The report demonstrates that two distinct isoforms of the ErbB4 receptor tyrosine kinase stimulate either proliferation or apoptosis by mechanisms involving differential transcriptional regulation of the PDGFRA gene. These data have implications for developing approaches to target ErbB4 signaling in cancer.
Abstract
The significance of ErbB4 in tumor biology is poorly understood. The ERBB4 gene is alternatively spliced producing juxtamembrane (JM-a and JM-b) and cytoplasmic (CYT-1 and CYT-2) isoforms. Here, signaling via the two alternative ErbB4 JM isoforms (JM-a CYT-2 and JM-b CYT-2) was compared. Fibroblasts expressing ErbB4 JM-a demonstrated enhanced ErbB4 autophosphorylation, growth, and survival. In contrast, cells overexpressing ErbB4 JM-b underwent starvation-induced death. Both pro- and antisurvival responses to the two ErbB4 isoforms were sensitive to an ErbB kinase inhibitor. Platelet-derived growth factor receptor-alpha (PDGFRA) was identified as an ErbB4 target gene that was differentially regulated by the two ErbB4 isoforms. The soluble intracellular domain of ErbB4, released from the JM-a but not from the JM-b isoform, associated with the transcription factor AP-2 and promoted its potential to enhance PDGFRA transcription. Survival of cells expressing JM-a was suppressed by targeting either PDGFR-α or AP-2, whereas cells expressing JM-b were rescued from cell death by the PDGFR-α agonist, PDGF-BB. These findings indicate that two alternative ErbB4 isoforms may promote antagonistic cellular responses and suggest that pharmacological inhibition of ErbB4 kinase activity may lead to either suppression or promotion of cellular growth.
INTRODUCTION
ErbB/HER receptors form the epidermal growth factor receptor (EGFR) subfamily of receptor tyrosine kinases including ErbB1 (EGFR, HER1), ErbB2 (c-Neu, HER2), ErbB3 (HER3), and ErbB4 (HER4). ErbB receptors consist of an extracellular ligand-binding ectodomain, a hydrophobic transmembrane domain, and an intracellular cytoplasmic domain with enzymatic tyrosine kinase activity. Several EGF-like growth factors, such as neuregulins (NRG), bind to ErbB receptors, stimulating receptor dimerization, conformational changes, and subsequent autophosphorylation of tyrosine residues. These phosphorylation events trigger activation of downstream signal transduction molecules that couple ErbBs to cellular responses, such as proliferation, differentiation, apoptosis, and survival. Overexpression and mutations of ErbBs have been associated with malignant growth. Moreover, drugs that target ErbB1 or ErbB2 have demonstrated clinical effect as cancer therapeutics. These drugs include anti-ErbB antibodies and small-molecular-weight tyrosine kinase inhibitors (Hynes and MacDonald, 2009).
Relatively little is known about the cancer biology of ErbB4. Overexpression of ErbB4 promotes breast cancer cell proliferation and transforms fibroblasts (Cohen et al., 1996; Tang et al., 1999; Maatta et al., 2006). In contrast, ErbB4 activation has also been suggested to induce differentiation of breast cancer cells, confer cell cycle arrest, or stimulate apoptosis (Chen et al., 1996; Jones et al., 1999; Ni et al., 2001; Williams et al., 2004; Vidal et al., 2005; Muraoka-Cook et al., 2006a,b). Interpretation of published results has been complicated by the existence of four structurally and functionally different ErbB4 isoforms. These isoforms are generated from a single ERBB4 gene by tissue-specific alternative splicing (Junttila et al., 2000, 2003). The isoforms are all functional and tyrosine phosphorylated upon treatment with NRG-1 (Maatta et al., 2006). ErbB4 juxtamembrane (JM) isoforms (JM-a and JM-b) differ in their susceptibility to proteolysis at the extracellular JM domains (Elenius et al., 1997). As a consequence, only JM-a isoforms with 23 unique amino acids within the JM domain undergo ectodomain shedding, whereas JM-b isoforms with 13 alternative amino acids at the JM domain do not (Elenius et al., 1997). ErbB4 cytoplasmic (CYT) isoforms differ at cytoplasmic tails by including (CYT-1) or not (CYT-2) a 16-amino acid stretch containing binding sites for phosphoinositide 3-kinase (PI3-K; Elenius et al., 1999; Kainulainen et al., 2000), as well as for WW domain-containing proteins such as Nedd-like ubiquitin ligases (Omerovic et al., 2007; Sundvall et al., 2008b; Feng et al., 2009; Zeng et al., 2009).
In addition to activating cascadical signaling pathways such as Ras–Raf–MAP/ERK kinase (Mek) –mitogen-activated protein kinase (MAPK) and PI3K/Akt (Kainulainen et al., 2000), the intracellular domains (ICD) of proteolytically processed ErbB4 isoforms have been shown to directly translocate into the nucleus and regulate transcription (Ni et al., 2001; Lee et al., 2002; Komuro et al., 2003; Williams et al., 2004; Maatta et al., 2006; Sardi et al., 2006). First, ErbB4 JM-a is cleaved by tumor necrosis factor-α converting enzyme (TACE) approximately eight amino acids N-terminal to the transmembrane domain (Elenius et al., 1997; Rio et al., 2000; Cheng et al., 2003). Subsequently, the remaining N-terminally truncated receptor undergoes regulated intramembrane proteolysis (RIP) by γ-secretase activity creating a soluble ICD fragment that may function as a transcriptional coregulator for several regulators of transcription, such as STAT5A, estrogen receptor-α, ETO2, and YAP-2 (Jones, 2008).
Previous findings have supported differential roles for different ErbB4 isoforms in cancer biology. CYT isoforms differ in subcellular targeting and stability (Maatta et al., 2006; Sundvall et al., 2007; Sundvall et al., 2008b) and have opposing effects on mouse mammary epithelium in vivo. ICD of CYT-1 type decreases mammary epithelial growth, whereas ICD of CYT-2 type causes epithelial hyperplasia (Muraoka-Cook et al., 2009). Only cleavable ErbB4 JM-a CYT-2 capable of releasing a soluble ICD promotes survival of myeloid cells and growth of breast cancer cells in vitro (Maatta et al., 2006). In addition, nuclear localization of ErbB4 epitope associates with worse survival than localization of ErbB4 at the cell surface (Junttila et al., 2005).
Here, we compared the transforming potential of two ErbB4 isoforms in stably transfected mouse NR6 fibroblasts, a well-characterized model of ErbB-induced transformation (Chazin et al., 1992; Carey et al., 2006). Consistent with earlier analyses with other models (Junttila et al., 2005; Maatta et al., 2006), the JM-a CYT-2 isoform promoted proliferation, survival, and anchorage-independent growth. However, cells expressing ErbB4 JM-b CYT-2 with an alternative JM domain stimulated, rather than suppressed, apoptosis. Both responses were sensitive to an ErbB kinase inhibitor, indicating that ErbB4 inhibition can either promote or suppress growth, depending on the isoform expressed by the target cell. Further analyses on the mechanisms underlying the responses demonstrated a role for differential regulation of the PDGFRA gene at the transcriptional level. These findings suggest that the two ErbB4 isoforms may stimulate opposite cellular responses and underline the importance of using isoform-specific reagents when analyzing the potential of ErbB4 as a cancer drug target.
MATERIALS AND METHODS
Stable Transfectants
NR6 mouse fibroblasts (Pruss and Herschman, 1977) were maintained in DMEM (Invitrogen, Carlsbad, CA) supplied with 10% fetal calf serum (FCS; PromoCell GmbH, Heidelberg, Germany). Cells were transfected with pcDNA3.1ErbB4JM-aCYT-2 or pcDNA3.1ErbB4JM-bCYT-2 expression constructs (Maatta et al., 2006) using Lipofectamine plus transfection reagent (Invitrogen) according to manufacturer's instructions. Stable clones were selected and cultured in the presence of 500 μg/ml G418 (geneticin; Calbiochem, La Jolla, CA). ErbB4 protein expression was analyzed as described below.
Chemical Inhibitors and RNA Interference
Inhibitors for ErbB receptors (AG 1478) and PDGFR (AG 1296) were purchased from Calbiochem. siRNAs targeting AP-2α (Hs_TFAP2A_5), Mek1 (Hs_MAP2K1_6), Erk1/2 (Hs_MAPK1_10), Sp1 (Hs_SP1_5), Oct-1 (Hs_POU2F1_2), or a nonsilencing control siRNA were purchased from Qiagen (Chatsworth, CA). siRNA sequences are listed in Supplemental Table 1. siRNA specific for ErbB4 JM-a and control siRNA specific for the JM-b isoform (not expressed in MCF-7 cells), have been previously described (Maatta et al., 2006). siRNAs were transfected to cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The knockdown efficiency of RNA interference was determined by real-time RT-PCR as described below.
Western Blotting and Protein Phosphorylation Analyses
For ErbB4 and PDGFR-α protein expression analysis, NR6 transfectants were lysed, and samples equivalent to 75 μg of total protein were separated in 8% SDS-PAGE gels followed by Western blotting with anti-ErbB4 antibody (sc-283; Santa Cruz Technology, Santa Cruz, CA) or polyclonal anti-PDGFR-α antibody (Cell Signaling Technology, Beverly, MA) as previously described (Kainulainen et al., 2000).
