Abstract
Cancer arises as the result of a natural selection process among cells of the body, favoring lineages bearing somatic mutations that bestow them with a proliferative advantage. Of the thousands of mutations within a tumor, only a small fraction functionally drive its growth; the vast majority are mere passengers of minimal biological consequence. Yet the presence of any mutation, independent of its role in facilitating proliferation, tags a cell’s clonal descendants in a manner that allows them to be distinguished from unrelated cells. Such markers of cell lineage can be used to identify the abnormal proliferative signature of neoplastic clonal evolution, even at a stage which predates morphologically recognizable dysplasia. This article focuses on molecular techniques for assessing cellular clonality in humans with an emphasis on how they may be used for early detection of tumorigenic processes. We discuss historical as well as contemporary approaches and consider ways in which powerful new genomic technologies might be harnessed to develop a future generation of early cancer diagnostics.
Keywords: Field cancerization, field effect, preneoplastic, somatic evolution, biomarker, field defect, clonal evolution, cancer genome
1. Introduction
Tumorigenesis is driven by somatic evolution [1–4]. Random mutations that arise during life and confer a growth advantage upon a cell will lead to that cell’s preferential multiplication within a tissue. New variants that emerge within the expanding population fuel further waves of selection and expansion that iteratively repeat until all the phenotypes of a mature cancer have been achieved [5]. The forces dictating this process are identical to the Darwinian principles that govern evolution among individual organisms. Many of the challenges to which a cancer cell must adapt stem from growth controls built into its own genome. In multicellular organisms, a common genome derived from the founding zygote serves as a contract among cells to restrict autonomous proliferation that would negatively impact the fitness of the organism as a whole. In a developing cancer, these genetically hardwired antineoplastic defense mechanisms are systematically mutated away until tumor cells gain the ability to proliferate indefinitely.
Mutation leading to selectable genetic diversity is at the heart of the evolutionary process. Just how this diversity arises in cancer remains uncertain, but recent cancer genome sequencing studies have indicated that tumors contain tens of thousands of mutations [6]. Methods by which healthy tissues suppress the accumulation of genetic errors include: maintaining a very low per-cell-division rate of mutation, minimizing the number of cell divisions that occur in long-lived stem cells, purging of mutated cells through programmed cell death and immune surveillance, and use of a hierarchical structure of cell division whereby most cells become terminally differentiated into non-reproducing entities without the ability to propagate further errors [7]. A long-standing controversy has existed over the question of which, if any, of these mechanisms must be disrupted to allow sufficient diversity to accumulate for a cancer to evolve [1, 8–11]. Other articles in this issue consider the evidence for and against a mutator phenotype in cancer. In this review we assume a neutral position on this complex issue and instead focus on ways by which the mutations that do arise can be used to identify the signature of clonal evolution. We discuss how heritable genetic changes of many varieties can serve as markers of cell lineages to track the emergence of new clones as an empirical metric of dysregulated cell growth for the purpose of early diagnosis of cancer.
2. Field cancerization: early clonal evolution
The term “field cancerization” first appeared in a 1953 publication by Slaughter et al [12] wherein it was observed that a field of subtly abnormal epithelium commonly surrounds oral cancers and that multiple distinct tumors often co-occur within these zones. They noted:
“this pattern of distribution is of interest because it suggests a regional carcinogenic activity of some kind, in which a preconditioned epithelium has been activated over an area in which multiple cell groups undergo a process of irreversible change towards cancer”
The authors reported that such fields routinely extended beyond the margins of the surgical resections they examined and posited that this fact might explain the high local recurrence rate of oral cancers following surgery. No biochemical explanation was offered to explain the fields, simply thoughtful phenomenological observations.
