Abstract
Pathogens are detected by pattern recognition receptors that, upon activation, orchestrate an appropriate immune response. The TLRs and the nucleotide-binding oligomerization domain-like receptors (NLRs) are prototypic pattern recognition receptors that detect extracellular and cytosolic pathogens, respectively. Listeria monocytogenes has both extracellular and cytosolic phases and is detected in the cytosol by members of the NLR family. These include two NLR members, NLRC4 and NLRP3, that, upon detection of cytosolic L. monocytogenes, induce the assembly of the inflammasome. Inflammasomes serve as platforms for the activation of the protease caspase 1, which mediates the processing and secretion of pro–IL-1β and pro–IL-18. We previously provided evidence that L. monocytogenes is also detected by a third inflammasome. We now use biochemical and genetic approaches to demonstrate that the third detector senses bacterial DNA and identify it as Aim2, a receptor that has previously been shown to detect viral DNA.
Macrophages have multiple classes of pattern recognition receptors that mediate a variety of immune responses. Activation of TLRs on the cell surface results in the induction and release of many cytokines, including TNF, IL-6, and IL-12 (1). TLRs also induce the production but not secretion of pro–IL-1β and pro–IL-18. Secretion of these molecules only occurs after proteolytic processing by caspase 1, a component of the nucleotide-binding oligomerization domain-like receptor (NLR)-induced inflammasome (2, 3). NLRs are cytosolic pattern recognition receptors that contain a protein interaction domain (either a caspase recruitment domain [CARD] or Pyrin domain), a nucleotide binding site, and leucine-rich repeats. Upon activation, NLRs oligomerize and recruit procaspase 1 via the CARD domain, either directly through CARD–CARD interactions or indirectly through pyrin–pyrin interactions with the adaptor protein apoptotic speck protein containing a CARD (ASC), whose CARD domain then recruits procaspase 1. The assembled inflammasome forms a platform that mediates caspase 1 activation.
Listeria monocytogenes is a Gram-positive bacterium acquired through ingestion of contaminated food. It infects and replicates within a variety of cells types, including macrophages. After postphagocytosis, L. monocytogenes uses listeriolysin O (LLO) to degrade the vacuolar membrane and escape into cytosol (4). Infection propagates as L. monocytogenes uses actin-based motility to facilitate its entry into a neighboring cell (5). Cytosolic L. monocytogenes activates caspase-1 via NLRP3 and NLRC4 (6, 7). NLRP3 is a pyrin-containing NLR that requires the adaptor ASC. Although the specific agonist has not been defined, NLRP3 is known to be activated when L. monocytogenes escapes the vacuole (8). In contrast to NLRP3, NLRC4 is a CARD-containing protein that can interact with caspase-1 in an ASC-independent manner (9, 10). NLRC4 is activated by flagellin, a component of the bacterial flagellum (10).
We previously presented data that suggested the presence of a third cytosolic receptor for L. monocytogenes (6). This sensor was ASC-dependent, suggesting that it contains a pyrin domain (6). We demonstrate in this study that Aim2, a recently identified cytosolic DNA receptor (11-14), is the third sensor. Aim2 binds DNA (or its synthetic analog, polyA:T) directly through its HIN200 domain and ASC via its pyrin domain. This results in the formation of an atypical inflammasome because Aim2 lacks an oligomerization and a leucine-rich repeat domain and thus is not an NLR. We further demonstrate that Aim2 detects cytosolic L. monocytogenes DNA.
Materials and Methods
Bacterial and mammalian cell culture
Bone marrow-derived macrophages were cultured in 50% DMEM with 30% FBS, 20% L929 cell supernatant, penicillin, and streptomycin for 6 d before infection. Wild-type (WT) L. monocytogenes 10403s and LLO-deficient (DLLO) strain were gifts from Dan Portnoy (University of California, Oakland, CA).
Macrophage infection
Macrophages were stimulated with 50 ng/ml ultrapure LPS (List Biological Laboratories, Campbell, CA) for 3 h at 37°C to induce pro–IL-1β expression. L. monocytogenes were grown to midlog at ambient temperature. At time of infection, cells were spun at 1000 rpm (250 × g) for 5 min to increase contact between macrophages and L. monocytogenes. Macrophages were then incubated for 1 h before the addition of LPS and gentamicin to kill extracellular bacteria. Cells were then incubated for 3 h before supernatants were harvested.
Bacterial lysate preparation
WT L. monocytogenes was grown to stationary phase at 37°C with shaking. Bacteria were pelleted, washed once in PBS, and resuspended in 50 ml PBS. Bacteria were then passed twice through a French press at 1000 psi to lyse the bacteria. Lysates were centrifuged at 17,000 rpm to remove insoluble fraction.