For analysis of protein phosphorylation status, cells were starved overnight in DMEM containing no FCS and stimulated for 10–40 min with or without recombinant human NRG-1β1 (50 ng/ml; R&D Systems, Minneapolis, MN). For analysis of ErbB4 and PDGFR-α tyrosine phosphorylation, 1 mg of total protein lysate was immunoprecipitated with an anti-ErbB4 antibody (sc-283) or anti-PDGFR-α antibody (Cell Signaling Technology) and analyzed by Western blotting using an anti-phosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY), as previously described (Maatta et al., 2006). Filters were reblotted with anti-ErbB4 (sc-283) or anti-PDGFR-α (Cell Signaling Technology) to control loading. For analysis of Erk, Akt, and p38 phosphorylation, samples equivalent to 75 μg of total protein were separated in 8 or 10% SDS-PAGE gels followed by Western blotting with anti-phospho-p44/42 MAPK (Thr202/Tyr204; Cell Signaling Technology), anti-phospho-Akt (Ser 473; Cell Signaling Technology), and anti-phospho-p38 MAPK (Thr180/Tyr182; Zymed Laboratories, South San Francisco, CA) antibodies. To control loading, membranes were stripped and reblotted with anti-p44/42 (Cell Signaling Technology), anti-Akt (sc-1618; Santa Cruz), anti-p38 MAPK (Zymed Laboratories) or anti-actin (sc-1616; Santa Cruz Technology) antibodies.
Kinase activity of ErbB4 JM-a CYT-2 and JM-b CYT-2 in NR6 cells was analyzed using in vitro kinase assays as previously described (Maatta et al., 2006).
Proliferation Analyses
For cell-counting experiments, NR6 transfectants were plated on 24-well plates at a density of 50,000 cells/well in DMEM containing 10% FCS. The following day, the media were replaced by DMEM containing 0–5% FCS supplemented with or without NRG-1 (50 ng/ml) or PDGF-BB (50 ng/ml; PeproTech, Rocky Hill, NJ). At days 2–5 after initial plating, cells were visualized and photographed under a phase-contrast microscope, washed with PBS, removed with trypsin, and counted under a microscope with hemocytometer. Analyses were carried out in triplicates. When inhibitors were used, they were added 24 h after plating in serum-free culture medium. Data were statistically analyzed using Student's t test.
For MTT proliferation assays, NR6 transfectants expressing ErbB4 JM-a CYT-2 or JM-b CYT-2 were plated onto 96-well plates in triplicates in DMEM containing 10% FCS. The next day, 150 nM siRNA targeting AP-2α or a nonsilencing control siRNA were transfected to cells. The number of viable cells was estimated 1, 2, or 3 d after initiation of the starvation using a CellTiter 96 nonradioactive cell proliferation assay (MTT; Promega, Madison, WI). Data were statistically analyzed using Student's t test.
Soft Agar Colony Formation Assay
Bottom layers (2 ml) composed of DMEM containing 0.5% agar (Bacto-agar; Difco, Detroit, MI), 10% FCS, and 1% l-glutamine-penicillin-streptomycin (GPS) solution were prepared into six-well plate wells. Top layers (1.2 ml) consisting of 30,000 NR6 transfectants and DMEM containing 0.33% agar, 10% FCS, 1% GPS, and 0 or 100 ng/ml NRG-1 were added onto the solidified bottom layers. After 3 d of incubation, 200 μl per well of fresh DMEM supplemented with 10% FCS and 1% GPS was added on wells to maintain humidity. Cells were incubated at 37°C for up to 7 wk and photographed under a phase-contrast microscope. Data were statistically analyzed using ANOVA (Dunnett T3 posthoc t tests).
Cell Cycle Analysis
NR6 transfectants were starved without serum for 72 h. Subsequently, both adherent and floating cells were harvested, washed with PBS, and fixed with 70% ethanol at −20°C for 20 min. Fixed samples were washed with PBS, and treated with RNAse A (0.15 mg/ml; Sigma, St. Louis, MO), and DNA was stained with propidium iodine (PI; 40 μg/ml; Sigma). DNA content per particle was determined with FACSCalibur (BD Bioscience, San Jose, CA). Data were analyzed using CellQuest Prosoftware (BD).
TUNEL and DAPI Staining
NR6 transfectants were plated onto 13-mm coverslips at a density of 70,000 cells per milliliter of DMEM containing 10% FCS. The next day the cells were washed with DMEM, and the media were replaced by DMEM containing no serum. Cells were starved without serum for 2 d, washed with PBS, and fixed with methanol. DNA strand breaks were stained with TUNEL (TdT-mediated dUTP nick end labeling) staining reagent according to manufacturer's instructions (In Situ Cell Death Detection Kit, TMR red; Roche, Indianapolis, IN). To analyze nuclear morphology, nuclei were stained with 0.5 μg/ml DAPI (4′,6-diamidino-2-phenylindole; Sigma) in PBS containing 3% bovine serum albumin for 10 min at room temperature. After staining, coverslips were washed with PBS and H2O and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Cells were photographed and counted under a fluorescence microscope. The percentage of condensed nuclei to all nuclei was determined from at least five arbitrary fields. Experiments were repeated at least three times.
cDNA Microarrays
For cDNA microarray analyses, NR6 transfectants were starved without serum for 8 h and treated with or without NRG-1 (50 ng/ml) for 2 h. Vector control cells without NRG-1 treatment were used as a reference sample. RNA was extracted with TRIzol (Invitrogen) and purified with RNeasy MinElute Cleanup Kit (Qiagen). The quality of the RNA samples was determined by spectrophotometry (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE), gel electrophoresis, and by analyzing β-actin expression by real-time RT-PCR (Iivanainen et al., 2003; Junttila et al., 2003). Only samples of uniform quality were chosen for cDNA microarray experiments. The samples were amplified with Ribo Amp RNA Amplification Kit (Arcturus Bioscience, Mountain View, CA).
Twenty micrograms of each RNA sample was labeled, and hybridization was carried out as previously described (Nikula et al., 2005), with the exception that hybridization chambers were purchased from TeleChem International (Sunnyvale, CA). The arrays contained ∼15,000 mouse genes from the National Institute on Aging NIH mouse library spotted in duplicate on poly-l-lysine coated glass slides (Finnish DNA Microarray Centre, Turku, Finland). Microarray data were extracted and normalized using Scan-Array Express software (v.2.1.8; Perkin-Elmer, Waltham, MA), as described (Nikula et al., 2005). The sample expression level at each spot on the array was compared with the expression level of the reference sample, and qualitatively poor spots were discarded. A gene was considered as differentially expressed only if data were obtained from both spots and there was at least a 2.0-fold change in the average expression level. The results were analyzed with Kensington Discovery Edition software (v.2.0.4; InforSense Cambridge, MA) and DAVID (Database for Activation, Visualition, and Integrated Discovery; Functional Annotation Bioinformatics Microarray Analysis; http://david.abcc.ncifcrf.gov/).
RT-PCR
cDNA microarray data were confirmed by RT-PCR. RNA was isolated and reverse-transcribed as described before (Junttila et al., 2003). PDGFRA transcripts were analyzed with primers listed in Supplemental Table 1. To control for mRNA quality, primers specific for β-actin (Iivanainen et al., 2003) were used. PCR reactions were carried out in a total volume of 30 μl, containing 0.67 μM primers, 3 μl of 10× Mg2+-free DyNAzyme buffer (Finnzymes, Espoo, Finland), 500 μM of each dNTP (Finnzymes), 1 U of DyNAzyme II EXT DNA Polymerase (Finnzymes), and 1 μl of template cDNA or water (negative control). PCR samples were denatured at 94°C for 3 min and subsequently cycled 25 times (β-actin) or 35 times (PDGFRΑ) through 1-min steps of annealing at 60°C, extension at 72°C, and denaturation at 94°C. PCR products were separated by electrophoresis using 2% agarose gels, stained with ethidium bromide, and visualized with UV light.
Real-time RT-PCR analysis of ErbB4 expression was carried out as described (Junttila et al., 2003, 2005), except that ABI Prism 7500 or 7900HT Sequence Detectors (Applied Biosystems. Foster City, CA) were used. For amplification of other transcripts, primers and probes listed in Supplemental Table 1 were used.
PDGFRA Promoter Assay
The effect of ErbB4 signaling on PDGFRA promoter activity was analyzed in MCF-7 cells, which endogenously express ErbB4 JM-a CYT-1 and JM-a CYT-2 isoforms (Maatta et al., 2006). The cells were plated on 24-well plates and cotransfected with pEGFP-C3 (polymeric enhanced green fluorescent protein [pEGFP]; Clontech, Palo Alto, CA) and constructs expressing luciferase reporter driven by PDGFRA promoter fragments of different sizes (pSLA4PDGFRαluc–441/+118, pSLA4PDGFRαluc–944/+118, and pSLA4PDGFRαluc–1253/+118; Afink et al., 1995) using Lipofectamine 2000 (Invitrogen). Starting 30 h after transfection, the cells were starved in the absence of serum for 16 h and stimulated for 2 h with 80 ng/ml NRG-1 or for 18 h with 10 μM retinoic acid (RA). The fluorescence signal of EGFP was used to normalize the luminescence signal generated by the luciferase reporters as previously reported (Sundvall et al., 2007). To assess the effect of AP-2 and ErbB4 ICD overexpression on PDGFRA promoter, vector control, pCMVHA-AP2α, pCMVHA-AP2γ (Aqeilan et al., 2004a), and/or pcDNA3.1ErbB4ICD2 (Sundvall et al., 2007), constructs were expressed with pSLA4PDGFRαluc–1253/+118 and pRL-TK Renilla luciferase (Stratagene, La Jolla, CA) reporter constructs in human embryonic kidney (HEK)-293T cells. Dual Luciferase Reporter Assay (Promega) was used to measure luciferase activities. For RNA interference analyses, siRNAs were transfected to cells as above 4 h after transfecting the luciferase and EGFP constructs. A minimum of six independent experiments was carried out for all promoter assays, and the statistical significance was determined with one-way ANOVA (Dunnett two-sided or Dunnett T3 posthoc t tests).