In the decades since, oral cancer-associated fields have been characterized through molecular means and found to frequently possess a subset of the genetic and epigenetic abnormalities occurring in the tumor itself [13–15]. Similar mutant fields have been identified surrounding a variety of other cancer types including esophageal [16, 17], lung [18, 19], bladder [20], breast [21] and colon in the setting of chronic ulcerative colitis [22–24], among others. In many cases, molecularly-definable fields exist in the absence of histological changes. These findings are remarkably in line with Slaughter’s original hypothesis and the currently held evolutionary model of cancer development whereby successive waves of mutation, selection and clonal expansion gradually accrue the genetic changes necessary for malignant transformation. By this model a field is simply an ancestral clone possessing a partial complement of the genotypes and phenotypes of a fully formed cancer.
Given that the spread of clonal cell populations over large areas of epithelium appears to be an early part of the developmental pathway of some cancer types, many investigators have asked whether the presence of such clones can be used to predict future tumor development. The most common approach taken for identifying fields has been to screen for molecular changes commonly found in tumors themselves and thought to drive their growth. This has typically entailed focusing on alterations affecting known proto-oncogenes or tumor suppressors including: point mutations, chromosomal rearrangements, deletions, amplifications, loss-of-heterozygosity events and/or epigenetic modifications. Among the best studied systems have been Barrett’s Esophagus, oral leukoplakia and ulcerative colitis, all highly cancer-predisposing conditions with well-established field intermediates and where tissue is relatively accessible and routinely sampled as part of clinical care. While the cancer-predictive value of mutant fields identified by this method has been remarkably good for some diseases [25], for many others, it has remained limited.
One possible explanation derives from the fact that a cancer’s evolution is a stochastic process and the genetic changes driving one tumor will not always be the same as those selected in others [3, 26]. Such logic has been substantiated by large-scale sequencing studies of breast and colon cancer exome “landscapes” indicating that, while a handful of genes (“mountains”) are commonly mutated across different tumors, a far greater number are mutated only infrequently (“hills”) [27]. The tactic of screening for mutations in common drivers will, by definition, be unable to detect clonal expansions driven by mutations in unpredicted genes or regulators elsewhere in the genome. Given that it is beyond our current abilities to know of all possible rare drivers, an alternative approach must be taken to identify such clones.
3. Identifying clonality through genetic passengers
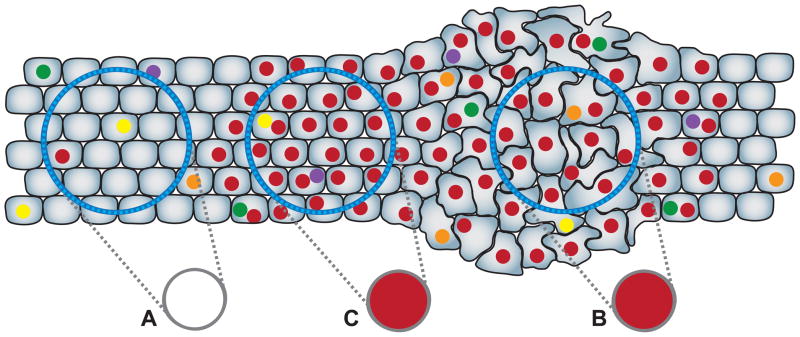
As cells divide throughout life, irrespective of cancer, mutations are continually introduced into their genomes at low frequency [28]. Most of these land outside of genes or regulatory regions and are likely to be functionally silent (neutral) but serve to indelibly mark each cell with a unique genetic fingerprint. The majority of such mutations will normally be undetectable by routine genotyping techniques because they are present in only one or a few cells whose signal is obscured by the vastly larger number of non-mutant genotypes in the surrounding tissue (Fig. 1A). If, however, a cell gains the ability to clonally proliferate as a result of one or more driver mutations, it will also carry along this larger number of neutral mutations to detectable level as chance passengers (Fig. 1B,C). Thus, the identification of any mutation in a tissue by conventional methods of aggregate DNA analysis, regardless of its functional status, is an indication that a clonal expansion has occurred. With few exceptions outside of the immune system, large clonal expansions arising in adults are abnormal and a signature of neoplasia.
