Here we analyze the nuclear trafficking dynamics of PML and its SUMO-dependent ubiquitin ligase, RNF4, in response to arsenic. We show that arsenic induces rapid reorganization of the cell nucleus by SUMO modification of nuclear body associated PML and uptake of the ubiquitin E3 ligase RNF4 leading to the ubiquitin-mediated degradation of PML.
Abstract
In acute promyelocytic leukemia (APL), the promyelocytic leukemia (PML) protein is fused to the retinoic acid receptor alpha (RAR). Arsenic is an effective treatment for this disease as it induces SUMO-dependent ubiquitin-mediated proteasomal degradation of the PML-RAR fusion protein. Here we analyze the nuclear trafficking dynamics of PML and its SUMO-dependent ubiquitin E3 ligase, RNF4 in response to arsenic. After administration of arsenic, PML immediately transits into nuclear bodies where it undergoes SUMO modification. This initial recruitment of PML into nuclear bodies is not dependent on RNF4, but RNF4 quickly follows PML into the nuclear bodies where it is responsible for ubiquitylation of SUMO-modified PML and its degradation by the proteasome. While arsenic restricts the mobility of PML, FRAP analysis indicates that RNF4 continues to rapidly shuttle into PML nuclear bodies in a SUMO-dependent manner. Under these conditions FRET studies indicate that RNF4 interacts with SUMO in PML bodies but not directly with PML. These studies indicate that arsenic induces the rapid reorganization of the cell nucleus by SUMO modification of nuclear body-associated PML and uptake of the ubiquitin E3 ligase RNF4 leading to the ubiquitin-mediated degradation of PML.
INTRODUCTION
The promyelocytic leukemia protein (PML) was originally identified in acute promyelocytic leukemia (APL), where it is fused to the retinoic acid receptor α (RARα) as a result of the t(15;17) chromosomal translocation (de Thé et al., 1990; de The et al., 1991; Kakizuka et al., 1991). Fusion of PML to RARα causes leukemia as it blocks differentiation of hematopoietic progenitor cells. In cells containing PML-RARα the fusion protein is dispersed into a large number of small nuclear bodies but in normal cells PML forms discrete nuclear structures named PML nuclear bodies (PML NBs), which harbor numerous other transiently or permanently localized proteins including Daxx, Sp100, SUMO-1, and p53 (Weis et al., 1994; Dyck et al., 1994; Bernardi and Pandolfi, 2007). Arsenic is successfully used to treat APL (Wang and Chen, 2008), but it is only recently that the mechanism of action of arsenic has been clarified. Arsenic leads to SUMO-dependent ubiquitin-mediated proteolysis of the PML-RAR fusion protein (Tatham et al., 2008; Lallemand-Breitenbach et al., 2008). This is mediated by Ring finger protein 4 (RNF4), also known as Small Nuclear RING Finger (SNURF), a member of the family of SUMO Targeted Ubiquitin Ligases (STUbLs) (Perry et al., 2008; Geoffroy and Hay, 2009). Originally identified as a coactivator of androgen receptor (Moilanen et al., 1998), human RNF4 encodes a protein of 194 aa that contains a RING domain (Hakli et al., 2004) and multiple SUMO interaction motifs (SIM) (Tatham et al., 2008). RNF4 interacts strongly with polySUMO chains due to the presence of multiple SIM in its N-terminal region (Tatham et al., 2008) and can be found associated with SUMO in PML bodies (Hakli et al., 2005). Indeed, RNF4 has the ability to ubiquitylate polySUMO chains conjugated to PML and target the protein for ubiquitin-mediated proteolysis (Tatham et al., 2008; Lallemand-Breitenbach et al., 2008). In cells depleted of RNF4, PML protein accumulates and arsenic treatment fails to induce the degradation of PML resulting in the accumulation of SUMO-modified PML in nuclear bodies (Tatham et al., 2008).
PML is a multifunctional protein that plays an essential role in regulating diverse cellular processes including tumor suppression, apoptosis, cell cycle regulation, senescence, transcriptional regulation, DNA damage, and genome stability (Bernardi and Pandolfi, 2007). In human cells, the primary transcript of the PML gene generates a number of alternatively spliced mRNAs, each of which encodes a distinct protein (Jensen et al., 2001). Seven PML isoforms have been classified based on C-terminal variation but many of the detailed studies have been performed with PML isoform III. PML, also known as TRIM19, contains the characteristic RBCC motif, which consists of a RING domain adjacent to two zinc-coordinating B-boxes and a coiled-coil domain. These motifs are required for the localization of PML within the PML nuclear body (NB) and the recruitment of other PML NBs-associated proteins (Bernardi and Pandolfi, 2007). Formation of PML NBs depends also on the posttranslational modification of PML with Small ubiquitin modifier (SUMO), a member of ubiquitin family. Indeed, PML is sumoylated on three lysine residues and PML mutants that cannot be sumoylated are unable to form proper PML NBs (Ishov et al., 1999). A requirement for SUMO modification in the genesis of PML NBs is also evident from genetic studies in cells derived from mouse blastocysts that are unable to carry out SUMO modification as a result of deletion of the gene encoding UBC9, the single E2 conjugating enzyme responsible for SUMO modification. These cells have dramatic defects in PML body organization, demonstrating that SUMO modification is required for the normal function of PML bodies (Nacerddine et al., 2005). This is supported by the observation that loss of SUMO-1 in mice affects PML expression levels and PML NB formation (Evdokimov et al., 2008). In addition, PML contains a phospho-regulated SUMO-interacting motif (SIM) in its C terminus that mediates interaction with other SUMOylated proteins and promotes their recruitment in PML NBs (Shen et al., 2006; Stehmeier and Muller, 2009). SUMO modification of PML protein is induced in many situations including stress responses, viral infection, and expression of proteins such as HDAC7 (Saitoh and Hinchey, 2000; Pampin et al., 2006; Gao et al., 2008). Increased SUMO modification of PML is evident after arsenic treatment and is accompanied by transit of PML from the nucleoplasm to PML NBs (Zhu et al., 1997; Lallemand-Breitenbach et al., 2001; Tatham et al., 2008; Lallemand-Breitenbach et al., 2008).
To analyze the dynamic interplay between PML, SUMO, and RNF4 in response to arsenic we have created fluorescent-tagged versions of these proteins and used them for real-time fluorescence microscopy to follow changes in trafficking, interactions, and ultimately degradation of PML. Our data demonstrate that after exposure of cells to arsenic PML undergoes rapid SUMO modification and is recruited into PML NBs independently of RNF4. This is followed by the accumulation of RNF4 in PML NBs, but Fluorescence Resonance Eneregy Transfer (FRET) experiments indicate that RNF4 was recruited by the SUMO modification on PML rather than directly by PML. Subsequent RNF4-mediated ubiquitylation leads to the proteasomal degradation of PML. Thus, the arsenic-induced SUMO modification of PML initiates a rapid reorganization of the cell nucleus leading to recruitment of PML and RNF4 into PML NBs and ultimately to the degradation of PML and dissolution of PML NBs.
MATERIALS AND METHODS
Plasmids and DNA Constructs
The enhanced yellow fluorescent protein (YFP) or cyan-fluorescent protein (CFP) were used to construct fluorescent-tagged versions of PML, RNF4, or SUMO-2 (See Table S1 in Supplemental Material). Human PMLIII (accession number AAB19601) or rat RNF4 (accession number NM_019182) were amplified by PCR from pcDNA3.1 to generate SalI and BamHI sites flanking the sequence. PCR products were then digested with the appropriate enzymes and inserted in place of H2B into either pBOS-H2B-YFP or pBOS-H2B-CFP vectors (Leung et al., 2004). These vectors contain the mammalian EF1α promoter to drive expression of the fluorescent-tagged protein and harbor the blasticidin resistance gene as a selection marker. Fluorescent-tagged RNF4 mutants (RNF4-CS1 and RNF4 mtSIM1, 2, 3, 4) were obtained according to the strategy described above. Human SUMO-2 full-length (accession number CAG46970) was amplified by PCR to generate EcoRI and BamHI sites flanking the sequence. PCR product was inserted in the C-terminal region of EGFP into the pEGFP-C1 plasmid (Clontech, Palo Alto, CA).