Confocal Microscopy
COS-7 cells were transiently transfected with constructs encoding hemagglutinin (HA)-tagged ErbB4 JM-b CYT-2 or the sole intracellular domain of the CYT-2-type, fixed 24 h after transfection, and stained with rat anti-HA antibody (epitope 12CA5; Zymed) and mouse monoclonal anti-AP-2α antibody (sc-25343; Santa Cruz Biotechnology), as previously described (Sundvall et al., 2008b).
Coimmunoprecipitation of ErbB4 with AP-2
Interaction between full-length or ICD of ErbB4 and AP-2 was determined by coimmunoprecipitation analysis of COS-7 cell transfectants, as previously described (Sundvall et al., 2008b). Constructs encoding HA-tagged ErbB4 JM-b CYT-2 or the sole intracellular domain of the CYT-2-type (Sundvall et al., 2007) and Myc-tagged AP-2α or AP-2γ (Aqeilan et al., 2004a) were used.
Glutathione S-transferase Pulldown Assay
To generate glutathione S-transferase (GST) fusion constructs encoding either ICD of the CYT-2-type (ICD2), N-terminally truncated ICD2 (ICD-ΔN, including amino acids 997-1292) or C-terminally truncated ICD2 (ICD-ΔC; amino acids 676-996), pcDNA3.1ErbB4ICD2 (Sundvall et al., 2007) was amplified with primers (Supplemental Table 1) including sites for restriction enzymes. The PCR products were ligated into a pGEX6p-1 vector (GE HealthcareWaukesha, WI) using SalI and NotI restriction sites. For GST pulldown assays, Myc-tagged AP-2γ (Aqeilan et al., 2004a) or Myc-tagged Wwox (Aqeilan et al., 2004b) constructs were expressed in COS-7 cells, and the interaction between AP-2γ or Wwox and the GST fusion proteins was analyzed as previously described (Sundvall et al., 2008b).
RESULTS
NR6 Transfectants Expressing ErbB4 JM-a CYT-2 and ErbB4 JM-b CYT-2
NR6 cells are a population of Swiss 3T3 fibroblasts that was originally selected for a lack of EGF response and are devoid of EGFR (Pruss and Herschman, 1977). NR6 cells also lack endogenous ErbB3 and ErbB4 and express only ErbB2 (data not shown). Moreover, NR6 cells have successfully been used to analyze transforming potential of ErbBs (Chazin et al., 1992; Carey et al., 2006). Because only nonadherent hematopoietic cells totally lack ErbB2 (Maatta et al., 2006), NR6 cells represent a model to analyze transforming activity of ErbB4 isoforms in a cell background with minimal endogenous ErbB expression.
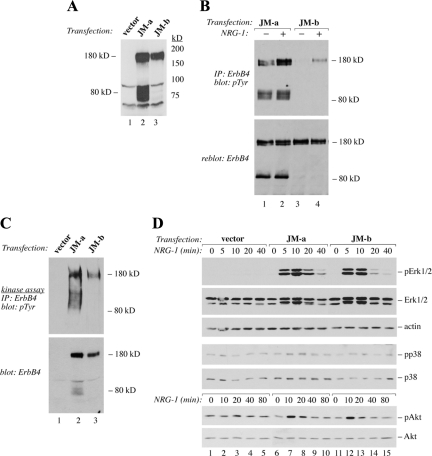
NR6 cells were stably transfected to overexpress two ErbB4 isoforms with the same CYT-2 domain but different JM domains (JM-a CYT-2 and JM-b CYT-2). The JM-a CYT-2 isoform was chosen as it has previously been shown to be the most potent ErbB4 isoform in stimulating ligand-independent survival of myeloid 32D cells (Maatta et al., 2006) and growth of MCF-7 breast cancer cells (Junttila et al., 2005; Maatta et al., 2006). As the ligand-independent responses to JM-a CYT-2 expression have been shown to be partially dependent on cleavage of a soluble ICD by RIP (Maatta et al., 2006), the noncleavable JM-b CYT-2 was included in the experiments for comparison. Several independent transfectant clones were established and screened for ErbB4 expression by Western blotting (Figure 1A; Supplemental Figure 1). In all ErbB4 transfectants a 180-kDa band representing the full-length ErbB4 was visible. An 80-kDa fragment representing the carboxy-terminal cleavage product of ErbB4 was constitutively generated in cells expressing ErbB4 JM-a CYT-2, but not in cells expressing ErbB4 JM-b CYT-2. Both ErbB4 isoforms were tyrosine-phosphorylated upon NRG-1 stimulation, but only JM-a CYT-2 demonstrated efficient constitutive phosphorylation in the absence of ligand stimulation (Figures 1B and 5B). Furthermore, the 80-kDa fragment of ErbB4 JM-a CYT-2 demonstrated high basal tyrosine phosphorylation (Figures 1B and 5B). These findings are consistent with our previous observations in other cell backgrounds (Maatta et al., 2006) and demonstrate that JM-a CYT-2 but not JM-b CYT-2 is constitutively processed to generate an 80-kDa phosphorylated carboxy-terminal fragment in NR6 cells.
Figure 1.
Expression and activity of ErbB4 isoforms in NR6 cells. (A) Expression of ErbB4 protein in NR6 transfectants was analyzed by Western blotting. (B) Cells were starved overnight without serum and stimulated with or without 50 ng/ml NRG-1 for 10 min. ErbB4 tyrosine phosphorylation was analyzed by immunoprecipitation with an anti-ErbB4 antibody followed by Western blotting with an anti-phosphotyrosine antibody. Membrane was reblotted with an anti-ErbB4 antibody. (C) For in vitro kinase assay, cell lysates were immunoprecipitated with an anti-ErbB4 antibody, incubated with ATP, and analyzed for ErbB4 tyrosine phosphorylation by immunoprecipitation with anti-ErbB4 antibody followed by Western blotting with an anti-phosphotyrosine antibody. (D) Cells were starved overnight and stimulated with or without NRG-1 for indicated time periods. Phosphorylation of Erk, Akt, and p38 was analyzed by Western blotting with phosphospecific antibodies. Subsequently, membranes were reblotted with anti-Erk, anti-Akt, anti-p38, or anti-actin antibodies.
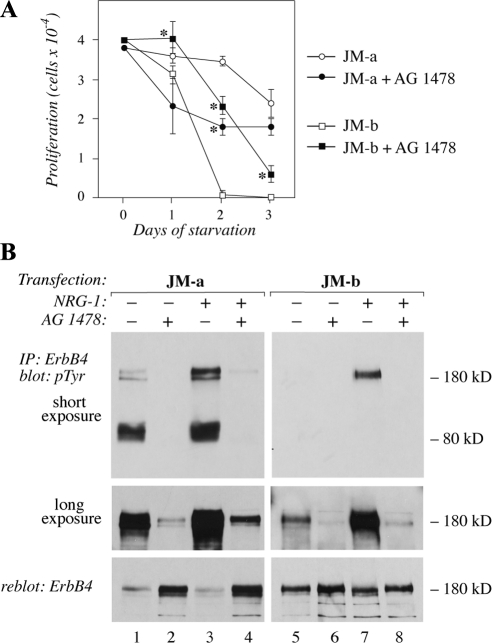
Figure 5.
The effect of inhibition of ErbB tyrosine kinase activity on cells expressing ErbB4 isoforms. (A) NR6 cells expressing JM-a CYT-2 or JM-b CYT-2 were plated onto 24-well plates in the presence of 10% FCS. The next day the medium was replaced with serum-free medium containing 0 or 10 μM of the ErbB kinase inhibitor AG 1478. Adherent cells were counted at indicated time points with hemocytometer. *p < 0.05 for a difference between AG 1478-treated and control cells. (B) Cells were starved overnight without serum, treated for 6 h with 0 or 10 μM AG 1478, and stimulated for 15 min with 0 or 50 ng/ml NRG-1. ErbB4 tyrosine phosphorylation was analyzed by immunoprecipitation with an anti-ErbB4 antibody followed by Western blotting with an anti-phosphotyrosine antibody. Two different exposures of the phosphotyrosine blot are shown. Membrane was reblotted with an anti-ErbB4 antibody. Binding of the anti-phosphotyrosine antibody 4G10 masks the epitope for the anti-ErbB4 sc-283, resulting in an apparently reduced ErbB4 signal for the heavily phosphorylated ErbB4 species in the reblot (lanes 1 and 3).