Figure 1.
Somatic mutations mark clonally derived populations. Every cell in the body has a unique genotype relative to the founding zygote as a result of random mutations (colored spots) that arise at low frequency during life. (A) Because conventional DNA sequencing of a tissue sample (blue circles) detects only the majority genotype (gray circles), most mutations will not be identified in normal tissues because they are present in only one or a few cells. If, however, a cell gains the ability to clonally proliferate and progeny remain spatially clustered, the unique mutational signature of the founder becomes manifest regardless of whether the cells are morphologically abnormal (B) or not (C). Screening for detectable mutations in non dysplastic, but at-risk tissues can be used to identify the abnormal proliferative signature of preneoplastic clones as a means detecting early cancer processes. Note that new random mutations arise in some cells during clonal expansion which are not detected. The mutations used to define a clone need not be the same ones functionally driving its expansion.
As a means of identifying preneoplastic clones, screening for neutral mutations has several advantages. First, the approach conceptually focuses on identifying the generic phenotype of abnormal growth patterns without the need for a priori knowledge of the heterogeneous genotypes that may induce it. The general concept can be easily transferred between cancer types having different characteristic drivers with little or no modification. Second, of all mutations carried by an emerging clone, a far greater number are passengers than drivers. Among the tens of thousands of mutations that have been identified in cancer genomes during the last three years, only a tiny subset is likely to be etiologically related [27, 29, 30]. Lastly, screening for mutations in strongly cancer-associated genes conceivably might even reduce the detectability of very early clones. Experimental introduction of powerful oncogenes, such as activated members of the ras pathway, induces growth arrest and senescence in otherwise untransformed human cells in vitro [31]. This suggests that the most primitive clones in vivo may be less likely to bear such lesions, having not yet accrued prior mutations to facilitate oncogene tolerance. The main drawback of relying on passenger mutations to identify clonal expansions is that they are not limited to a handful of defined loci, but occur scattered throughout the genome. Mutagenesis, however, is not completely random; replication errors occur in certain hotspots orders of magnitude more frequently than elsewhere.
In this article we focus on techniques for identifying clones using neutral passengers and defer discussion of suspected driver-based methods to other excellent reviews [32–35]. For purposes herein, “neutral” is loosely defined as genetic or stable epigenetic changes identified based on methods not requiring knowledge of positively selectable loci. It is of course impossible to know with absolute certainty that mutation of a given site will not affect cell phenotype, but the spirit of the definition is to distinguish targeting of likely passengers from that of probable drivers. For the sake of brevity we limit the scope of our discussion to methods that are theoretically generalizable across multiple cell-types in the body. For example, we do not consider the elegant technique of using somatic rearrangement of Band T-cell receptors as clonal markers in blood cancers [36], nor methods involving detection of random genomic integration sites of organ-specific viruses [37]. We begin with methods of historical importance involving X-chromosome inactivation before proceeding to approaches utilizing different varieties of mutational hotspots.
4. X-linked genes and Lyonization
The earliest molecular evidence suggesting the monoclonal origin of a cancer came from studies by Linder and Gartler [38] based on heterozygous alleles of the X-linked G6PD gene. In the primitive blastocyst stage of female embryonic development, one X-chromosome per cell is randomly inactivated through hypermethylation, a process known as Lyonization [39]. Using the fact that different isoforms of the G6PD protein migrate with different motilities during gel electrophoresis, Linder and Gartler showed that, whereas G6PD from normal myometrium in heterozygous females appeared as two distinct bands (corresponding to both allelic isoforms), protein from 27 leiomyoma samples universally contained only a single band. They interpreted this observation to mean that normal uterus contains a fine mixture of cells with different X-alleles inactivated whereas tumors must be clonally derived from a single cell bearing a just one expressed allele.