Antibodies
Antigen affinity purified sheep SUMO-1, sheep SUMO-2, chicken RNF4, and chicken PML antibodies were prepared in house. Mouse anti–β-actin (Sigma, St. Louis, MO) and mouse anti-GFP (clones 7.1 and 13.1, Roche, Indianapolis, IN) were obtained from commercial sources. Rabbit anti-RNF4, mouse anti-PML 5E10, rabbit anti-PML, mouse anti-p53 (DO-1), and mouse anti-nucleolin antibodies were a kind gifts from Jorma Palvimo (University of Kuopio, Finland), Roel van Driel (University of Amsterdam, The Netherlands), Anne Dejean (Institute Pasteur, Paris, France), David Lane (University of Dundee, Scotland), and Ara Hovanessian (University of Paris, France), respectively.
Cell Culture and Immunofluorescence Analysis
HeLa cells were cultured in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (GIBCO BRL, Grand Island, NY) in a 10% CO2 atmosphere at 37°C. To establish PML-YFP stable cell line, HeLa cells were grown in 10-cm Petri dishes to 50% of confluence and were transfected the next day with the fluorescent-tagged PML-YFP plasmid using Fugene 6 (Roche) according to the manufacturer's directions. Two days posttransfection, 1 μg/ml blasticidin was added to select cells stably expressing the fusion protein. Fluorescent cells were then sorted using a FACS Vantage SE cell sorter with DIVA option (BD Biosciences, San Jose, CA) with a helium-neon laser at 488 nm. The same protocol of transfection was used to establish the HeLa YFP-SUMO-2 cell line, except that 100 μg/ml neomycin was used to select resistant clones.
For immunofluorescence experiments, cells were seeded onto coverslips and were fixed with 4% paraformaldehyde for 10 min, at room temperature. Cells were then permeabilized with 0.2% Triton for 10 min and were blocked in PBS containing 5% BSA and 0.1% Tween for 30 min. Primary and secondary antibodies diluted in PBS with 1% BSA and 0.1% Tween were successively added on the cells for 1h. Cells were then stained 0.1 μg/ml DAPI and were mounted in Vectashield mounting medium.
Cell Cycle Analysis
Hela cells or HeLa PML-YFP cell line were trypsinized and washed with PBS before fixation with cold 70% ethanol for 2 h at 4°C. Cells were centrifuged at 1000 rpm for 5 min and washed twice in PBS. Cells were incubated in PBS containing 50 mg/ml propidium iodide (PI), 50 μg/ml RNAseA and 0.1% (vol/vol) Triton X-100 for 30 min. The intensity of DNA staining (FLH-2) was determined by flow cytometry on a FACS Calibur (Becton Dickson, San Diego, CA) to determine the percentage of cells in G1, S, and G2/M phases.
Protein Extraction and Western Blot Analysis
Cells were lysed in 2× Laemmli lysis buffer (150 mM Tris-HCL (pH 6.8), 5% (wt/vol) SDS, 25% (vol/vol) glycerol, and 0.01% (wt/vol) bromophenol blue) supplemented with 10 mM iodoacetamide. Lysates were clarified by sonication for 10 s on low power (Branson digital sonifier 450) and protein concentration was determined by using the BCA assay kit (Thermo Scientific). β-mercaptoethanol was added to a final concentration of 0.75 M into each sample. Total cellular protein was subjected to SDS-polyacrylamide gels electrophoresis (SDS-PAGE) and transferred onto Hybond membrane for Western blot analysis.
Immunoprecipitation
For immunoprecipitation of PML-YFP protein, nuclear extracts were prepared from HeLa cells stably expressing PML-YFP grown in 10-cm Petri dishes. After scraping into the medium, cells were washed twice in PBS containing 100 mM iodoacetamide and were collected by centrifugation at 1500 rpm for 5 min. Cells were resuspended in ice cold buffer A (10 mM HEPES pH 7.9, 1.5 mM MgCl2,10 mM KCl, complete protease inhibitor cocktail tablets (Roche), 200 mM iodoacetamide) and were disrupted by using a syringe to pass them through a narrow (26 gauge) needle. Nuclei were separated from the cytoplasmic fraction by centrifugation at 3200 rpm for 5 min at 4°C, washed twice with buffer A, and resuspended in RIPA buffer (50 mM Tris pH 6.8, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 10 mM iodoacetamide). Nuclear extract was sonicated and clarified by centrifugation at 13,000 rpm for 1 min. The nuclear supernatant was precleared with Sepharose beads before incubation with GFP-trap beads (Chromotek) for 2 h at 4°C. Beads were collected by centrifugation at 3200 rpm and washed twice with RIPA buffer. Proteins were eluted by adding 2× SDS lysis buffer and analyzed by Western blotting.
Treatment of Cells with Cycloheximide
To inhibit protein synthesis, HeLa cells were grown in six-well plates for 24 h before addition of fresh medium containing 50 μg/ml cycloheximide. Cells were harvested at different times after cycloheximide treatment and subjected to SDS-PAGE and Western blot analysis to detect RNF4, p53 and actin proteins. To carry out real-time imaging, 50 μg/ml cycloheximide and 1 μM arsenic in Leibovitz's L-15 medium (Invitrogen) supplemented with 10% fetal bovine serum was added to PML-YFP cells seeded in Labtek chamber slides.
Quantitative Reverse Transcription-PCR
To quantify endogenous Rnf4 transcripts during arsenic treatment, total RNA was extracted from HeLa cells stably expressing PML-YFP using an SV total RNA isolation kit (Promega). Total cDNAs were amplified with a SuperScript II reverse transcriptase kit (Invitrogen), and real-time quantitative PCR was performed using a TaqMan system for 40 cycles of 15 s at 95°C and 1 min at 60°C with an ABI 7700 instrument (Applied Biosystems). TaqMan probes used for detection of human RNF4 and the internal control actin were purchased from Applied Biosystems (Hs00231302_m1 RNF4, Hs99999903_ml ACTB).
Live-Cell Imaging in Real Time and Measurement of Fluorescence Intensities
For live-cell imaging experiments, HeLa cells were seeded onto Lab-Tek II four-chamber slides (Nalgen Nunc) and transfected the next day with the appropriate fluorescent-tagged proteins using Fugene 6 (Roche). HeLa cells stably expressing fluorescently tagged fusion proteins were seeded in the same way. When required, double-stranded small interfering RNA (siRNA) for RNF4 or control scrambled sequence (Dharmacon, Lafayette, CO) were introduced at 5 nM final concentration using Lipofectamine RNAiMAX (Invitrogen). Two days later, cells were changed into Leibovitz's L-15 medium (Invitrogen) supplemented with 10% fetal bovine serum. Images were collected using the DeltaVision microscope system (Applied Precision) with an Olympus IX70 microscope and a cooled CCD camera (Coolsnap HQ, Photometrics). Temperature was maintained at 37°C using an environmental chamber (Solent Scientific, Segensworth, UK). Stacks of 20 sections with z-step set to 400 nm were collected in different channels with 35–50 ms exposure with a Plan Apo 60 × 1.40 numerical aperture objective lens (Olympus). A single z-section was captured by differential interference contrast microscopy (DIC) to visualize the position of PML NB within the cells during arsenic treatment. Collected images were deconvolved with SoftWoRx (Applied Precision), and z-stack projections were made before merging with the optical section. Because PML structures were mobile, a region of interest (ROI) containing one PML body has been defined at each time point to measure fluorescence intensities using OMERO Beta-4.3 software. The intensities in a random region outside the nucleoplasm divided by the region of interest was taken as the value to which the intensities within the PML nuclear body were compared. Data were collected from around 20 cells.