ErbB4 JM-a CYT-2 Has Efficient Autokinase Activity
Differential tyrosine phosphorylation of the two isoforms could result from differences in intrinsic kinase activities between the two proteins. To address the relative kinase activities, both isoforms were immunoprecipitated from NR6 cells and subjected to in vitro kinase assay in the presence of ATP. Both isoforms demonstrated basal autokinase activity (Figure 1C). However, the activity of the 180-kDa JM-a CYT-2 was on average twice the activity observed for 180-kDa JM-b CYT-2, after normalizing for ErbB4 protein expression. In addition, enhanced autokinase-stimulated phosphotyrosine content of the 80-kDa JM-a CYT-2 fragment was observed. However, chemical inhibition of ErbB4 cleavage by blocking either TACE or γ-secretase activity did not reduce the basal 180-kDa JM-a CYT-2 tyrosine phosphorylation, suggesting that proteolytic production of an active 80-kDa fragment was not necessary for phosphorylation of the full-length receptor (data not shown). These findings demonstrate a difference between kinase activities of the two ErbB4 isoforms.
Signal Transduction Pathways Activated by JM-a CYT-2 and JM-b CYT-2
Differences in the activation patterns and kinetics of signaling pathways, such as Mek/Erk pathway associate with different cellular responses, such as proliferation versus differentiation (Marshall, 1995). To test whether isoform-specific characteristics associated with differential intracellular signaling, NR6 transfectants were stimulated with NRG-1 and analyzed by Western blotting using phospho-specific antibodies for the MAPKs Erk1/2, and p38, as well as Akt (Figure 1D). Both isoforms were capable of activating Erk1/2, although the phosphorylation stimulated by JM-a CYT-2 seemed to be sustained somewhat longer. Neither of the isoforms activated p38. In line with earlier findings (Kainulainen et al., 2000), activation of Akt, a downstream target of PI3-K, was relatively weak by both of the CYT-2 isoforms lacking the direct PI3-K docking site (Elenius et al., 1999). These findings suggest that, with the exception of differences in the kinetics of Erk activation, the two ErbB4s did not significantly differ in stimulating some of the major signal transduction pathways involved in ErbB-regulated proliferation and survival.
ErbB4 JM-a CYT-2 Promotes Proliferation and Anchorage-independent Growth
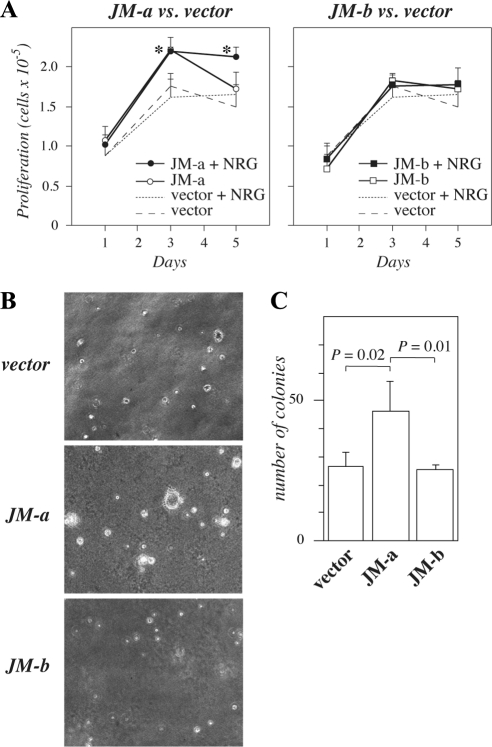
To test whether JM-a CYT-2 and JM-b CYT-2 expression affected proliferation, growth curves were generated for transfectants maintained in DMEM containing 5% FCS. An increase in proliferation was evident in cells expressing JM-a CYT-2 when compared with vector transfectants (Figure 2A). JM-b CYT-2 expression had no effect on cell numbers in the presence of 5% FCS (Figure 2A). To analyze the potential of the isoforms to induce anchorage-independent growth, transfectants were seeded in soft agar. Cells expressing JM-a CYT-2 formed significantly more colonies than cells expressing JM-b CYT-2 or vector control cells (Figure 2, B and C). These data suggest that ErbB4 JM-a CYT-2, but not JM-b CYT-2, promotes growth of NR6 cells in soft agar.
Figure 2.
Growth of NR6 cells expressing ErbB4 isoforms. (A) NR6 transfectants were plated onto 24-well plates in the presence of 10% FCS. The next day the medium was replaced with medium containing 5% FCS with or without 50 ng/ml NRG-1. Adherent cells were counted at indicated time points with hemocytometer. *p < 0.05 for a difference between ErbB4 overexpression versus vector in the presence of NRG-1. (B and C) Cells were cultured in soft agar for 7 wk. Representative images (B) and quantitation of colonies (C) are shown.
ErbB4 JM-b CYT-2 Promotes Starvation-induced Cell Death
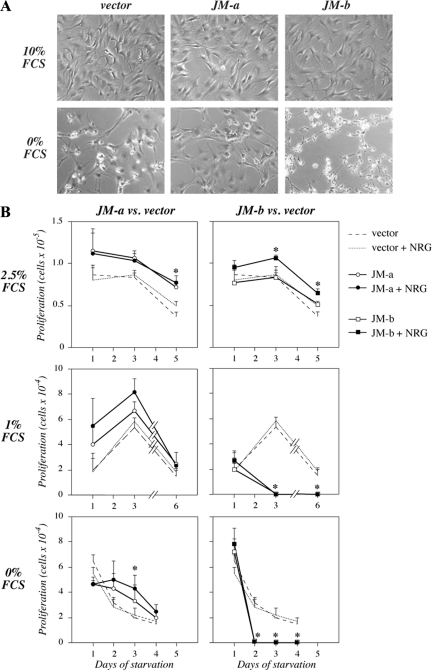
To compare the effect of ErbB4 isoform expression on cells progressing in cell cycle or induced into quiescence, NR6 transfectants were grown in the presence or absence of serum. All transfectants displayed normal fibroblast morphology when cultured in 10% FCS (Figure 3A). However, cells expressing JM-a CYT-2 preserved their normal morphology longer than vector control cells under serum-free conditions. Interestingly, cells expressing JM-b CYT-2 seemed to lose their spindle-shaped morphology and detach from the culture plates clearly before the vector control cells (Figure 3A). Consistent with the morphological differences, JM-a CYT-2 expression stimulated resistance to starvation-induced decline in cell numbers (Figure 3B). In contrast, expression of JM-b CYT-2 induced a rapid loss of all adherent cells when grown in medium containing 1 or 0% FCS (Figure 3B). Similar results were obtained with independent clones and when the cells were treated with different concentrations of the ErbB4 ligand NRG-1 (Supplemental Figure 2). Taken together, these data suggest that ErbB4 JM isoforms confer opposite cellular responses to serum deprivation: JM-a CYT-2 promotes resistance but JM-b CYT-2 sensitivity to cell death.
Figure 3.
The effect of serum starvation on cells expressing ErbB4 isoforms. (A) NR6 transfectants were cultured for 2 d in medium containing 0 or 10% of FCS and photographed. (B) NR6 transfectants were plated onto 24-well plates in the presence of 10% FCS. The next day the medium was replaced with medium containing 0, 1, or 2.5% FCS with or without 50 ng/ml NRG-1. Adherent cells were counted at indicated time points with hemocytometer. *p < 0.05 for a difference between ErbB4 overexpression versus vector in the presence of NRG-1.
Cells Expressing ErbB4 JM-b CYT-2 Have Characteristics of Apoptosis
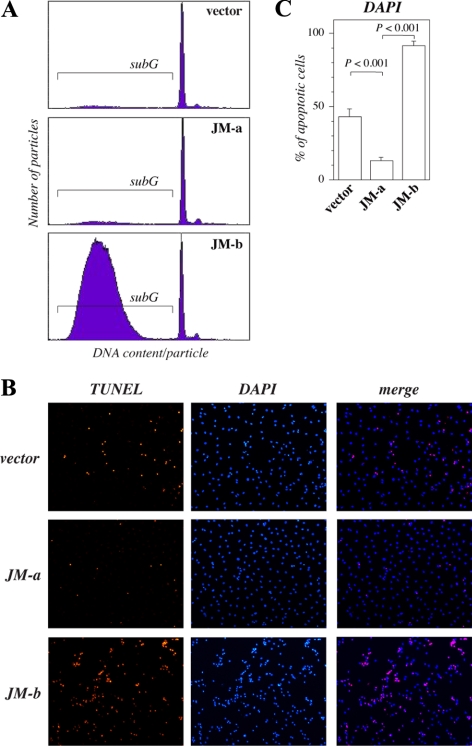
To characterize the mechanism leading to facilitated death of cells expressing ErbB4 JM-b CYT-2 under serum deprivation, DNA content of transfectants was analyzed by fluorescent-activated cell sorting (FACS). After 3 d of starvation, the subG1 population with small DNA content was considerably greater in cells expressing JM-b CYT-2 when compared with other transfectants (Figure 4A), consistent with apoptosis. Within 2 d after serum deprivation the nuclei in 43% of the vector control cells gained morphological characteristics of apoptosis (Figure 4, B and C). The percentage of cells with apoptotic changes was more than doubled to 91% in cells expressing JM-b CYT-2, but was only 13% in cells expressing JM-a CYT-2 (Figure 4, B and C). Condensed nuclei demonstrated by bright DAPI staining were also almost exclusively TUNEL positive (Figure 4B). These data indicate that cells expressing ErbB4 JM-b CYT-2 die because of apoptosis-like mechanisms in response to serum deprivation.
