuclear speckles in regulating gene expression.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Drs. J. Caceres (U1-70K plasmid), M. Carmo-Fonseca (GFP-U2AF65), S. Murphy (7SK plasmid), and R. Reddy (7SK plasmid) for providing reagents and Dr. C. Frank Bennett (ISIS Pharmaceuticals, Carlsbad, CA) for antisense DNA oligonucleotides against 7SK RNA. We thank Drs. R. Ileng Kumaran and Supriya G. Prasanth for helpful suggestions and technical assistance. This work was supported by grants from NIGMS (42694-21) and NIH/NCI 5PO1CA013106-38 (to D.L.S.) and UIUC start-up funds (to K.V.P.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0105) on September 29, 2010.
REFERENCES
- Amaral P. P., Dinger M. E., Mercer T. R., Mattick J. S. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Barboric M., Kohoutek J., Price J. P., Blazek D., Price D. H., Peterlin B. M. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M., Lenasi T., Chen H., Johansen E. B., Guo S., Peterlin B. M. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C., Bonnet F., Nguyen V. T., Labas V., Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C., Spiluttini B., Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
- Blazek D., Barboric M., Kohoutek J., Oven I., Peterlin B. M. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005;33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J. Transcription: surprising role for an elusive small nuclear RNA. Curr. Biol. 2002;12:R147–R149. doi: 10.1016/s0960-9822(02)00711-x. [DOI] [PubMed] [Google Scholar]
- Bregman D. B., Du L., van der Zee S., Warren S. L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubulya P. A., Prasanth K. V., Deerinck T. J., Gerlich D., Beaudouin J., Ellisman M. H., Ellenberg J., Spector D. L. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers S. A., Price J. P., Cooper J. J., Li Q., Price D. H. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Carter K. C., Taneja K. L., Lawrence J. B. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Carmichael G. G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., Hutchinson J. N., Sara S. A., Ensminger A. W., Fox A. H., Chess A., Lawrence J. B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F. F. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Costa F. F. Non-coding RNAs: lost in translation? Gene. 2007;386:1–10. doi: 10.1016/j.gene.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Dinger M. E., Amaral P. P., Mercer T. R., Pang K. C., Bruce S. J., Gardiner B. B., Askarian-Amiri M. E., Ru K., Solda G., Simons C., Sunkin S. M., Crowe M. L., Grimmond S. M., Perkins A. C., Mattick J. S. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger M. E., Pang K. C., Mercer T. R., Crowe M. L., Grimmond S. M., Mattick J. S. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diribarne G., Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- Egloff S., Van Herreweghe E., Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell Biol. 2006;26:630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C., Pheasant M., Mattick J. S. The amazing complexity of the human transcriptome. Eur. J. Hum. Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gurney T., Jr, Eliceiri G. L. Intracellular distribution of low molecular weight RNA species in HeLa cells. J. Cell Biol. 1980;87:398–403. doi: 10.1083/jcb.87.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaland R. E., Herrmann C. H., Rice A. P. siRNA depletion of 7SK snRNA induces apoptosis but does not affect expression of the HIV-1 LTR or P-TEFb-dependent cellular genes. J. Cell Physiol. 2005;205:463–470. doi: 10.1002/jcp.20528. [DOI] [PubMed] [Google Scholar]
- Habets W., Hoet M., Bringmann P., Luhrmann R., van Venrooij W. Autoantibodies to ribonucleoprotein particles containing U2 small nuclear RNA. EMBO J. 1985;4:1545–1550. doi: 10.1002/j.1460-2075.1985.tb03815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. L., Smith K. P., Byron M., Lawrence J. B. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N., Jahchan N. S., Hong E., Li Q., Bayfield M. A., Maraia R. J., Luo K., Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C. H., Mancini M. A. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- Hogg J. R., Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Deerinck T. J., Ellisman M. H., Spector D. L. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. N., Ensminger A. W., Clemson C. M., Lynch C. R., Lawrence J. B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S. M., Tsukamoto T., Salghetti S. E., Tansey W. P., Sachidanandam R., Prasanth K. V., Ried T., Shav-Tal Y., Bertrand E., Singer R. H., Spector D. L. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Willingham A. T., Gingeras T. R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Krueger B. J., et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W., Benecke B. J. Structural and functional analysis of a human 7 S K RNA gene. J. Mol. Biol. 1987;195:31–41. doi: 10.1016/0022-2836(87)90325-1. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Spector D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lin R., Maeda S., Liu C., Karin M., Edgington T. S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2006 doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Lin S., Coutinho-Mansfield G., Wang D., Pandit S., Fu X. D. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Fu X. D. SR proteins and related factors in alternative splicing. Adv. Exper. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Long J. C., Caceres J. F. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Markert A., Grimm M., Martinez J., Wiesner J., Meyerhans A., Meyuhas O., Sickmann A., Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Izaguire-Sierra M., Praveen K., Rajendra T. K. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Ward D. C. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. RNA regulation: a new genetics? Nat. Rev. Genet. 2004a;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mattick J. S. The functional genomics of noncoding RNA. Science. 2005;309:1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- Mattick J. S. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S., Makunin I. V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Sunkin S. M., Mehler M. F., Mattick J. S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels A. A., Bensaude O. RNA-driven cyclin-dependent kinase regulation: when CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners. Biotech. J. 2008;3:1022–1032. doi: 10.1002/biot.200800104. [DOI] [PubMed] [Google Scholar]
- Michels A. A., Fraldi A., Li Q., Adamson T. E., Bonnet F., Nguyen V. T., Sedore S. C., Price J. P., Price D. H., Lania L., Bensaude O. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels A. A., Nguyen V. T., Fraldi A., Labas V., Edwards M., Bonnet F., Lania L., Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem. and cell biology. 2008;129:5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., Spector D. L. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- Muratani M., Gerlich D., Janicki S. M., Gebhard M., Eils R., Spector D. L. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 2002;4:106–110. doi: 10.1038/ncb740. [DOI] [PubMed] [Google Scholar]
- Murphy S., Di Liegro C., Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Nguyen V. T., Kiss T., Michels A. A., Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- O'Keefe R. T., Mayeda A., Sadowski C. L., Krainer A. R., Spector D. L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K. C., Dinger M. E., Mercer T. R., Malquori L., Grimmond S. M., Chen W., Mattick J. S. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Pang K. C., Stephen S., Dinger M. E., Engstrom P. G., Lenhard B., Mattick J. S. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrast P. S., Wang C., Hernandez N., Huang S. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol. Biol. Cell. 2002;13:915–929. doi: 10.1091/mbc.01-08-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B. M., Price D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pheasant M., Mattick J. S. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- Platani M., Goldberg I., Lamond A. I., Swedlow J. R. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Prasanth S. G., Xuan Z., Hearn S., Freier S. M., Bennett C. F., Zhang M. Q., Spector D. L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Sacco-Bubulya P. A., Prasanth S. G., Spector D. L. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. V., Spector D. L. Encyclopedia of Life Sciences. Chichester: John Wiley & Sons, Ltd:; 2005. The cell nucleus. http://www.els.net/ [doi: 10.1038/npg.els.0001337] [Google Scholar]
- Prasanth K. V., Spector D. L. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Sacco-Bubulya P., Spector D. L. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 2002;156:425–436. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M., Abdellatif M., Oh H., Xie M., Bagella L., Giordano A., Michael L. H., DeMayo F. J., Schneider M. D. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- Sasaki Y. T., Ideue T., Sano M., Mituyama T., Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S. M., Fusco D., Janicki S. M., Spector D. L., Singer R. H. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L. SnapShot: cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Spector D. L., Schrier W. H., Busch H. Immunoelectron microscopic localization of snRNPs. Biol. Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M. E., Wilusz J. E., Amaral P. P., Mattick J. S., Spector D. L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M., Barciszewska M. Z., Erdmann V. A., Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim. Biophys. Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Taft R. J., Pang K. C., Mercer T. R., Dinger M., Mattick J. S. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Van Herreweghe E., Egloff S., Goiffon I., Jady B. E., Froment C., Monsarrat B., Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham A. T., Gingeras T. R. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Wilusz J. E., Freier S. M., Spector D. L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E., Sunwoo H., Spector D. L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhu Q., Luo K., Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yik J. H., Chen R., Nishimura R., Jennings J. L., Link A. J., Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Yik J. H., Chen R., Pezda A. C., Samford C. S., Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik J. H., Chen R., Pezda A. C., Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- Zhong X. Y., Wang P., Han J., Rosenfeld M. G., Fu X. D. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Yik J. H. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976;8:19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.