Microscopy
Immortalized NLRP3/NLRC4DKO macrophages expressing ASC-V5 were treated with LPS. Log-phase L. monocytogenes was stained with Hoechst or DAPI stain at 50 μg/ml for 15 min at 37°C then washed five times in PBS. Macrophages were then infected with L. monocytogenes. After 4 h of infection, ASC was visualized with anti-V5. Listeria surface Ags were stained with anti-Listeria Ab (Virostat 4201, Virostat, Portland, ME).
Retroviral transduction of short hairpin RNA
Short hairpin RNA (shRNA) from Open Biosystems (Huntsville, AL; Aim2A: RMM1766-6343887; Aim2B: RM1776-9344596; Aim2C: RMM1766-9353444; control TLR5: RMM1766-96882065) was cloned into the LMP retroviral vector and transduced into immortalized WT or NLRP3/NLRC4DKO macrophages. Transduced cells were selected with puromycin.
Results and Discussion
Cytosolic L. monocytogenes activates IL-1β secretion independent of NLRP3 and NLRC4
We previously demonstrated that cytosolic L. monocytogenes induces inflammasome activation via three signaling pathways, including NLRP3, NLRC4, and a third unknown sensor pathway, with a combination of bacterial mutants and gene deletion macrophages (6). By using the aflagellate ΔflaA mutant L. monocytogenes (to eliminate NLRC4 detection), we demonstrated that NLRP3−/− macrophages responded to L. monocytogenes, whereas ASC−/− macrophages did not. This indicated that the third sensor acts through ASC. To assess the relative contribution of each detector, we generated NLRP3/NLRC4DKO mice to specifically remove the two established sensors without altering the ASC adaptor protein or the infecting L. monocytogenes. In agreement with our previous data, NLRP3 mediated detection was predominant at 1 h postinfection (Fig. 1A), and the contribution of NLRC4 increased over time (Fig. 1B, 1C). By using NLRP3/NLRC4DKO cells, we determined that the third sensor induced IL-1β processing and secretion late, similar to NLRC4 (Fig. 1B–D). Further, we verified that this third sensor acts through ASC, as there was no processing or secretion of IL-1β in NLRC4/ASCDKO cells (Fig. 1). These results demonstrate that cytosolic L. monocytogenes is detected by a third sensor that signals through ASC and activates caspase 1.
FIGURE 1.
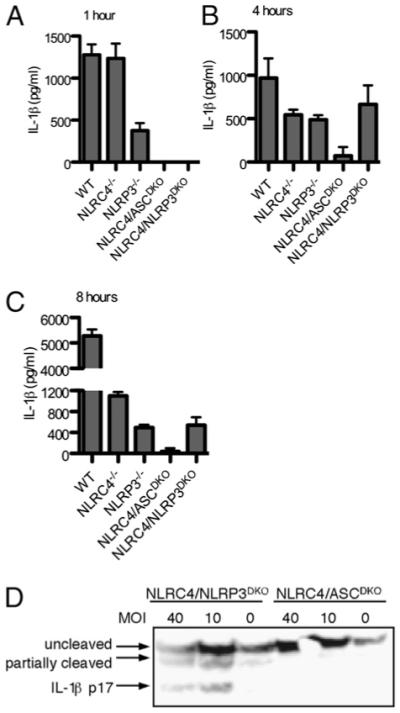
A third sensor activates IL-1β secretion following L. monocytogenes infection. LPS-primed macrophages were infected with L. monocytogenes at a multiplicity of infection 5 for 1 h (A), 4 h (B), or 8 h (C), and IL-1β secretion was determined by ELISA. D, IL-1β processing was determined by Western blot 4 h postinfection.
The third sensor detects L. monocytogenes DNA in the cytosol. To identify the agonist that activates this third sensor, we transfected extracts from lysed L. monocytogenes into macrophages and examined the IL-1β response (Fig. 2). Macrophages secreted IL-1β in response to crude soluble extracts from L. monocytogenes after transfection into the cytosol. To further investigate the class of compound that was detected, we treated the extracts with either DNase or trypsin. Although protein digestion did not alter the IL-1β secretion observed, DNA digestion completely abrogated the response (Fig. 2A). We confirmed that DNA was the active component of the L. monocytogenes lysate by demonstrating that transfected genomic DNA purified from L. monocytogenes induced IL-1β secretion by WT and NLRP3/NLRC4DKO macrophages (Fig. 2B). The secretion of IL-1β occurred in a dose-dependent manner. In agreement with our infection data, this detection pathway requires ASC signaling as ASC/NLRC4DKO macrophages showed no IL-1β secretion (Fig. 2B). These data indicate that NLRP3/NLRC4DKO macrophages can respond to L. monocytogenes DNA that accesses the cytosolic compartment of macrophages.
FIGURE 2.