Figure 4.
Starvation-induced death of NR6 cells expressing ErbB4 isoforms. (A) Cells were cultured for 72 h in the absence of FCS, and their DNA contents were analyzed by PI staining and FACS. The proportion of cells containing subG1 quantities of DNA per particle is indicated. (B) Cells were stained with TUNEL (red) and DAPI (blue) after starvation for 2 d and photographed under a fluorescence microscope. (C) The percentage of condensed nuclei stained with DAPI was calculated from five arbitrary fields per cell line under a microscope.
Chemical ErbB Kinase Inhibitor Reverses Both ErbB4 Isoform-specific Responses to Serum Deprivation
To test whether ErbB4 kinase activity was necessary for the two opposite cellular responses stimulated by the two ErbB4 isoforms upon starvation, NR6 transfectants were cultured under serum-free conditions in the presence and absence of a chemical ErbB kinase inhibitor AG 1478. Addition of AG 1478 significantly inhibited the growth of cells expressing JM-a CYT-2 (Figure 5A). No significant effect on growth was observed in vector control cells (data not shown). Significantly, adding AG 1478 into cells expressing JM-b CYT-2 partially rescued them from apoptosis (Figure 5A). The effect of AG 1478 on blocking both basal and ligand-induced tyrosine phosphorylation of both ErbB4 isoforms was demonstrated by Western analysis (Figure 5B). These findings suggest that both the pro- and antisurvival responses were at least partially dependent on ErbB4 kinase activity. The observation also indicates that inhibition of ErbB4 kinase activity may result in either promotion or suppression of apoptosis, depending on the isoforms present on the targeted cell. Similar findings with AG 1478 were also made in analyses of MCF-7 breast cancer cell transfectants expressing either JM-a CYT-2 or JM-b CYT-2 isoforms (data not shown).
Regulation of PDGFRA Expression by ErbB4 Isoforms
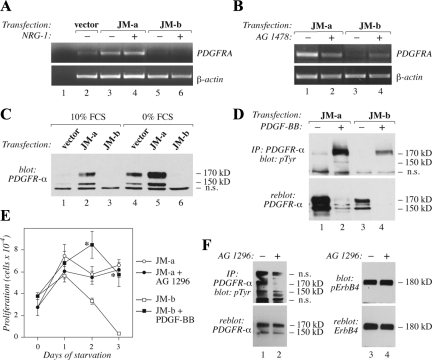
To identify molecular mechanisms underlying the different cellular responses promoted by the two ErbB4 isoforms, a cDNA microarray including 15,000 mouse cDNAs was screened for genes regulated in an ErbB4 isoform-specific manner. The cDNA array analysis indicated that ErbB4 JM-a CYT-2 and ErbB4 JM-b CYT-2 indeed regulated different sets of genes (Supplemental Table 2). One of the identified genes, PDGFRA, was up-regulated by JM-a CYT-2 but down-regulated by JM-b CYT-2. The finding was confirmed by RT-PCR analysis of PDGFRA mRNA (Figure 6A) and Western analysis of PDGFR-α protein (Figure 6C). Consistent with the regulation taking place at the level of transcription, no differences in the relative degradation rate of the PDGFR-α protein, or the expression of the known PDGFR E3 ubiquitin ligase, Cbl, were observed between the transfectants (data not shown).
Figure 6.
Regulation of PDGFRA expression by ErbB4 isoforms. (A) NR6 transfectants were cultured for 8 h in the absence of serum, stimulated for 2 h with or without 50 ng/ml NRG-1, and analyzed for PDGFRA mRNA expression by RT-PCR. (B) NR6 transfectants were treated with or without 10 μM AG 1478 for 8 h in the absence of serum. PDGFRA mRNA expression was analyzed by RT-PCR. (C) Cells were cultured overnight in the presence or absence of 10% FCS. PDGFR-α protein expression was analyzed by Western blotting with anti-PDGFR-α antibody. n.s., nonspecific band also present in the vector control lanes. (D) Cells were starved overnight without serum and stimulated for 10 min with 0 or 50 ng/ml PDGF-BB. PDGFR-α tyrosine phosphorylation was analyzed by immunoprecipitation with an anti-PDGFR-α antibody followed by Western blotting with an anti-phosphotyrosine antibody. Membrane was reblotted with an anti-PDGFR-α antibody. (E) NR6 transfectants were plated onto 24-well plates. The next day the medium was replaced with serum-free medium containing or not 20 μM of PDGFR inhibitor AG 1296 or 50 ng/ml PDGF-BB. Adherent cells were counted at indicated time points with hemocytometer. *p < 0.05 for a difference between AG 1296– or PDGF-BB–treated and control cells. (F) NR6 cells expressing JM-a CYT-2 were cultured for 16 h in the presence or absence of 20 μM AG 1296 in serum-free medium. PDGFR-α tyrosine phosphorylation was analyzed by immunoprecipitation with an anti-PDGFR-α antibody followed by Western blotting with an anti-phosphotyrosine antibody. Membrane was reblotted with an anti-PDGFR-α antibody. ErbB4 tyrosine phosphorylation was analyzed by Western blotting with a phospho-specific anti-ErbB4 antibody followed by reblotting with an anti-ErbB4 antibody.
Because the antagonistic cellular responses stimulated by the two ErbB4 isoforms were both sensitive to AG 1478, the effect of the kinase inhibitor was also tested on isoform-regulated PDGFRA mRNA expression (Figure 6B). As expected, AG 1478 reduced PDGFRA mRNA expression in cells expressing JM-a CYT-2 but induced PDGFRA expression in cells expressing JM-b CYT-2. Analysis of PDGFR-α protein expression in transfectants cultured overnight in the presence or absence of FCS also demonstrated that starvation alone induced PDGFR-α expression, and expression of JM-a CYT-2 facilitated this induction, whereas expression of JM-b CYT-2 suppressed it (Figure 6C). These data suggest that ErbB4 isoforms regulate different sets of genes and identify PDGFRA as one gene that is regulated in opposite directions by ErbB4 JM-a CYT-2 and JM-b CYT-2.
PDGFR-α Has a Functional Role on the Pathway Leading to Different Cellular Responses Downstream of the Two ErbB4 Isoforms
The amount of tyrosine phosphorylated PDGFR-α followed the changes in total PDGFR-α expression in the transfectants (Figure 6D), suggesting functional significance. To further evaluate the functional contribution of PDGFR-α in regulating different cellular responses to the two ErbB4 isoforms, serum-starved NR6 transfectants were treated with the PDGFR kinase inhibitor AG 1296 or with the PDGFR ligand PDGF-BB (Figure 6E). AG 1296 reduced the number of cells expressing JM-a CYT-2, whereas PDGF-BB rescued cells expressing ErbB4 JM-b CYT-2 from starvation-induced death. The effect of AG 1296 on blocking the tyrosine phosphorylation of PDGFR-α but not of ErbB4 JM-a was confirmed by Western analysis (Figure 6F).
Taken together, these findings indicate that ErbB4 isoforms may mediate opposite cellular responses. The data also suggest that differential regulation of PDGFRA gene is a central mechanism involved in isoform-specific regulation of cell behavior in fibroblasts.
Regulation of PDGFRA Promoter Activity by ErbB4
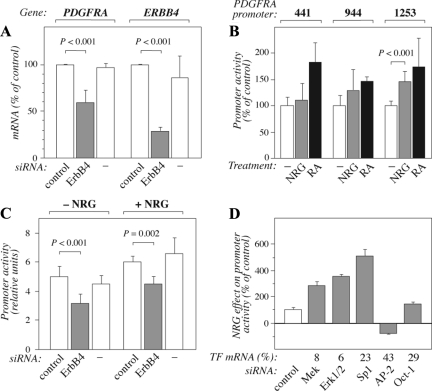
Targeting ERBB4 by RNA interference (73% knockdown as estimated by real-time RT-PCR from parallel samples) significantly suppressed the expression of PDGFRA mRNA in SK-N-MC neuroblastoma cells that naturally overexpress constitutively active ErbB4 JM-a isoforms (Figure 7A), demonstrating transcriptional regulation of PDGFRA expression also by endogenous ErbB4. To address the mechanism by which stimulation of endogenous ErbB4 JM-a regulates PDGFRA promoter activity, MCF-7 breast cancer cells (that express JM-a CYT-1 and JM-a CYT-2; Maatta et al., 2006) were transfected with luciferase reporter gene constructs encoding PDGFRA promoter fragments of different sizes. NRG-1 stimulation enhanced the promoter activity of the longest construct (1253 base pairs upstream of the transcription start site) by ∼50% (Figure 7B). An increase ranging between 45 and 85% was achieved with all the promoter constructs when cells were stimulated with 10 μM retinoic acid, a positive control known to promote PDGFRA expression (Figure 7B; Wang et al., 1990). The NRG-1–induced PDGFRA promoter activity in MCF-7 cells was blocked to the level of nonstimulated control by siRNA targeting endogenous ErbB4 (JM-a isoform-specific siRNA, 63% knockdown) but not by control siRNA (Figure 7C).