Modern variations on this technique rely on directly interrogating the genetic polymorphisms that distinguish the maternal and paternal X-chromosomes. One approach uses methylation-specific restriction endonucleases [40], or methylation specific PCR [41] to differentiate between active and inactive alleles of genetically variable loci. Among the most commonly interrogated sites are the CAG repeat portion of the human androgen receptor (HUMARA) and the phosphoglycerate kinase (PGK) gene. Another approach assesses which polymorphic variant of an X-linked gene is transcribed using RT-PCR or other methods [42].
As a technique for studying the basic clonal features of cancer, the X-chromosomal approach has proved useful for more than four decades. A variety of studies have demonstrated that the embryonic “patch” size defined by a particular inactivated X-chromosome is relatively large in some human tissues [43–46]. Others have used this observation to support the intriguing possibility that some tumors bearing a single active X-chromosome could be polyclonally derived from a modest number of cells within an embryonic patch [47–49]. This large patch size also means, however, that as a general method for identifying new clonal expansions as an indicator of early neoplasia, screening for homogeneous expression from a single X-allele may yield false positives in some organs if the area sampled is too small. Another significant shortcoming is the fact that the method is limited to patients who are both female and heterozygous at the particular X-linked loci being screened.
5. Microsatellite slippage
Microsatellites, also known as short tandem repeats or STRs, are iteratively repeated elements of 1–6 basepairs that make up approximately 3% of the human genome [50]. These sites represent the prototypical example of mutational hotspots, resulting from biochemical properties that make them difficult to accurately traverse by replicative polymerases [51]. Their meiotic mutability is well recognized to contribute to the generational progression of triplet repeat expansion diseases including Fragile X Syndrome, Huntington’s Disease and Spinal Muscular Atrophies, among others [52]. Relative instability of tract length within the germline makes them highly polymorphic in the population, and thus useful markers for linkage analysis [53], forensics [54] and phylogenetic inference of human evolution and migration [55]. Haplotype diversity at these loci additionally makes them useful for loss-of-heterozygosity (LOH) analysis in both cancers and preneoplastic fields [56].
Polymerase errors made when copying across microsatellites are predominantly corrected by the cell’s mismatch repair (MMR) system. Hereditary nonpolyposis coli, also known as Lynch syndrome, is an inherited deficiency in MMR associated with a >80% lifetime risk of colorectal and other cancers [57]. Somatic loss of MMR activity is also observed in 10–15% of sporadic colorectal cancers in patients without hereditary disorders of DNA repair [58]. On a biochemical level, deficiency of MMR elevates microsatellite slippage rates between 100 and 1000-fold [59], making MMR- tumors one of the most definitive examples of a cancer-associated mutator phenotype [8]. The enormous number of passenger mutations arising in MMR- cancers annotates their genomes with an especially thorough record of the past. Tsao and colleagues capitalized on this unique phenomenon by using slipped microsatellite loci as a molecular clock to study the mitotic age of MMR- tumors [60]. A similar concept was recently used to phylogenetically map the cell lineages of tumor metastases in an MMR-compromised mouse [61].
Even with intact MMR, microsatellites exhibit mitotic frameshift rates several orders of magnitude above that of non-repetitive sequences [62, 63]. Length altering microsatellite mutations have been identified in a variety of non-MMR deficient cancers and adjacent tissues [64]. In Barrett’s Esophagus they have been observed in fields that temporally precede adenocarcinoma [65]. The detection of low-frequency microsatellite slippage in cancers or preneoplastic fields has often been reported as “microsatellite instability” [66–68]. While this wording may not be precisely correct, given that the detection of a mutation is not absolute evidence that the rate of mutation is necessarily elevated [69], the ubiquity of slipped alleles speaks to their potential usefulness as clonal markers. A concern, however, is that many studies which have used microsatellite slippage to identify expansions have only been able to detect a fraction of known clonal entities defined by other types of mutations. This is not wholly unexpected given that mutational marking is stochastic: the probability of being able to detect a clonal population is a function of the number of sites screened, the per-cell-division rate of mutation at these sites and the number of divisions undergone by a cell lineage prior to the last expansion bottleneck. Improved sensitivity should thus always be attainable by assessing a larger number of markers sites and those of greater mutability.