Fluorescence Recovery after Photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were carried out on the same slides and microscope described above. Entire PML NBs were bleached with a 488-nm argon laser at 50% laser intensity to reduce the YFP signal by 90%. A nucleoplasmic region outside PML NB was also bleached as a control. Images were taken before the bleach pulse and image acquisition after bleaching was adjusted according to the recovery of the fluorescent proteins by using 0.05% laser transmission intensity to minimize scan bleaching. As PML recovers slowly after photobleaching, 300 images were collected at 1-min intervals over a 10 min period whereas 32 images were captured for RNF4 within 11 s. Projection of z-sections were made only for PML. The fluorescence intensity was measured in bleached PML NBs and in control areas outside the nuclear dots and quantified with OMERO Beta-4.3 software. The fluorescence intensity in the bleached PML was normalized to the nonbleached signal after substraction of the background signal. Data were collected from ten PML NBs.
Fluorescence Resonance Energy Transfer Acceptor Photobleaching
Fluorescence resonance energy transfer (FRET) was measured by acceptor photobleaching method as previously described (Ellis et al., 2008). The FRET experiment was conducted on a DeltaVision microscope system (Applied Precision) fitted with a quantifiable laser module, including a 20-mW 532-nm CW laser, suitable for photobleaching YFP without cobleaching CFP. Fluorescence of the donor (CFP) was observed by using argon laser 436/10 nm excitation and 480/40 band-pass emission filters, whereas fluorescence of acceptor (YFP) was observed using argon laser 532 nm excitation and 580/70 band-pass emission filters. Images were collected with a Plan-Apochromat 100 × 1.35 numerical aperture lens (Olympus) and a cooled CCD camera (CoolSnap HQ; Photometrics).
Photobleaching of YFP fluorescence was achieved by irradiation of the region of interest containing one PML NB with the 532 nm excitation filter at 50% laser intensity. A nucleoplasmic region of the same area was bleached as a negative control, resulting in minimal CFP bleaching (0–2%) which has not been taken into account for the calculation of FRET efficiency. CFP fluorescence was measured before (CFPbefore) and after (CFPafter) the YFP bleaching by collecting a total of 23 images. Pixel intensities of the different regions were analyzed with OMERO software and apparent FRET efficiency was expressed as the percent increase of prebleach CFP fluorescence in the region of interest compared with that observed after YFP photobleaching, according to the equation: FRET efficiency [%] = (CFPafter − CFPbefore) × 100/CFPafter, previously described by Stanek et al. (Stanek and Neugebauer, 2004). P value was calculated with the two-tailed homoscedastic t test comparing the FRET efficiencies with and without As2O3 treatment.
RESULTS
Establishment of YFP-PML HeLa Stable Cell Line
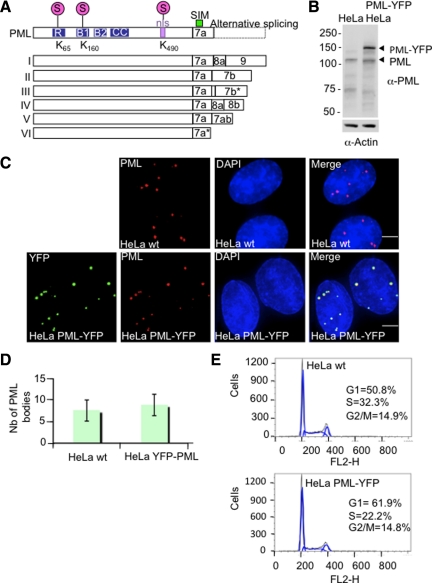
We have shown previously that the RING domain containing protein RNF4 targets SUMO modified PML for ubiquitin-mediated proteolysis after exposure of cells to arsenic (Tatham et al., 2008). To investigate the cellular mechanisms involved in arsenic-induced PML degradation, we generated a HeLa cell line stably expressing a fluorescent version of PML that allows the degradation of PML to be quantified in real-time. Although multiple nuclear isoforms of PML have been reported (Figure 1A), isoform III is the only one which has been extensively studied upon arsenic treatment. We therefore created a fusion of the PML III isoform with yellow fluorescent protein (YFP) and selected HeLa cells stably expressing the fusion protein. Cells expressing levels of PML-YFP comparable to that of endogenous PML were obtained by FACS. Analysis of the PML-YFP cells and the parental HeLa cells by immunofluorescence after staining with a PML antibody and by YFP fluorescence revealed that the PML-YFP HeLa cells displayed similar numbers of PML bodies and that PML-YFP was efficiently incorporated into PML subnuclear bodies (Figure 1, C and D). Western blot analysis of cell extracts with an anti-PML antibody revealed that PML-YFP was expressed at levels similar to that of endogenous PML and was of the predicted molecular weight (Figure 1B). FACS analysis after propidium iodide staining indicated that the PML-YFP and parental HeLa cells had a comparable cell cycle profile (Figure 1E).
Figure 1.
Establishment and characterization of HeLa PML-YFP stable cell line. (A) Schematic showing the nuclear isoforms of PML that share the same N-terminal region but differ in their C termini due to the alternative splicing of exons 7–9. Nuclear PML isoforms I to V harbor three SUMOylation sites (K65, K160, and K490), a SUMO-interacting motif (SIM), a nuclear localization signal (nls), and a RBCC motif containing a RING finger domain (R) adjacent to two zinc coordinating B-boxes and a coiled-coil domain (CC). PML VI differs from the other PML isoforms due the absence of a SIM domain. (B) Whole cell extracts from parental HeLa cells and HeLa PML-YFP were analyzed by SDS-PAGE followed by Western Blot using a rabbit anti-PML antibody. An anti-actin antibody was used as a control of loading. (C) HeLa were stably transfected with a plasmid expressing PML-YFP placed under the control of EF1 promoter. Endogenous PML in parental HeLa cells (wt) was detected with a chicken antibody to PML and PML in cells stably expressing the PML-YFP fusion protein was detected by immunofluorescence using a mouse anti-PML antibody whereas expression of the fluorescent-tagged PML protein was visualized by YFP fluoresence. DNA was stained with DAPI. Bar, 5 μm. (D). Graph showing the number of PML bodies in HeLa wt and HeLa PML-YFP. (E) Cell cycle analysis of HeLa and HeLa PML-YFP cell lines was determined by flow cytometry. The percentage of cells in G1, S and G2/M phase was determined by measuring the intensity of DNA staining with propidium iodide (FL2-H).
Kinetics of Arsenic-Induced RNF4-Mediated PML Degradation
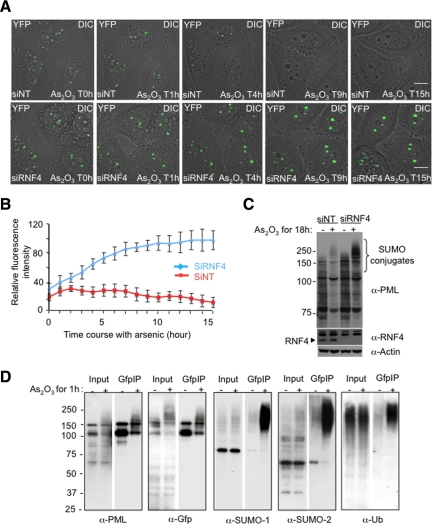
To establish the kinetics of arsenic-induced degradation of PML in the presence or absence of RFN4, PML-YFP HeLa cells were transfected with either a pool of siRNA against human RNF4 or a pool of nontargeting SiRNA as a negative control. Depletion of RNF4 was confirmed by Western blotting using a rabbit anti-RNF4 antibody (Figure 2C). To follow the degradation of PML in real time, PML-YFP HeLa cells were exposed to arsenic (As2O3) and YFP fluorescence quantified at 15-min intervals over an 18-h period by fluorescence microscopy (Figure 2A, full movie is available in Supplemental Figure S1, A and B). The position of PML NBs within the cells during arsenic treatment was monitored by differential interference contrast microscopy (DIC) and fluorescence intensity of multiple individual PML bodies was quantified at each time point (Figure 2B).