Figure 7.
Regulation of PDGFRA transcription by ErbB4 activity. (A) SK-N-MC human neuroblastoma cells were treated for 48 h with or without control or ErbB4-specific siRNAs. Relative PDGFRA and ERBB4 mRNA expression was analyzed by real-time RT-PCR and normalized to the expression of the reference gene β-actin. (B) MCF-7 breast cancer cells were transfected with the PDGFRA promoter luciferase reporter constructs pSLA4PDGFRαluc–441/+118 (441), pSLA4PDGFRαluc–944/+118 (944), or pSLA4PDGFRαluc–1253/+118 (1253) together with a plasmid encoding EGFP. Cells starved for 16 h were treated for 2 h with or without 80 ng/ml NRG-1 to activate ErbB4 or for 18 h with 10 μM retinoic acid (RA) as a positive control known to stimulate PDGFRA promoter activity. Columns represent relative luciferase activity of promoter constructs normalized by the EGFP fluorescence signal (no treatment = 100% for each experiment). (C) MCF-7 cells expressing pSLA4PDGFRαluc–1253/+118 were treated for 48 h with or without control or ErbB4-specific siRNAs, starved for 16 h, and treated for 2 h with 0 or 80 ng/ml NRG-1. Columns represent relative luciferase activity of promoter constructs normalized by the EGFP fluorescence signal. (D) MCF-7 cells expressing pSLA4PDGFRαluc–1253/+118 were treated for 48 h with control siRNA or siRNAs targeting the indicated transcription factors (TF), starved for 16 h and treated for 2 h with 0 or 80 ng/ml NRG-1. PDGFRA promoter activity was determined as in C. The values represent the effect of NRG-1 treatment as percentages of the control siRNA sample. The transcription factor mRNA expression levels, as determined by real-time RT-PCR, are shown relative to control siRNA treatment (TF mRNA %).
Previous reports have indicated that PDGFRA promoter contains putative binding sites for transcription factors such as Sp-1, AP-2, and Oct-1 (Afink et al., 1995; Kawagishi et al., 1995). To determine factors involved in ErbB4-mediated PDGFRA regulation, the effects of specific siRNAs were tested on NRG-1–stimulated PDGFRA promoter activity in MCF-7 cells (Figure 7D). siRNA targeting Oct-1 did not significantly affect the induction of the promoter activity by NRG-1, whereas AP-2α siRNA suppressed it compared with nonspecific control siRNA (Figure 7D). In contrast, the effect of NRG-1 was enhanced in the presence of siRNA targeting Sp1 by about fivefold (Figure 7D). Because Sp1-mediated suppression of PDGFRA promoter activity in response to fibroblast growth factor-2 (FGF-2) has been shown to involve the Mek/Erk-pathway (Bonello and Khachigian, 2004), siRNAs targeting Mek1 or Erk1/2 were also tested. Down-regulation of either Mek1 or Erk transcripts led to a similar about threefold induction in NRG-1–induced PDGFRA promoter activity (Figure 7D). Taken together these data suggest that, although the net effect of stimulating endogenous PDGFRA promoter by activating ErbB4 JM-a was positive, the promoter was regulated by both repressive as well as enhancing signals.
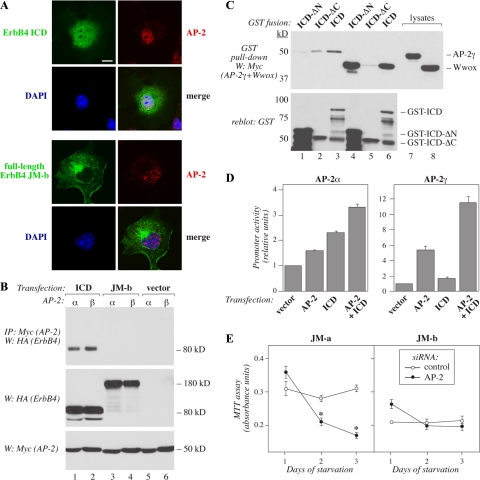
AP-2 Solely Interacts with Soluble ICD of ErbB4
As AP-2 was identified as a transcription factor positively regulating PDGFRA promoter in cells expressing ErbB4 JM-a (Figure 7D), and the cleavable ErbB4 isoform was unique in promoting PDGFRA transcription (Figure 6A), a possible colocalization of AP-2 with the soluble ErbB4 ICD was addressed. Confocal microscopy of COS-7 transfectants demonstrated a partial colocalization of ErbB4 ICD, but not of full-length noncleavable ErbB4 JM-b, with AP-2α in the nuclei (Figure 8A). Furthermore, both AP-2α as well as AP-2γ associated with ErbB4 ICD but not with full-length ErbB4 JM-b in coprecipitation experiments (Figure 8B). GST pulldown experiments confirmed an interaction between AP-2γ and the N-terminal kinase region of ErbB4 ICD (Figure 8C, lanes 1–3). As a control demonstrating a previously characterized interaction (Aqeilan et al., 2005), Wwox was shown to bind to the C-terminal part of ICD (Figure 8C, lanes 4–6). Consistent with the effects of AP-2–specific RNA interference on PDGFRA promoter activity (Figure 7D), both AP-2α and AP-2γ increased the potential of ErbB4 ICD to stimulate PDGFRA promoter activity in HEK-293T transfectants (Figure 8D). Interestingly, coexpression of AP-2 with nonstimulated full-length ErbB4 did not result in enhanced promoter activity (data not shown).
Figure 8.
Selective and functional association of soluble ErbB4 ICD with AP-2. (A) COS-7 cells were transfected with pcDNA3.1ICD2-HA or pcDNA3.1ErbB4JM-bCYT-2-HA, stained with anti-HA (green) and anti-AP-2α (red) antibodies. The nuclei were stained with DAPI (blue). The cells were visualized by confocal microscopy. Bar, 10 μm. (B) COS-7 cells were transiently transfected with constructs encoding HA-tagged ErbB4 JM-b CYT-2 or ErbB4 ICD2 and Myc-tagged AP-2α or AP-2-γ. Lysates were immunoprecipitated with anti-Myc antibody and anti-HA antibody was used in Western blotting. Expression levels were controlled by Western analysis with anti-HA and anti-Myc antibodies. (C) GST fusion proteins including the whole intracellular domain of the CYT-2-type (ICD), N-terminally truncated ICD (ICD-ΔN), or C-terminally truncated ICD (ICD-ΔC) were incubated with lysates of COS-7 cells transiently expressing Myc-tagged AP-2γ or Wwox. Material precipitating with glutathione Sepharose beads was analyzed by Western blotting with anti-Myc antibody. Membranes were reblotted with anti-GST antibody. (D) HEK-293T cells were cotransfected with the PDGFRA promoter-luciferase construct pSLA4PDGFRαluc–1253/+118 and a construct encoding Renilla luciferase together with an empty vector, constructs encoding the ErbB4 ICD of CYT-2-type (ICD), AP-2α, or AP-2γ, or a combination of constructs encoding ICD and AP-2. Columns represent relative luciferase activity of the PDGFRA promoter construct normalized by fluorescence signal from the Renilla luciferase. (E) NR6 cells stably expressing JM-a CYT-2 or JM-b CYT-2 were transfected with siRNA targeting AP-2α or with a nonsilencing control siRNA. After siRNA transfection the cells were starved for 3 d, and the number of viable cells was determined with MTT assay. *p < 0.05.
To address the functional significance of AP-2 on promoting cellular growth downstream of the different ErbB4 isoforms, number of viable serum-starved NR6 transfectants was estimated by MTT assays on 96-well plates in the presence of siRNA targeting AP-2α or nonsilencing siRNA control. The siRNA targeting AP-2α (42% knockdown) suppressed the number of cells expressing JM-a CYT-2 but had no significant effect on cells expressing JM-b CYT-2 (Figure 8E), consistent with the inability to JM-b CYT-2 to partially colocalize (Figure 8A) and associate with (Figure 8B) with AP-2.
These findings indicate that AP-2 promotes PDGFRA transcription downstream of ErbB4 JM-a by directly interacting with the soluble ICD in the nucleus. These data also provide a mechanistic explanation for the observation that only the cleavable JM-a CYT-2 isoform promoted PDGFRA transcription.
DISCUSSION
ErbB1 and ErbB2 receptors have successfully been used as cancer drug targets in the clinic (Hynes and MacDonald, 2009). However, the biological role of ErbB4 and its potential applicability as a cancer drug target has remained unclear (Sundvall et al., 2008a; Hollmen and Elenius, 2010). Currently, there is no consensus about the cellular responses stimulated via ErbB4. Several lines of evidence suggest that ErbB4 induces differentiation (Chen et al., 1996; Jones et al., 1999) or apoptosis (Arasada and Carpenter, 2005; Naresh et al., 2006). However, ErbB4 has also been documented to promote proliferation and tumor growth (Tang et al., 1999; Alaoui-Jamali et al., 2003; Hollmen et al., 2009). One possible explanation for the different conclusions is that the ERBB4 gene is spliced into four functionally unique isoforms (Junttila et al., 2003; Maatta et al., 2006; Muraoka-Cook et al., 2009), and most available data has been produced using undefined reagents or analyzing different isoforms.