The rate of mitotic microsatellite slippage depends on the repeat type, length and other less predictable factors involving adjacent sequence context, transcriptional status and chromatin structure [63, 70]. Values for different loci are quite variable, ranging from less than 10−6 to nearly 10−4 in normal human cells in culture [62, 63]. Monomeric repeats of polydeoxyguanosine [poly(dG) tracts] are particularly unstable, with long tracts on the upper end of this range. Several years ago our group developed a technique for constructing fate maps of mouse development by phylogenetically analyzing the mutational relationships of hundreds of poly(dG) tracts among many individual cells [71, 72]. We recently adapted our experimental pipeline to screen for poly(dG) slippage mutations as a biomarker of preneoplastic clones in ulcerative colitis (UC) [23]. We genotyped 28 non-coding poly(dG) repeats of 12 or more residues in DNA from non-dysplastic colon biopsies of 19 individuals with UC by multiplex capillary fragment analysis. Half of these patients had cancer or advanced dysplasia in other portions of their colon and half had no histologically identifiable malignancies. Of the mutations found, 97% occurred in the cancer group. Whereas only one biopsy from one non-cancer individual bore a mutant marker, every single cancer-affected patient had at least one clonal field detectable by mutations in non-dysplastic colonic mucosa as much as 80 cm away from the cancer site. Of the thousands of genotypings carried out, only about 1% were mutant relative to the germline, yet approximately 2/3rds of all nondysplastic biopsies taken from the cancer group carried a mutation in at least one of the 28 markers, indicating a clonal derivation. This study illustrates the critical importance of high-throughput screening when relying upon random passenger mutations for clone detection. There are more than 3300 comparable poly(dG) tracts in the human genome and efforts by our group to be able to simultaneously screen the majority of these with even higher-throughput methods are ongoing.
6. Mitochondrial mutations
A heritable genetic component of nearly all human cells that is often overlooked is that of the mtDNA. The mitochondrial genome is a 16.5 kb circular loop of DNA that is replicated by organelle-specific machinery independently of the cell cycle. Each cell contains multiple mitochondria and each mitochondria may contain up to ten genomes, bringing the copy number of mtDNA genomes per cell into the hundreds for some tissues [73]. As with nuclear mutations, mtDNA mutations are passed on to daughter cells during cell division, thus serving as a marker of cell lineage. Yet because mitochondria appear to lack the complex DNA repair machinery of the nucleus and mtDNA is continually exposed to reactive oxygen intermediates from the electron transport chain and is unprotected by histones, the per-base-pair mutation rate is significantly higher than that of the nuclear genome [74].
There have been many reports of mitochondrial mutations in cancers of all varieties [75], and more recently, in fields surrounding tumors themselves. Sidransky’s group sequenced the complete mitochondrial genome from lung tumors and histologically normal respiratory epithelium in the lungs of long-time smokers [76]. Multiple mtDNA mutations were found in non-dysplastic mucosa which were identical to those from nearby cancers. In another study, the same group looked at tumors, negative surgical margins and peripheral lymphocytes from 50 patients with squamous cell carcinoma of the head and neck that recurred after an initial surgery [77]. Of these patients, approximately half had at least one mtDNA mutation in the cancer relative to the control tissue, and of these, approximately half were also found in the histologically negative margins.