Figure 2.
Effect of RNF4 on arsenic-induced PML degradation in real time. (A) Time lapse experiments were performed on HeLa PML-YFP stable cells transfected with a nontarget siRNA (siNT) or a siRNA against RNF4 (siRNF4) and exposed to 1 μM arsenic for 18h. PML-YFP was imaged in real time by fluoresence microscopy over 15 h by collecting a stack of 20 sections with the YFP channel (green) and one image with the differential interference contrast (DIC) every 15 min. The projected z-sections collected in YFP channel were merged to the respective DIC image to monitor the position of PML NB within the cells. Bar, 5 μM. (B) Fluorescence intensity of PML bodies was quantified by defining a region of interest containing one PML body and comparing it a region in the nucleoplasm. Relative fluorescence intensity represents the difference of intensities between these two regions. The graph shows mean values from at least 10 cells. (C) Whole cell extracts from PML-YFP HeLa cells transfected with siRNA to RNF4 or a nontarget siRNA were analyzed by SDS PAGE followed by Western Blotting with a chicken anti-PML antibody to show the accumulation of PML in the absence of RNF4. Depletion of RNF4 was controlled with a rabbit anti-RNF4 antibody. (D) Nuclear extracts from PML-YFP HeLa cells either untreated or exposed to 1 uM arsenic for 1 h were incubated with GFP-trap beads and bound proteins collected. Proteins were eluted from the beads and analyzed by SDS-PAGE followed by Western blotting with antibodies against PML, GFP, SUMO-1, SUMO-2, and ubiquitin to evaluate proteins bound to PML-YFP. The input represents 10% of the nuclear lysate.
As we have previously reported (Tatham et al., 2008) the number and the size of PML bodies were increased in cells depleted of RNF4 (Figure 2A and Supplemental Figure S1D). This indicates that in the PML-YFP HeLa cells the level of endogenous RNF4 is sufficient to regulate the level and localization of both endogenous PML and PML-YFP.
In cells treated with arsenic, the fluorescence intensity of nuclear body associated PML was increased within one hour (Figure 2, A and B). This is consistent with a previous report (Lallemand-Breitenbach et al., 2001) showing that arsenic induces a rapid SUMO-dependent recruitment of nucleoplasmic PML into nuclear bodies. This recruitment was independent of RNF4 and suggests that RNF4 is not involved in the recruitment of PML into nuclear bodies at early stage of the arsenic response. After 4 h of exposure to arsenic, the fluorescence intensity of PML-YFP in cells containing RNF4 declines, until at 15 h the signal is almost undetectable (Figure 2, A and B). Western blot analysis reveals that this is a consequence of arsenic-induced degradation of PML (Figure 2C). However, in RNF4-depleted cells, PML-YFP fluorescence in nuclear bodies increased progressively in response to arsenic leading to the accumulation of PML-YFP in large nuclear bodies (Figure 2, A and B). Finally, we have used HeLa clone 6 cells (Tatham et al., 2008), which are depleted of RNF4, to establish another cell line expressing a cyan fluorescent version of PML isoform III (PML-CFP). When PML-CFP HeLa clone 6 (RNF4 depleted) and PML-YFP HeLa cells are mixed and grown on the same coverslip then exposed to arsenic this allows the simultaneous analysis of PML degradation in the presence and absence of RNF4 under identical conditions. This confirms (Supplemental Figure S1C) that RNF4 induces the degradation of PML-YFP in response to arsenic, while in cells lacking RNF4, arsenic induces the continuous accumulation of PML-CFP in nuclear bodies. To analyze the posttranslational modifications that initiate these processes PML-YFP HeLa cells were exposed to arsenic for 1 h and PML-YFP proteins collected by immunoprecipitation. Analysis of such immunoprecipitates by Western blotting indicated that 1 h after exposure of cells to arsenic, PML-YFP displayed an increased apparent molecular weight as a consequence of extensive modification by SUMO-1, SUMO-2, and ubiquitin (Figure 2D).
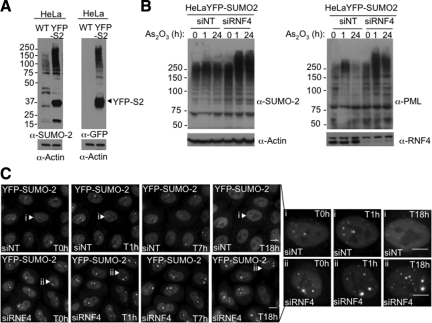
Kinetics of SUMO-2 Redistribution in Response to Arsenic
To better understand the role of SUMO in the nuclear dynamics of PML, the distribution of SUMO-2 during arsenic treatment, in the presence or the absence of RNF4, was followed by real-time imaging (Figure 3). HeLa cells stably expressing YFP-SUMO-2 were transfected with either a pool of siRNA against human RNF4 or a pool of nontargeting siRNA as a negative control (Figure 3, A and B). After 48 h, cells were incubated with arsenic (As2O3) over a 15-h period to follow the localization of SUMO-2 (Figure 3B). In the presence of RNF4, YFP-SUMO-2 is initially present both in the nucleoplasm and in PML bodies. After exposure of cells to arsenic the PML body associated YFP-SUMO-2 is lost and YFP-SUMO-2 redistributes to the nucleoplasm (Figure 3C). In the absence of RNF4, YFP-SUMO-2 is found in large nuclear bodies and after exposure of cells to arsenic YFP-SUMO-2 further accumulates in these large brightly staining bodies and also begins to accumulate adjacent to nucleoli (Figure 3C and Supplemental Figure S2), as shown by immunostaining with an antinucleolin antibody. In some YFP-SUMO-2 expressing cells depletion of RNF4 results in the formation of “peanut” shaped nuclei, the origin of which is at present unclear.
Figure 3.
Subcellular localization of SUMO-2 during arsenic treatment. (A) Characterization of HeLa YFP-SUMO-2 cell line. HeLa cells were stably transfected with a plasmid expressing a YFP-SUMO-2 fusion protein. Whole cells extracts from both parental HeLa and stable YFP-SUMO-2 cells were analyzed by SDS-PAGE followed by Western blot with rabbit anti-SUMO-2 and mouse anti-GFP antibodies. (B) HeLa YFP-SUMO-2 stable cell line was transfected with siRNA to RNF4 or a control nontargeting si RNA (NT) for 48 h before adding 1 μM of arsenic. Whole cell extracts were analyzed by SDS-PAGE followed by Western Blotting with rabbit anti-SUMO-2 and chicken anti-PML antibodies. Depletion of RNF4 was controlled with a rabbit anti-RNF4 antibody. (C) Subcellular localization of SUMO-2 during arsenic treatment. YFP-SUMO-2 HeLa cells were transfected with siRNA to RNF4 or a control nontargeting siRNA (NT) and YFP fluorescence measured for 18 h after the addition of 1 μM of arsenic. Images shows a projection of z-sections collected into YFP channel at selected time points. Bar, 5 μM.