Here, two of the ErbB4 isoforms, JM-a CYT-2 and JM-b CYT-2, were overexpressed in NR6 cells, and their signaling responses were compared. JM-a CYT-2 has previously been shown to promote survival and proliferation in myeloid and breast cancer cells (Junttila et al., 2005; Maatta et al., 2006). JM-a CYT-2 promoted survival also in NR6 fibroblasts upon serum deprivation, but interestingly, JM-b CYT-2 induced cell death. Consistent with differential roles also in vivo, JM-a isoforms are frequently present in various types of epithelial carcinomas at relatively high quantities, whereas the JM-b isoforms are typically absent (Junttila et al., 2005; Hollmen and Elenius, 2010; data not shown). In addition, there is a selective up-regulation of the JM-a isoforms in pediatric ependymoma when compared with normal brain tissue (Gilbertson et al., 2002). These findings demonstrate that ErbB4 isoforms may have significantly different biological activities and indicate a selection pressure favoring expression of the survival-promoting JM-a isoform in malignant cells.
The known functional difference between the JM-a CYT-2 and JM-b CYT-2 isoforms is that the 23 unique amino acids in the extracellular juxtamembrane region of JM-a provide a proteinase cleavage site that is missing from the alternative 13 amino acids of JM-b (Elenius et al., 1997). As a consequence, only isoforms of the JM-a type may signal via a mechanism involving a two-step proteolytic cleavage generating a soluble ICD (Ni et al., 2001; Maatta et al., 2006). In accordance, only NR6 transfectants expressing JM-a CYT-2 demonstrated efficient ligand-independent accumulation of a highly tyrosine-phosphorylated and kinase active 80-kDa proteolytic fragment. These observations implicate a role for the release of the soluble ICD solely from the JM-a CYT-2 isoform as a mechanism causally involved in the differential cellular functions promoted by the two isoforms. Addressing the significance of ErbB4 cleaving enzymes on the cellular responses of NR6 transfectants using inhibitors of either TACE or γ-secretase was precluded by the significant background effects of these compounds on the parental NR6 cell line (data not shown). Consistent with the role of JM-a isoform cleavage in promoting growth, however, an antibody specifically recognizing the JM-a isoform and preventing its cleavage suppresses the growth of breast cancer cells (Hollmen et al., 2009).
To further characterize the molecular mechanisms underlying the functional differences between the cleavable and noncleavable ErbB4 isoforms, gene expression patterns of NR6 transfectants were compared using cDNA microarrays. The analysis indicated PDGFRA as one of the target genes that was differentially regulated by JM-a CYT-2 and JM-b CYT-2. Experiments with a chemical inhibitor of the PDGFR-α kinase that suppressed the survival effect of JM-a CYT-2 further suggested a functional link between PDGFR-α up-regulation and ErbB4 JM-a CYT-2 expression. In addition, PDGF-BB, an agonist of PDGFR-α, rescued cells from JM-b CYT-2–induced death. Interestingly, FCS used to supplement cell culture media is known to be a rich source of PDGF ligands (Ross et al., 1974). This may indicate that the survival effects of ErbB4 isoforms were only observed after serum starvation as the lack of medium-derived PDGF sensitized cells to regulated PDGFR expression. Serum starvation alone also up-regulated PDGFR-α expression in the vector control NR6 cells, putatively as an adaptation to low extracellular ligand concentration (Lih et al., 1996), and this up-regulation was further enhanced by the presence of ErbB4 JM-a CYT-2, but was reversed by JM-b CYT-2. Previously NRG-1 has been shown to inhibit PDGF-BB–stimulated vascular smooth muscle cell functions (Clement et al., 2007), but a direct role of ErbB4 in regulation of PDGFR has not been described. Our data indicate PDGFRA as one of the target molecules differently regulated by ErbB4 isoforms and suggest a major role for it in ErbB4 isoform specific signaling responses leading to distinct behavior of the NR6 transfectants.
PDGFRA promoter assays in the presence and absence of siRNAs targeting transcription factors with suggested interactions with the PDGFRA promoter (Afink et al., 1995; Kawagishi et al., 1995) identified AP-2 as a factor positively regulating PDGFRA transcription. The specific association of AP-2 with the cleaved ICD derived from ErbB4 JM-a was indicated as the soluble ICD but not full-length ErbB4 1) partially colocalized with AP-2 in the nucleus, 2) interacted with AP-2 in coprecipitation and GST pull-down assays, and 3) had a synergistic effect with AP-2 on enhancing PDGFRA promoter activity. Moreover, 4) siRNA targeting AP-2 efficiently blocked the survival of cells expressing the cleavable JM-a CYT-2 but not of cells expressing JM-b CYT-2. Both AP-2α and AP-2γ associated with ErbB4 ICD, although the transcriptional synergism between the ICD and AP-2γ seemed to be stronger compared with AP-2α. In contrast, full-length ErbB4 JM-b not capable of releasing a soluble ICD fragment, did not demonstrate colocalization or association with AP-2, and the viability of cells expressing the JM-b isoform was not significantly affected by siRNA targeting AP-2. These findings support a model in which the survival-promoting effect of the JM-a CYT-2 isoform involves up-regulation of PDGFRA transcription via a unique and direct interaction of the released ICD fragment in the nucleus with AP-2. The observations also imply that the survival-promoting activity of the soluble ICD may, in the case of cleavable ErbB4 JM-a, counterbalance death-promoting signaling pathways, such as the Mek/Erk pathway, stimulated by all full-length ErbB4 forms at the cell surface. Indeed, our unpublished observations indicate that targeting of Mek significantly rescues cells expressing ErbB4 JM-b from starvation-induced death.
In conclusion, we provide evidence of opposite cellular functions promoted by alternatively spliced JM isoforms of ErbB4. Recently, similar observations of dramatically different cellular responses were reported for another pair of ErbB4 isoforms, the CYT isoforms, which promote either proliferation or differentiation in the mouse mammary gland (Muraoka-Cook et al., 2009). Our findings with the AG 1478 compound indicated that a wide-spectrum ErbB kinase inhibitor could promote either cell death or survival depending on the type of ErbB4 isoform present. These data underline the importance of investigating the basic cancer biology of ErbB4 isoforms. Moreover, the findings indicate that differential consequences of blocking different ErbB4 isoforms should be taken into consideration when developing novel diagnostic tests and therapeutics for targeting ErbB receptors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maria Tuominen, Minna Santanen, and Mika Savisalo for excellent technical assistance and Ilkka Paatero for help with RNA interference experiments. This work has been supported by the Academy of Finland, Emil Aaltonen Foundation, EU-Marie Curie Reintegration Grant, Finnish Cancer Organizations, Finnish Medical Society Duodecim, Foundation for the Finnish Cancer Institute, Jenny ja Antti Wihuri Foundation, K. Albin Johansson Foundation, Sigrid Jusélius Foundation, Paulo Foundation, Turku University Central Hospital, and Turku University Foundation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0332) on October 13, 2010.