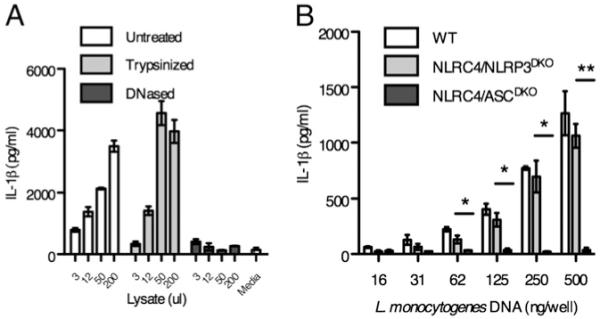
L. monocytogenes DNA induces IL-1β secretion. A, L. monocytogenes lysate was mock treated or digested with trypsin or DNase before delivery to LPS-primed NLRP3/NLRC4DKO macrophages by lipid transfection. IL-1β secretion was measured after 2 h. Indicated microliters of lysate were transfected into LPS primed macrophages. B, Genomic DNA from L. monocytogenes was purified and delivered to LPS-primed macrophages by lipid transfection. IL-1β secretion was determined after 4 h. *p < 0.05; **p < 0.01.
L. monocytogenes DNA colocalizes with ASC specks
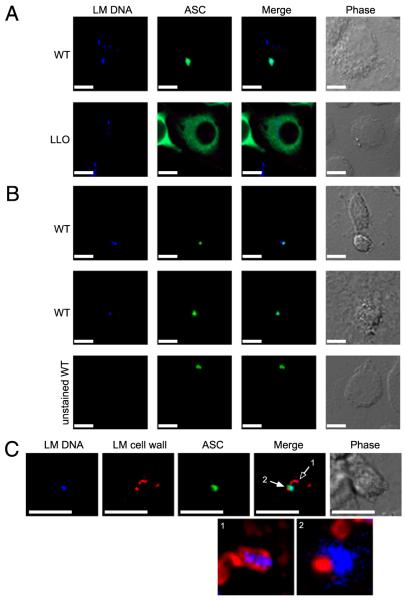
Upon NLR activation, components of the inflammasome, including ASC, are recruited into large complexes within cells termed specks (15). The vast majority of the soluble ASC within a macrophage is recruited into the speck, and this complex is the site of caspase 1 and IL-1β processing (15). Cytosolic polyA: T induces ASC speck formation (12), and we therefore examined whether L. monocytogenes DNA colocalizes with the ASC speck during infection of macrophages with the bacteria. NLRC4/NLRP3DKO macrophages expressing ASC-V5 were infected with DNA dye (DAPI or Hoescht) labeled L. monocytogenes. We observed that 48% of ASC specks colocalized with detectable L. monocytogenes DNA (Fig. 3A, 3C). In contrast, ΔLLO mutants did not induce speck formation (Fig. 3A). As a control, specks induced by unstained L. monocytogenes were not fluorescent in the blue channel (Fig. 3B). To confirm that the DNA observed in the speck was released from bacteria, we visualized the L. monocytogenes cell wall in macrophages with ASC specks (Fig. 3C). DNA within intact bacteria was contained within the bacterial cell wall and did not colocalize with ASC (Fig. 3C, open arrow). DNA associated with lysed bacteria was proximal to but not contained by the bacterial cell wall and colocalized with ASC specks (Fig. 3C, closed arrow). As controls, we confirmed that DAPI staining does not alter bacterial fitness and that the ASC-V5 construct does not compromise inflammasome activation (Supplemental Fig. 1). These data suggest that a subset of L. monocytogenes lyses within the macrophage and that the ensuing release of DNA induces inflammasome activation.
FIGURE 3.
L. monocytogenes DNA colocalizes with ASC specks. NLRP3/NLRC4DKO macrophages expressing ASC-V5 were infected with L. monocytogenes. A, Hoechst-stained WT or ΔLLO L. monocytogenes DNA (blue) and ASC (green). B, DAPI-stained or unstained L. monocytogenes DNA (blue) and ASC (green). C, DAPI-stained L. monocytogenes DNA (blue), L. monocytogenes cell wall (red), and ASC (green). Enlarged merged images of L. monocytogenes DNA with cell wall from (1) an intact bacterium and (2) a lysed bacterium are shown without ASC green overlay. Corresponding phase images are shown. Scale bars, 15 μm. Original magnification ×60.
Aim2 is the third sensor of cytosolic L. monocytogenes
Based upon recent reports that Aim2 is the sensor for viral DNA in the cytosol and that it signals through ASC (11-14), we hypothesized that Aim2 might also be detecting L. monocytogenes DNA during infection of macrophages. We functionally knocked down Aim2 signaling by stable expression of shRNA (Supplemental Fig. 2A, 2B). Aim2 shRNA, but not an irrelevant shRNA, abrogated IL-1β secretion in NLRP3/NLRC4DKO macrophages infected with L. monocytogenes (Fig. 4A). In WT macrophages, the Aim2 deficiency was compensated by intact NLRP3 and NLRC4 signaling (Fig. 4B), consistent with our previous results (Fig. 1). As a control, Aim2 knockdown did not alter IL-1β secretion following infection with Salmonella typhimurium, which is detected by NLRC4 (Supplemental Fig. 2C). Thus, the Aim2 inflammasome detects L. monocytogenes during bacterial infection of macrophages.