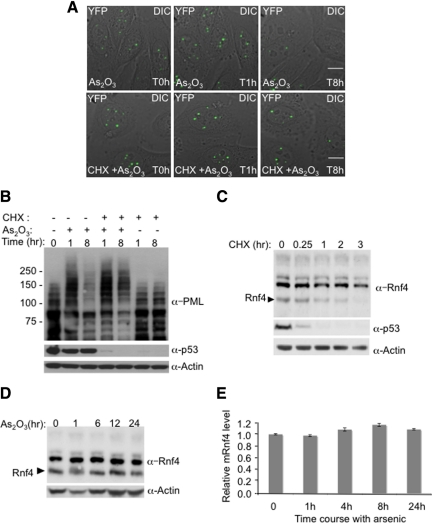
Continued Protein Synthesis Is Required for Arsenic-Induced Degradation of PML
It has been previously reported that arsenic-induced apoptosis could be blocked by the protein synthesis inhibitor cycloheximide (Park et al., 2001). To establish whether cycloheximide blocked arsenic-induced PML degradation, PML-YFP HeLa cells were incubated with either arsenic or with arsenic and cycloheximide. YFP fluorescence was followed by fluorescence microscopy and the levels of PML were monitored by Western blot (Figure 4, A and B). We also monitored the levels of the rapidly turning over protein, p53 (Figure 4B). As expected arsenic induced degradation of PML-YFP but in the presence of cycloheximide arsenic treatment did not fully induce PML-YFP degradation (Figure 4, A and B), suggesting that the continued synthesis of a specific protein(s) was required for arsenic-induced PML degradation. As RNF4 is required for arsenic-induced PML degradation, the levels of this protein were monitored by Western blotting after treatment of the cells with cycloheximide (Figure 4C). Treatment with cycloheximide led to decreased levels of RNF4, until after 3 h RNF4 was undetectable (Figure 4C). By comparison, p53 levels were reduced after 15 min and were almost undetectable 1 h after exposure to cycloheximide (Figure 4, B and C). Reduction in the levels of RNF4, as a consequence of cycloheximide treatment, is a likely explanation for the inhibition of arsenic-induced degradation of PML by cycloheximide. This is consistent with the observation that cycloheximide did not block the initial RNF4-independent accumulation of PML-YFP in nuclear bodies. As RNF4 levels are critical for arsenic-induced PML degradation, we monitored the levels of RNF4 protein by Western blotting and mRNA by quantitative PCR. Over the 24-h period of arsenic treatment neither RNF4 protein nor mRNA levels were altered (Figure 4, D and E).
Figure 4.
Inhibition of RNF4 synthesis blocks arsenic-induced PML degradation. (A and B) Arsenic fails to induce the degradation of PML when protein translation is inhibited. HeLa PML-YFP stable cells were incubated with 1 μM arsenic and 50 μg/ml cycloheximide and analyzed by fluorescence microscopy for 16h (A). A stack of 20 z-sections was collected every 15 min in the YFP channel (green), and one image was taken with differential interference contrast (DIC). The projected z-stacks were merged to the respective DIC image. Bar, 5 μM. The level of PML and p53 proteins were analyzed during arsenic and cycloheximide treatment by SDS PAGE followed by Western Blotting with chicken PML and mouse p53 antibodies (B). (C) Monitoring of RNF4 and p53 protein levels during cycloheximide treatment. PML-YFP HeLa cells were incubated with 50 μg/ml cycloheximide, and whole cell extracts were analyzed at different times by SDS PAGE followed by Western Blotting with rabbit RNF4 and mouse p53 antibodies. (D) Arsenic does not alter the level of RNF4 protein. Total cells extracts from HeLa PML-YFP exposed to arsenic for 24 h were analyzed by SDS PAGE followed by Western blot with a rabbit anti-RNF4 antibody to follow the level of RNF4 during arsenic treatement. (E) Quantification of Rnf4 transcripts during arsenic treatment. Total RNA from HeLa PML-YFP stable cells exposed to 1 μM arsenic for 24 h was extracted and real-time quantitative PCR was performed with specific gene primers for RNF4 and actin. The graph shows the relative mRNF4 level after normalization to actin transcripts.
Arsenic Induced SUMO-Dependent Recruitment of RNF4 to PML Nuclear Bodies
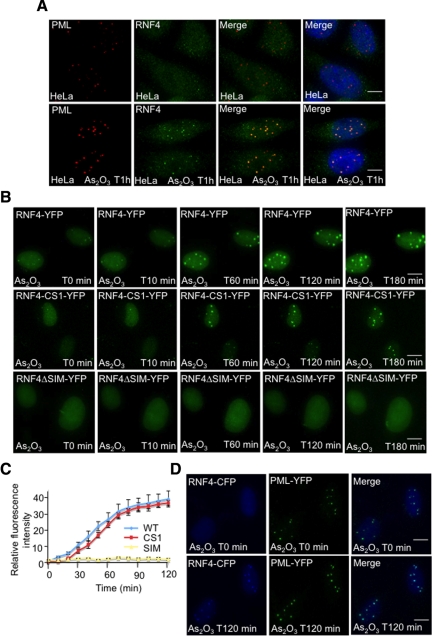
Having shown that RNF4 was essential to induce PML degradation, it was important to understand how PML is targeted by RNF4. Immunofluorescence microscopy was used to establish the subcellular localization of endogenous RNF4 and how this is changed after arsenic treatment. In the absence of arsenic, RNF4 is diffusely distributed throughout the nucleus, but after exposure of cells to arsenic RNF4 accumulates in PML bodies (Figure 5A). To follow this recruitment of RNF4 into PML bodies in real time, cells expressing RNF4-YFP were exposed to arsenic and the subcellular localization of RNF4 followed by fluorescence microscopy. RNF4-YFP is rapidly recruited into nuclear bodies in response to arsenic (Figure 5B and Supplemental Figure S2A). This recruitment of RNF4-YFP into nuclear bodies was not dependent on the E3 ubiquitin ligase activity of RNF4, as RNF4 containing a RING domain mutation was recruited into nuclear bodies with similar kinetics to that of RNF4 wt (Figure 5, B and C). However, recruitment of RNF4-YFP into nuclear bodies was SUMO-dependent, as a version of RNF4 lacking functional SUMO interactions motifs failed to be recruited into nuclear bodies in response to arsenic (Figure 5, B and C and Supplemental Figure S2B). To demonstrate that the nuclear bodies into which RNF4 was recruited were indeed PML bodies, RNF4-CFP was introduced into PML-YFP HeLa cells and the localization of the fluorescent proteins following arsenic treatment was monitored by fluorescence microscopy (Figure 5D and Supplemental Figure S2, C and D). RNF4-CFP rapidly accumulated in the same nuclear bodies that were marked by PML-YFP, indicating that arsenic induced the recruitment of RNF4 into PML nuclear bodies.
Figure 5.
Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. (A and B) Subcellular localization of RNF4 during arsenic treatment. (A) HeLa cells were exposed to 1 uM arsenic for 60 min and the localization of endogenous PML and RNF4 determined by immunofluorescence using 5E10 mAb to PML and an affinity purified chicken antibody to RNF4. (B) HeLa cells were transfected with fluorescent-tagged RNF4 plasmids expressing either wild type RNF4-YFP or protein mutated in the RING domain of RNF4 (RNF4-YFP-CS1) or RNF4 with all 4 SIMs mutated (RNF4-delta SIM). YFP fluorescence (green) was monitored in real-time microscopy after arsenic treatment by collecting a stack of 20 z-sections every 10 min for 3 h. Images represent a projection of the z-sections. (C) Quantification of RNF4-YFP fluorescence was determined by measuring the difference of fluorescence intensity between one PML body and a region outside the nucleoplasm. The graph shows means values of the relative fluorescence intensity after normalization from 10 cells. (D) Arsenic-induced recruitment of RNF4 in HeLa PML-YFP stable cells. PML-YFP HeLa cells were transfected with a RNF4-CFP plasmid and fluorescence monitored for 2 h after addition of arsenic. Images shows the recruitment of RNF4-CFP (blue) within PML bodies (green). Bar, 5 μM.