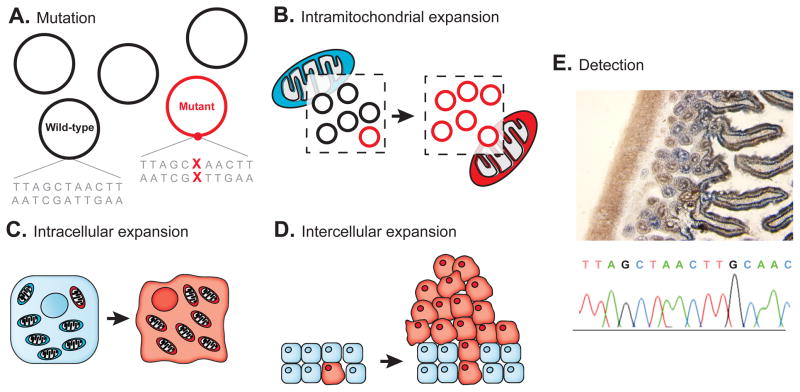
An elegant technique, developed over the last several years, uses spontaneously-arising mutations in the mitochondrially-encoded cytochrome c oxidase gene (COX1) to directly visualize cell lineage relationships in situ. Cytochrome c oxidase comprises the last step of the electron transport chain (complex IV) and mutations that disrupt its activity can be identified as blue (versus brown) staining cells in tissue sections using duel epitope histochemistry (Fig. 2E) [78]. Patches of blue cells indicate clonally-derived populations and the relationship between adjacent COX1- patches can be further delineated by microdissection and DNA sequencing for the causative mutation. The group which developed the method has used it to identify the stem cell compartment at the base of colonic crypts and shown that patches of genetically related crypts form and increase in size with age [79]. More recently they have located the putative stem cell compartment for regenerative units in stomach [80], small intestine [81], skin and pancreas [78] and liver [82] as well as characterized the clonality of cirrhotic liver nodules [83]. Conceivably such an approach might be used to identify early neoplastic clones in at-risk tissues by direct staining of biopsies.
Figure 2.
The unique features of mitochondrial mutations as a marker of cell lineage. To become detectable by routine sequencing techniques: (A) a mutant mitochondrial genome must (B) replicate and displace other genomes from its organelle and then (C) this mitochondria must outcompete or transform other mitochondria within its cell prior to (D) the marked cell clonally expanding within a tissue. (E) Dual color histochemical staining for functional loss of cytochrome c oxidase (blue coloration) can identify a subset of mutations in the mitochondrially-encoded COX1 gene for direct visualization of clonal populations in situ. Adapted from [75]. Panel E courtesy of Dr. Lawrence Loeb, University of Washington, Seattle.
While a number of features of the mitochondrial genome render it uniquely suitable for lineage mapping, several drawbacks also exist. Neither standard sequencing, nor functional staining for respiratory chain defects, is able to identify mtDNA mutations that are not present in a substantial fraction of the mitochondrial genomes within a cell. To become detectable, a mutant genome must first overtake other genomes within its organelle and then this mitochondria must outcompete or transform other mitochondria within a cell, rendering the mutation homoplasmic, prior to the marked cell clonally expanding within a tissue (Fig. 2).
The way by which homoplasmy occurs remains largely unclear. Mitochondria continually fuse and divide within the cell and potentially recombine or cross-correct their DNA [84]. Partitioning of mitochondria in the cytoplasm of dividing daughter cells may be both regulated and influenced by stochastic factors, making inheritance more complex than the binary division through which the nuclear genome segregates. The extent to which drift or selection influences the emergence of particular mitochondrial variants is also unknown. Unlike in the nuclear genome, the majority of mtDNA is coding and a sizable percentage of random mutations will be expected to alter protein sequence [73]. In cancer, some mtDNA mutations have been found more frequently than expected by chance [75] and others have been strongly associated with proliferative phenotypes. For example, Ishikawa and colleagues [85] demonstrated that the presence of a single mtDNA point mutation can render cells highly metastatic. A recent study by the developers of the COX1- staining methodology suggests that, at least in colon, these mutations have a small but noticeable effect on cell proliferation and apoptosis [86]. Such reports bring into question the neutrality of some mitochondrial mutations as lineage markers. On the other hand, many hundreds of synonymous (non-protein changing) mtDNA variants have also been reported in cancer and mutations of all types occur heteroplasmically in different normal tissues [87]. Modeling studies have also suggested that the phenomenon of homoplasmy can be expected to occur by drift alone [88, 89]. Just as in the nuclear genome, both passenger and driver mtDNA mutations are associated with clonal proliferation, and teasing apart which is which remains challenging.