REFERENCES
- Afink G. B., Nister M., Stassen B. H., Joosten P. H., Rademakers P. J., Bongcam-Rudloff E., Van Zoelen E. J., Mosselman S. Molecular cloning and functional characterization of the human platelet-derived growth factor alpha receptor gene promoter. Oncogene. 1995;8:1667–1672. [PubMed] [Google Scholar]
- Alaoui-Jamali M. A., Song D. J., Benlimame N., Yen L., Deng X., Hernandez-Perez M., Wang T. Regulation of multiple tumor microenvironment markers by overexpression of single or paired combinations of ErbB receptors. Cancer Res. 2003;13:3764–3774. [PubMed] [Google Scholar]
- Aqeilan R. I., Donati V., Palamarchuk A., Trapasso F., Kaou M., Pekarsky Y., Sudol M., Croce C. M. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;15:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- Aqeilan R. I., Palamarchuk A., Weigel R. J., Herrero J. J., Pekarsky Y., Croce C. M. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004a;22:8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- Aqeilan R. I., et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc. Natl. Acad. Sci. USA. 2004b;13:4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R. R., Carpenter G. Secretase-dependent tyrosine phosphorylation of Mdm2 by the ErbB-4 intracellular domain fragment. J. Biol. Chem. 2005;35:30783–30787. doi: 10.1074/jbc.M506057200. [DOI] [PubMed] [Google Scholar]
- Bonello M. R., Khachigian L. M. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J. Biol. Chem. 2004;4:2377–2382. doi: 10.1074/jbc.M308254200. [DOI] [PubMed] [Google Scholar]
- Carey K. D., Garton A. J., Romero M. S., Kahler J., Thomson S., Ross S., Park F., Haley J. D., Gibson N., Sliwkowski M. X. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;16:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- Chazin V. R., Kaleko M., Miller A. D., Slamon D. J. Transformation mediated by the human HER-2 gene independent of the epidermal growth factor receptor. Oncogene. 1992;9:1859–1866. [PubMed] [Google Scholar]
- Chen X., Levkowitz G., Tzahar E., Karunagaran D., Lavi S., Ben-Baruch N., Leitner O., Ratzkin B. J., Bacus S. S., Yarden Y. An immunological approach reveals biological differences between the two NDF/heregulin receptors, ErbB-3 and ErbB-4. J. Biol. Chem. 1996;13:7620–7629. [PubMed] [Google Scholar]
- Cheng Q. C., Tikhomirov O., Zhou W., Carpenter G. Ectodomain cleavage of ErbB-4, characterization of the cleavage site and m80 fragment. J. Biol. Chem. 2003;40:38421–38427. doi: 10.1074/jbc.M302111200. [DOI] [PubMed] [Google Scholar]
- Clement C. M., Thomas L. K., Mou Y., Croslan D. R., Gibbons G. H., Ford B. D. Neuregulin-1 attenuates neointimal formation following vascular injury and inhibits the proliferation of vascular smooth muscle cells. J. Vasc. Res. 2007;4:303–312. doi: 10.1159/000101776. [DOI] [PubMed] [Google Scholar]
- Cohen B. D., Kiener P. A., Green J. M., Foy L., Fell H. P., Zhang K. The relationship between human epidermal growth-like factor receptor expression and cellular transformation in NIH3T3 cells. J. Biol. Chem. 1996;48:30897–30903. doi: 10.1074/jbc.271.48.30897. [DOI] [PubMed] [Google Scholar]
- Elenius K., Choi C. J., Paul S., Santiestevan E., Nishi E., Klagsbrun M. Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene. 1999;16:2607–2615. doi: 10.1038/sj.onc.1202612. [DOI] [PubMed] [Google Scholar]
- Elenius K., Corfas G., Paul S., Choi C. J., Rio C., Plowman G. D., Klagsbrun M. A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem. 1997;42:26761–26768. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- Feng S. M., Muraoka-Cook R. S., Hunter D., Sandahl M. A., Caskey L. S., Miyazawa K., Atfi A., Earp H. S., 3rd The E3 ubiquitin ligase WWP1 selectively targets HER4 and its proteolytically derived signaling isoforms for degradation. Mol. Cell. Biol. 2009;3:892–906. doi: 10.1128/MCB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson R. J., et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin. Cancer Res. 2002;10:3054–3064. [PubMed] [Google Scholar]
- Hollmen M., Elenius K. Potential of ErbB4 antibodies for cancer therapy. Future Oncol. 2010;1:37–53. doi: 10.2217/fon.09.144. [DOI] [PubMed] [Google Scholar]
- Hollmen M., Maatta J. A., Bald L., Sliwkowski M. X., Elenius K. Suppression of breast cancer cell growth by a monoclonal antibody targeting cleavable ErbB4 isoforms. Oncogene. 2009;10:1309–1319. doi: 10.1038/onc.2008.481. [DOI] [PubMed] [Google Scholar]
- Hynes N. E., MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;2:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Iivanainen E., et al. Angiopoietin-regulated recruitment of vascular smooth muscle cells by endothelial-derived heparin binding EGF-like growth factor. FASEB J. 2003;12:1609–1621. doi: 10.1096/fj.02-0939com. [DOI] [PubMed] [Google Scholar]
- Jones F. E. HER4 intracellular domain (4ICD) activity in the developing mammary gland and breast cancer. J. Mammary Gland Biol. Neoplasia. 2008;2:247–258. doi: 10.1007/s10911-008-9076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. E., Welte T., Fu X. Y., Stern D. F. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 1999;1:77–88. doi: 10.1083/jcb.147.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila T. T., Laato M., Vahlberg T., Soderstrom K. O., Visakorpi T., Isola J., Elenius K. Identification of patients with transitional cell carcinoma of the bladder overexpressing ErbB2, ErbB3, or specific ErbB4 isoforms: real-time reverse transcription-PCR analysis in estimation of ErbB receptor status from cancer patients. Clin. Cancer Res. 2003;14:5346–5357. [PubMed] [Google Scholar]
- Junttila T., Sundvall M., Lundin M., Lundin J., Tanner M., Härkönen P., Joensuu H., Isola J., Elenius K. Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 2005;4:1384–1393. doi: 10.1158/0008-5472.CAN-04-3150. [DOI] [PubMed] [Google Scholar]
- Junttila T., Sundvall M., Määttä J., Elenius K. Erbb4 and its isoforms: selective regulation of growth factor responses by naturally occurring receptor variants. Trends Cardiovasc. Med. 2000;7:304–310. doi: 10.1016/s1050-1738(01)00065-2. [DOI] [PubMed] [Google Scholar]
- Kainulainen V., Sundvall M., Määttä J., Santiestevan E., Klagsbrun M., Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J. Biol. Chem. 2000;12:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- Kawagishi J., Kumabe T., Yoshimoto T., Yamamoto T. Structure, organization, and transcription units of the human alpha-platelet-derived growth factor receptor gene, PDGFRA. Genomics. 1995;2:224–232. doi: 10.1006/geno.1995.9883. [DOI] [PubMed] [Google Scholar]
- Komuro A., Nagai M., Navin N. E., Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;35:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Jung K. M., Huang Y. Z., Bennett L. B., Lee J. S., Mei L., Kim T. W. Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 2002;8:6318–6323. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
- Lih C. J., Cohen S. N., Wang C., Lin-Chao S. The platelet-derived growth factor alpha-receptor is encoded by a growth-arrest-specific (gas) gene. Proc. Natl. Acad. Sci. USA. 1996;10:4617–4622. doi: 10.1073/pnas.93.10.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatta J. A., Sundvall M., Junttila T. T., Peri L., Laine V. J., Isola J., Egeblad M., Elenius K. Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol. Biol. Cell. 2006;1:67–79. doi: 10.1091/mbc.E05-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;2:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Muraoka-Cook R. S., et al. Heregulin-dependent delay in mitotic progression requires HER4 and BRCA1. Mol. Cell. Biol. 2006a;17:6412–6424. doi: 10.1128/MCB.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook R. S., Sandahl M., Husted C., Hunter D., Miraglia L., Feng S. M., Elenius K., Earp H. S., 3rd. The intracellular domain of ErbB4 induces differentiation of mammary epithelial cells. Mol. Biol. Cell. 2006b;9:4118–4129. doi: 10.1091/mbc.E06-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook R. S., Sandahl M. A., Strunk K. E., Miraglia L. C., Husted C., Hunter D. M., Elenius K., Chodosh L. A., Earp H. S., 3rd. ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol. Cell Biol. 2009;18:4935–4948. doi: 10.1128/MCB.01705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh A., Long W., Vidal G. A., Wimley W. C., Marrero L., Sartor C. I., Tovey S., Cooke T. G., Bartlett J. M., Jones F. E. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;12:6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- Ni C. Y., Murphy M. P., Golde T. E., Carpenter G. gamma-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;5549:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Nikula T., West A., Katajamaa M., Lonnberg T., Sara R., Aittokallio T., Nevalainen O. S., Lahesmaa R. A human immunochip cDNA microarray provides a comprehensive tool to study immune responses. J. Immunol. Methods. 2005;1–2:122–134. doi: 10.1016/j.jim.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Omerovic J., et al. The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J. 2007;11:2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- Pruss R. M., Herschman H. R. Variants of 3T3 cells lacking mitogenic response to epidermal growth factor. Proc. Natl. Acad. Sci. USA. 1977;9:3918–3921. doi: 10.1073/pnas.74.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio C., Buxbaum J. D., Peschon J. J., Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J. Biol. Chem. 2000;14:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- Ross R., Glomset J., Kariya B., Harker L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc. Natl. Acad. Sci. USA. 1974;4:1207–1210. doi: 10.1073/pnas.71.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi S. P., Murtie J., Koirala S., Patten B. A., Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;1:185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- Sundvall M., Iljin K., Kilpinen S., Sara H., Kallioniemi O. P., Elenius K. Role of ErbB4 in breast cancer. J. Mammary Gland Biol. Neoplasia. 2008a;2:259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- Sundvall M., Korhonen A., Paatero I., Gaudio E., Melino G., Croce C. M., Aqeilan R. I., Elenius K. Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc. Natl. Acad. Sci. USA. 2008b;11:4162–4167. doi: 10.1073/pnas.0708333105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M., Peri L., Maatta J. A., Tvorogov D., Paatero I., Savisalo M., Silvennoinen O., Yarden Y., Elenius K. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene. 2007;48:6905–6914. doi: 10.1038/sj.onc.1210501. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Concepcion X. Z., Milan M., Gong X., Montgomery E., Lippman M. E. Ribozyme-mediated down-regulation of ErbB-4 in estrogen receptor-positive breast cancer cells inhibits proliferation both in vitro and in vivo. Cancer Res. 1999;20:5315–5322. [PubMed] [Google Scholar]
- Vidal G. A., Naresh A., Marrero L., Jones F. E. Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J. Biol. Chem. 2005;20:19777–19783. doi: 10.1074/jbc.M412457200. [DOI] [PubMed] [Google Scholar]
- Wang C., Kelly J., Bowen-Pope D. F., Stiles C. D. Retinoic acid promotes transcription of the platelet-derived growth factor alpha-receptor gene. Mol. Cell. Biol. 1990;12:6781–6784. doi: 10.1128/mcb.10.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. C., Allison J. G., Vidal G. A., Burow M. E., Beckman B. S., Marrero L., Jones F. E. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 2004;3:469–478. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Xu J., Harris R. C. Nedd4 mediates ErbB4 JM-a/CYT-1 ICD ubiquitination and degradation in MDCK II cells. FASEB J. 2009;6:1935–1945. doi: 10.1096/fj.08-121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.