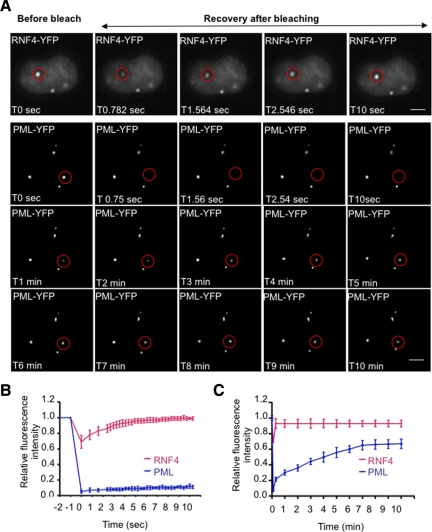
To establish the kinetics of RNF4 and PML trafficking through PML bodies we analyzed the mobility of RNF4 and PML III by Fluorescence Recovery After Photobleaching (FRAP). RNF4-YFP shuttles very quickly between PML bodies and nucleoplasm as it takes <10 s to fully recover after photobleaching of PML bodies (Figure 6, A and B). In contrast, recruitment of PML-YFP into PML bodies from the nucleoplasm is rather slow as full recovery after photobleaching takes almost 10 min (Figure 6, A–C). This slow uptake of PML isoform III into nuclear bodies is entirely consistent with previously published data (Weidtkamp-Peters et al., 2008).
Figure 6.
Comparison of RNF 4 and PML mobility. (A) The mobility of RNF4 and PML III was analyzed by Fluorescence Recovery After Photobleaching (FRAP) on living HeLa PML-YFP stable cells or HeLa cells transfected with RNF4-YFP. Circled areas (red) containing one PML NB were bleached to background level and the fluorescence recovery of RNF4 and PML was monitored for 11 s and 10 min, respectively. A single image was taken for RNF4 at selected time point, whereas a stack of 20 sections was collected every minute for PML. Images for PML represent a projection of z-sections. Bar, 5 μm. (B and C) Quantification of FRAP experiments. The fluorescence intensity in the bleached area was normalized to the change in fluorescence intensity outside the nucleus and in an unbleached PML NB. The graphs (B and C) show the mean values and standard deviations of relative prebleach and postbleach intensities of ten PML Nbs which was plotted over time in sec (B) or in min (C).
Role of RNF4 in the Recruitment of PML into Nuclear Bodies
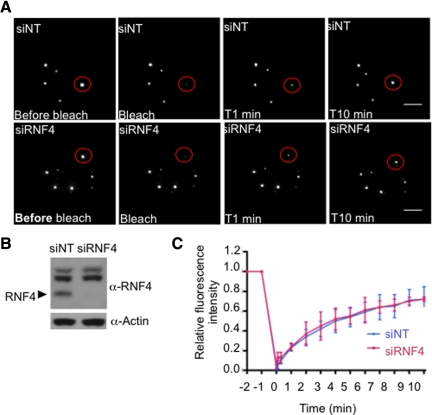
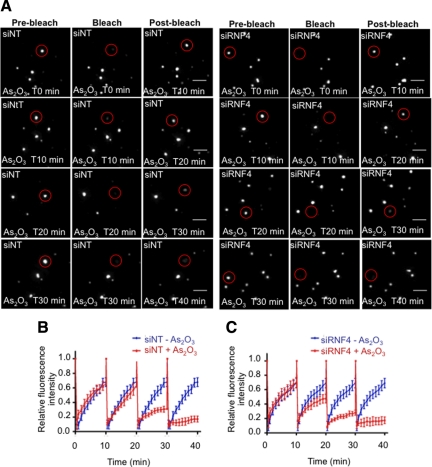
Arsenic treatment promotes recruitment of nucleoplasmic PML into PML nuclear bodies. In cells where RNF4 has been depleted, PML is already concentrated into PML bodies. To establish the role of RNF4 in the nuclear dynamics of PML we used FRAP. Initially, we monitored the dynamics of PML in the presence and the absence of RNF4 in PML-YFP HeLa cells. A region containing a single PML body was bleached to background level and fluorescence recovery was monitored for 10 min. Fluorescence recovery was similar in PML-YFP HeLa cells that had been depleted of RNF4 by siRNA compared with the same cells treated with a nontargeting siRNA (Figure 7, A–C). Thus it appears that RNF4 does not affect the trafficking of PML III between nucleoplasm and PML bodies. To establish the role of RNF4 in the partitioning of PML between the nucleoplasm and nuclear bodies in response to arsenic, we treated PML-YFP HeLa cells with siRNA to RNF4 or with a nontargeting control siRNA and used FRAP to monitor the uptake of PML-YFP into nuclear bodies at various times after exposure of the cells to arsenic (Figure 8A). While arsenic progressively decreases the uptake of PML-YFP into nuclear bodies, this is entirely independent of RNF4 as the rate of recovery is indistinguishable in cells treated with siRNA to RNF4 and cells treated with a nontargeting siRNA (Figure 8, B and C).
Figure 7.
RNF4 does not affect PML trafficking between PML and nucleoplasm. (A–C) Effect of RNF4 on PML mobility. (A) FRAP experiments were performed on HeLa PML-YFP stable cells transfected with a nontarget siRNA (siNT) or an siRNA against RNF4 (siRNF4). Images represent projection of z-sections of PML NBs before and after photobleaching over a period of 10 min. Bar, 5 μM. (B) Depletion of RNF4 was controled by SDS PAGE followed by Western blot with rabbit RNF4 antibody. (C) The graph shows the mean values and standard deviations of relative prebleach and postbleach intensities of ten PML Nbs as described in Figure 6.
Figure 8.
Effect of RNF4 on PML mobility during arsenic treatment. (A–C) RNF4 does not affect PML mobility during arsenic treatment. (A) PML fluorescence recovery after photobleaching was determined after arsenic treatment in HeLa PML-YFP stable cells transfected with a nontarget siRNA (siNT) or a siRNA against RNF4 (siRNF4). PML NBs were bleached after a period of 10, 20, 30, or 40 min of incubation with arsenic and fluorescence recovery was monitored over a period of 10 min for each time point. Images represent projection of z-sections of PML NBs before and after photobleaching over a period of 10 min. Bar, 5 μM. (B and C) The graphs show the mean values and standard deviations of relative prebleach and postbleach intensities of PML NBs from five different experiments.
In Vivo Interaction of RNF4 with SUMO-2 during Arsenic Treatment
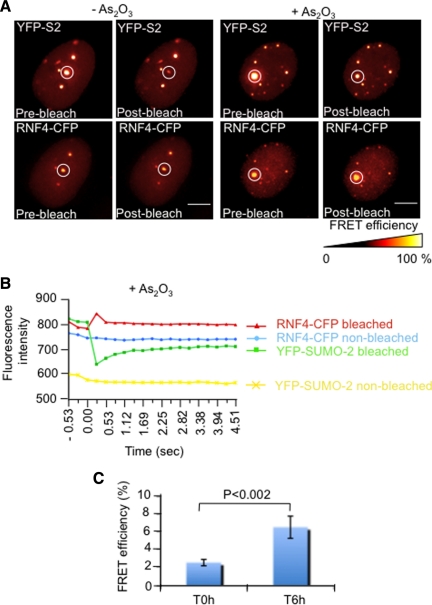
To establish that PML bodies were the sites of interaction between RNF4 and SUMO-2, we performed Fluorescence Resonance Energy transfer (FRET) in living cells (Figure 9). FRET is based on the ability of a higher-energy donor fluorophore (CFP) to transfer energy to a lower-energy acceptor molecule (YFP). In practice, FRET can be measured by photobleaching the acceptor molecule such that energy from the donor can no longer be transferred to the acceptor. This leads to an increase in fluorescence of the donor molecule. Therefore, YFP-SUMO-2 and RNF4-CFP were coexpressed in HeLa cells and a region of interest containing YFP-SUMO-2 within a PML body was irridiated without bleaching RNF4-CFP (Figure 9, A and B). Fluorescence intensity of the donor molecule (RNF4-CFP) was then measured in nontreated cells or in cells treated with arsenic for 6 h. Interestingly, an increase in fluorescence of RNF4-CFP was only detected when cells were treated with arsenic, suggesting that the formation of long polySUMO-2 chains upon arsenic treatment represents a scaffold for RNF4 to interact with SUMO-2 (Figure 9, A–C). In the absence of arsenic, the CFP signal remained constant, indicating that RNF4 does not stably interact with SUMO-2 in PML bodies but diffuses quickly between nucleoplasm and PML bodies. Also, photobleaching of YFP-SUMO-2 within the nucleoplasm in cells either nontreated or treated with arsenic did not lead to an increase of RNF4-CFP fluorescence, strongly suggesting that RNF4 interacts stably with SUMO-2 only in PML bodies (data not shown). This is consistent with our observation showing the recruitment of RNF4 in real time upon arsenic treatment. Attempts to use FRET to demonstrate an interaction between RNF4 and PML either in the absence or presence of arsenic were unsuccessful, irrespective of whether the fluorescent protein was fused to the C or N terminus of RNF4 and PML (data not shown). These data suggest that RNF4 interacts with polySUMO chains attached to PML, rather than directly with PML.