FIGURE 4.
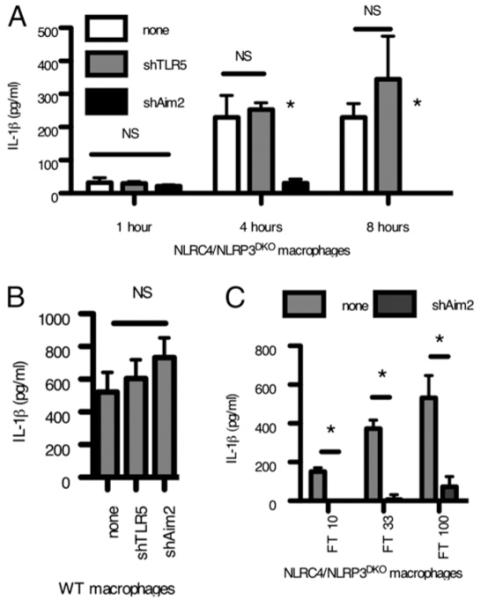
Aim2 responds to cytosolic bacteria. Control or Aim2 shRNA expressing NLRP3/NLRC4DKO (A) or WT macrophages (B) were generated by retroviral transduction and infected with L. monocytogenes at a multiplicity of infection of 10, and IL-1β secretion was determined after 4 h. C, Control or Aim2 shRNA expressing NLRP3/NLRC4DKO macrophages were infected with Francisella novicida for 8 h, and IL-1β secretion was determined. *p < 0.05.
Multiple detection pathways that result in caspase-1 activation following L. monocytogenes infection indicate that the detection of pathogens in the cytosolic compartment is a critical component of immunosurveillance. NLRP3 is temporally activated first, likely detecting vacuolar rupture, followed by NLRC4 and Aim2 activation as flagellin monomers and bacterial DNA are released into the cytosol. For Aim2 detection, our data suggest that a small percentage of cytosolic bacteria lose cell wall integrity and release their DNA into the macrophage cytosol. Damage to the bacterial cell wall may be a result of vacuolar acidification and lysosomal fusion before vacuolar escape or may be due to as yet undefined antimicrobial mechanisms within the macrophage cytosol. Detection of cytosolic bacteria by Aim2 is likely a common mechanism to detect cytosolic bacteria; we demonstrate that Francisella novicida, a bacterial pathogen that also escapes the vacuole, is also detected via the Aim2 pathway (Fig. 4C). While this manuscript was under review, other groups also reported that Aim2 detects Listeria and Francisella (16-20). Previously, the scope of infections known to be detected by Aim2 was limited to viruses, and these results implicate Aim2 in the response to cytosolic bacterial pathogens.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI065878, AI052286, AI032972, and AI025032.
Abbreviations used in this paper
- ASC
apoptotic speck protein containing a caspase recruitment domain
- CARD
caspase recruitment domain
- LLO
listeriolysin O
- NLR
nucleotide-binding oligomerization domain-like receptor
- shRNA
short hairpin RNA
- WT
wild-type
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Brodsky IE, Monack D. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin. Immunol. 2009;21:199–207. doi: 10.1016/j.smim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Schnupf P, Portnoy DA. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Pizarro-Cerdá J, Cossart P. Subversion of cellular functions by Listeria monocytogenes. J. Pathol. 2006;208:215–223. doi: 10.1002/path.1888. [DOI] [PubMed] [Google Scholar]
- 6.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J. Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuji NM, Tsutsui H, Seki E, Kuida K, Okamura H, Nakanishi K, Flavell RA. Roles of caspase-1 in Listeria infection in mice. Int. Immunol. 2004;16:335–343. doi: 10.1093/intimm/dxh041. [DOI] [PubMed] [Google Scholar]
- 8.Cassel SL, Joly S, Sutterwala FS. The NLRP3 inflammasome: a sensor of immune danger signals. Semin. Immunol. 2009;21:194–198. doi: 10.1016/j.smim.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutterwala FS, Flavell RA. NLRC4/IPAF: a CARD carrying member of the NLR family. Clin. Immunol. 2009;130:2–6. doi: 10.1016/j.clim.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao EA, Andersen-Nissen E, Warren SE, Aderem A. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 2007;29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 11.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 15.Bryan NB, Dorfleutner A, Rojanasakul Y, Stehlik C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 2009;182:3173–3182. doi: 10.4049/jimmunol.0802367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.