Figure 9.
In vivo interaction of RNF4-CFP with YFP-SUMO-2. (A) Fluorescence Resonance Energy Transfer (FRET) acceptor photobleaching was done on HeLa cells cotransfected with plasmids expressing RNF4-CFP and YFP-SUMO-2 either untreated or treated with 1 μΜ arsenic. Images shows sequential CFP and YFP fluorescence collected before and after photobleaching of a circled area (red) containing one PML NB. Bar 5 μM. (B) The graph shows the CFP and YFP fluorescence intensities of bleached and nonbleached nuclear areas of similar-sizes from cells exposed to arsenic for 6 h. (C) FRET efficiencies during arsenic treatment. The graph shows the mean values and SD of the FRET efficiency from five independent experiments. FRET efficiency is represented as the percent of increase of prebleach CFP fluorescence after YFP photobleaching of one PML NB.
DISCUSSION
The fate of PML protein, and the nuclear bodies in which it resides, have been extensively studied as fusion of the PML protein to the RARα is the cause of Acute Promyelocytic Leukemia. An effective treatment for this disease is the administration of arsenic, which results in the proteasomal degradation of the PML-RAR fusion protein. Immediately after arsenic treatment, the PML protein becomes polySUMO modified and it is this modification that recruits the SUMO-specific ubiquitin E3 ligase RNF4 (Tatham et al., 2008; Lallemand-Breitenbach et al., 2008). Subsequent ubiquitylation of SUMO-modified PML results in its proteasomal degradation. To gain insight into the details of PML degradation we established HeLa cells that express PMLIII-YFP at close to endogenous levels. When these cells are exposed to arsenic the PML-YFP fusion protein undergoes SUMO-dependent ubiquitin-mediated proteolysis and is degraded over a similar timescale to that of the endogenous protein. Here we have used real-time fluorescence microscopy to establish the dynamics of PML and RNF4 recruitment into PML bodies when cells are exposed to arsenic. This revealed that RNF4 does not affect residence time of PML in PML NBs, as the recovery of PML III-YFP into NBs after photobleaching does not change when RNF4 expression is ablated by siRNA (Figure 7). Exposure of cells to arsenic leads to a rapid decrease in the mobility of PML-YFP, which was again independent of the presence of RNF4 (Figure 8). This “fixing” of PML into PML NBs occurred <60 min after addition of arsenic and was coincident with a dramatic increase in the SUMO modification status of PMLIII-YFP (Figure 2). Recently, it has been proposed that PML immobilization in nuclear bodies could arise from the arsenic-induced cross-linking of PML, as a consequence of direct arsenic-mediated, cross-linking of cysteine residues in PML, and oxidation-induced formation of disulphide bonds. It is thought that these changes promote multimerization of PML, leading to enhanced recruitment of Ubc9 and thus increased SUMOylation of PML (Jeanne et al., 2010). As the form of PML that cannot be SUMO modified displays a very low residence time in PML bodies (Weidtkamp-Peters et al., 2008), it seems likely that the increased residence time of PML in NBs after arsenic treatment is a consequence of the observed PML SUMO modification.
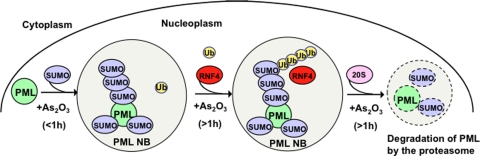
In the absence of arsenic, RNF4-CFP appears to be predominantly nucleoplasmic although a small proportion appears to be localized to PML NB (Figure 5). After addition of arsenic, RNF4-CFP is recruited from the nucleoplasm into PML bodies with similar kinetics to the recruitment of PMLIII-YFP into nuclear bodies (Figure 5) and paralleling the rapid arsenic-induced SUMO modification of PML (Figure 2). Immunofluorescence studies with an antibody to RNF4 indicated that the endogenous RNF4 protein also undergoes a similar arsenic-dependent relocalization from the nucleoplasm to PML nuclear bodies (Figure 5A). This arsenic-induced recruitment of RNF4-CFP into PML nuclear bodies is entirely SUMO-dependent, because a form of RNF4 that is unable to bind SUMO is not recruited in response to arsenic (Figure 5). As RNF4 binds specifically to polySUMO chains, these data suggest that arsenic-induced modification of PML with polySUMO chains occurs in PML nuclear bodies. This contention is supported by the FRET experiments showing that FRET between YFP-SUMO-2 and RNF4-CFP is detected in PML bodies, but not in the nucleoplasm (Figure 9). Although the sites at which polySUMO chains are formed have not been previously localized, these data are consistent with previously published work suggesting that PML bodies are sites of SUMO modification (Lallemand-Breitenbach et al., 2001; Quimby et al., 2006). While FRET allowed the detection of a robust interaction between RNF4 and SUMO-2 in PML bodies after arsenic treatment we were unable to detect any interaction between RNF4-CFP and PMLIII-YFP using this approach, despite using different combinations of plasmid constructs to express YFP or CFP at the amino or carboxy terminus of the proteins (data not shown). These data are in line with the known protein interaction properties of RNF4 where the conserved, multiple SIMs in its N terminus bind to polySUMO chains (Tatham et al., 2008). While it is possible that our inability to detect FRET between RNF4 and PML is due to an unfavorable steric arrangement of the CFP and YFP fluorophores, a more likely interpretation of the data are that RNF4-CFP does not interact with PMLIII-YFP, but binds to the long polySUMO chains on PML, thus increasing the distance between the YFP and CFP fluorophores out with the range at which a FRET signal can be detected. However these data do not appear to be consistent with a recent report (Percherancier et al., 2009) in which BRET was used to detect an interaction between PML luciferase and YFP-RNF4 upon addition of luciferase substrate. This interaction increased after arsenic addition but was dependent on PML SUMOylation and may be explained by an indirect interaction via SUMO and the greater distance over which BRET can be monitored. We have previously demonstrated that RNF4 was responsible for ubiquitylation of SUMO-modified PML (Tatham et al., 2008). Thus, the direct result of RNF4 recruitment into NBs by polySUMO-modified PML is likely to be the ubiquitylation of polySUMO chains attached to PML within the NBs. As the ultimate consequence of arsenic treatment is the degradation of PML, the most likely site of PML degradation is within the PML body. A number of observations are consistent with this hypothesis. It has previously been reported that the PML protein accumulates in PML bodies in the presence of proteasome inhibitors (Bailey and O'Hare, 2005) and that arsenic-induced degradation of PML is blocked by proteasome inhibition, with the PML protein accumulating in PML NBs (Lallemand-Breitenbach et al., 2008). Immunofluorescence studies have revealed that components of the proteasome are present in PML bodies (Lallemand-Breitenbach et al., 2001; Fabunmi et al., 2001; Lafarga et al., 2002; Rockel et al., 2005). Most dramatically, siRNA-mediated depletion of RNF4, the ubiquitin E3 ligase responsible for PML degradation, leads to the arsenic-induced accumulation of PML in PML NBs (Figure 2). On the basis of our findings reported here we propose that arsenic leads to release of an activity that generates polySUMO chains on NB-associated PML. This could be due to inhibition of SUMO-specific proteases, activation of a distinct SUMO E3 ligase such as Ran BP2 (Saitoh et al., 2006), or activation of an intrinsic SUMO E3 ligase associated with PML only observed in yeast so far (Quimby et al., 2006). Interestingly, it has been shown recently that low doses of H2O2 increase SENP3 levels that in turn reduce the number of PML bodies due to the deconjugation of SUMO-2/3 from PML (Han et al., 2010). However, it remains to be determined whether arsenic interferes with the expression of specific SUMO proteases. On the basis of these data we have formulated a working model for the mechanism of action of arsenic (Figure 10). Before arsenic treatment PML shuttles in, and out, of nuclear bodies. However, upon the addition of arsenic nuclear body-associated PML undergoes rapid polySUMOylation which “fixes” the modified PML in the nuclear bodies. In the absence of RNF4 there is continued movement of PML into nuclear bodies where it undergoes arsenic-induced polySUMO modification. However, as it cannot be ubiquitylated it is not degraded and the polySUMO chain fixes it in the nuclear bodies, precluding its return to the nucleoplasm. In the presence of RNF4 the polySUMO chains attached to PML are ubiquitylated, and this assembly is targeted to nuclear body-associated proteasomes where PML undergoes degradation. Although we have not addressed this question directly it seems likely that the polySUMO, polyUbiquitin hybrid chains are removed from PML before its degradation.
Figure 10.
Model for the subnuclear trafficking of PML and RNF4 in response to arsenic. Details of the model are described in the text.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sam Swift for technical assistance in live cell imaging, David Lleres and Sam Swift for hepful advice for FRET and FRAP analysis, Patricia Le Baccon for movies arrangement, and Mounira Chelbi-Alix and Centre National de la Recherche Scientifique for the provision of facilities to allow M.-C.G. to complete this work. M.-C.G. was supported by a EU RUBICON fellowship. K.J.W. was supported by a postgraduate fellowship for clinicians from the Wellcome Trust. This work was supported by Cancer Research UK.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-05-0449) on October 13, 2010.
REFERENCES
- Bailey D., O'Hare P. Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem. J. 2005;392:271–281. doi: 10.1042/BJ20050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R., Pandolfi P. P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- de The H., Chomienne C., Lanotte M., Degos L., Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- de The H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- Dyck J.A., Maul G. G., Miller W. H., Chen J. D., Kakizuka A., Evans R. M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Ellis J. D., Lleres D., Denegri M., Lamond A. I., Caceres J. F. Spatial mapping of splicing factor complexes involved in exon and intron definition. J. Cell Biol. 2008;181:921–934. doi: 10.1083/jcb.200710051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov E., Sharma P., Lockett S. J., Lualdi M., Kuehn M. R. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- Fabunmi R. P., Wigley W. C., Thomas P. J., DeMartino G. N. Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J. Cell Sci. 2001;114:29–36. doi: 10.1242/jcs.114.1.29. [DOI] [PubMed] [Google Scholar]
- Gao C., Ho C. C., Reineke E., Lam M., Cheng X., Stanya K. J., Liu Y., Chakraborty S., Shih H. M., Kao H. Y. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol. Cell. Biol. 2008;28:5658–5667. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy M. C., Hay R. T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- Hakli M., Karvonen U., Janne O. A., Palvimo J. J. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp. Cell Res. 2005;304:224–233. doi: 10.1016/j.yexcr.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Hakli M., Lorick K. L., Weissman A. M., Janne O. A., Palvimo J. J. Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett. 2004;560:56–62. doi: 10.1016/S0014-5793(04)00070-5. [DOI] [PubMed] [Google Scholar]
- Han Y., et al. SENP3-mediated de-conjugation of SUMO2/3 from PML is correlated with accelerated cell proliferation under mild oxidative stress. J. Biol. Chem. 2010;285:12906–12915. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanne M., Lallemand-Breitenbach V., Ferhi O., Koken M., Le Bras M., Duffort S., Peres L., Berthier C., Soilihi H., Raught B., de The H. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell. 2010;18:88–98. doi: 10.1016/j.ccr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Jensen K., Shiels C., Freemont P. S. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- Kakizuka A., Miller W. H., Jr, Umesono K., Warrell R. P., Jr, Frankel S. R., Murty V. V., Dmitrovsky E., Evans R. M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- Lafarga M., Berciano M. T., Pena E., Mayo I., Castano J. G., Bohmann D., Rodrigues J. P., Tavanez J. P., Carmo-Fonseca M. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell. 2002;13:2771–2782. doi: 10.1091/mbc.E02-03-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen A. M., Poukka H., Karvonen U., Hakli M., Janne O. A., Palvimo J. J. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. 1998;18:5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Pampin M., Simonin Y., Blondel B., Percherancier Y., Chelbi-Alix M. K. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J. Virol. 2006;80:8582–8592. doi: 10.1128/JVI.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. W., Choi Y. J., Jang M. A., Baek S. H., Lim J. H., Passaniti T., Kwon T. K. Arsenic trioxide induces G2/M growth arrest and apoptosis after caspase-3 activation and bcl-2 phosphorylation in promonocytic U937 cells. Biochem. Biophys. Res. Commun. 2001;286:726–734. doi: 10.1006/bbrc.2001.5416. [DOI] [PubMed] [Google Scholar]
- Percherancier Y., Germain-Desprez D., Galisson F., Mascle X. H., Dianoux L., Estephan P., Chelbi-Alix M. K., Aubry M. Role of SUMO in RNF4-mediated promyelocytic leukemia protein (PML) degradation: sumoylation of PML and phospho-switch control of its SUMO binding domain dissected in living cells. J. Biol. Chem. 2009;284:16595–16608. doi: 10.1074/jbc.M109.006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. J., Tainer J. A., Boddy M. N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Poukka H., Aarnisalo P., Santti H., Janne O. A., Palvimo J. J. Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor-mediated transcription by different mechanisms. J. Biol. Chem. 2000;275:571–579. doi: 10.1074/jbc.275.1.571. [DOI] [PubMed] [Google Scholar]
- Quimby B. B., Yong-Gonzalez V., Anan T., Strunnikov A. V., Dasso M. The promyelocytic leukemia protein stimulates SUMO conjugation in yeast. Oncogene. 2006;25:2999–3005. doi: 10.1038/sj.onc.1209335. [DOI] [PubMed] [Google Scholar]
- Rockel T. D., Stuhlmann D., von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 2005;118:5231–5242. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Uchimura Y., Tachibana T., Sugahara S., Saitoh H., Nakao M. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp. Cell Res. 2006;312:1418–1430. doi: 10.1016/j.yexcr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D., Neugebauer K. M. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmeier P., Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell. Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Wang Z. Y., Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S., Lenser T., Negorev D., Gerstner N., Hofmann T. G., Schwanitz G., Hoischen C., Maul G., Dittrich P., Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- Weis K., Rambaud S., Lavau C., Jansen J., Carvalho T., Carmo-Fonseca M., Lamond A., Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Zhu J., Koken M. H., Quignon F., Chelbi-Alix M. K., Degos L., Wang Z. Y., Chen Z., de The H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.