Regardless of what factors contribute to homoplasmy, the process appears to take considerable time. Greaves et. al. showed that in normal colon, homogeneously COX1- staining crypts do not appear until after the age of 40 and that these divide to form small clusters of related crypts that increase in size with age [79]. Presumably crypt division begins at a younger age but is not yet histochemically visible because the COX1- genotype has not had sufficient time to propagate to homoplasmy within the crypt stem cells. This is one illustration of a general consideration for all markers of cell lineage that is discussed further below–the inability to detect a clonal marker is not de facto evidence for the absence of a clone. The temporal delay of homoplasmy makes mitochondrial mutations a potentially problematic tool for identifying early neoplastic clones in the young. Emerging sequencing technologies [87] and other techniques capable of high resolution mutation analysis [74] are beginning to permit detailed investigation of low-frequency, heteroplasmic mtDNA mutations, which may partially obviate this concern in the future.
7. Epimutations
Another sometimes forgotten source of molecular information that is heritably transmitted during cell division is DNA methylation. Following genome replication, DNA methyltransferases copy the methylcytosine profile of the parent molecule to the newly synthesized daughter strand. While a relatively accurate process (approximately 1–2 mistakes introduced per 105 residues copied [90]), it remains considerably more error-prone than DNA replication itself (rates variably estimated to be from 10−9-10−11 per base per cell division in normal tissues [91]). De novo methylation tends to increase with age [92], most specifically, mitotic age [93]. Thus, just as with DNA mutations, methylation error patterns serve as a record of somatic cell ancestry.
Silencing of gene expression through hypermethylation is a common phenomenon observed in cancer [94]. There have been a variety of reports of similar epigenetic changes in non-dysplastic tissue surrounding tumors. Shen et al identified methylation of the MGMT gene promoter in normal-appearing tissue flanking sporadic colorectal cancers [95]. Ushijima and colleagues have demonstrated promoter methylation of both protein-coding genes and microRNAs in non-cancerous mucosa around gastric cancers associated with helicobacter infection [96]. Similar epigenetic changes have been observed in tissues adjacent to cancers in liver [97], esophagus [98, 99], lung [100], breast [101], kidney [102], and bladder [103], among others.
Interpreting the results of many of these studies in terms of clonality is complicated. While “fields” of methylation changes certainly exist around some cancers, it is difficult to know that these necessarily represent clonal entities from which the cancer evolved. The predominantly used technique (methylation-specific PCR) can detect epigenetic changes in a small percentage of cells in a population and offers a general assessment of methylation across a CpG island rather than a readout of specific methylated bases that would be needed for rigorous lineage assessment. Without this information, it is conceivable that such signals might result from aggregate changes of multiple small clones or individual cells induced by a common environmental factor such as inflammation. An additional consideration is that most studies have targeted promoter regions of tumor-associated genes, which raises the issues of non-neutral lineage markers considered previously.
A lineage mapping approach developed by Shibata’s group to circumvent some of these complications, entails bisulfite conversion and direct sequencing across CpG islands in portions of the genome not transcribed in the tissue type under study [104]. The technique retrieves the exact base-by-base pattern of spontaneously methylated CpG sites in densely clustered regions, yielding multiple data points from loci presumably under neither negative or positive selection. So far this group has used the accumulation of epimutations as a molecular clock to study dynamics of replicative units in the colon [90, 105], small intestine [106] and endometrium [107]. They have recently used the method to investigate how the spatial distance within colorectal cancers compares with epigenetic lineage distances and demonstrated that the terminal outgrowth of these tumors represent relatively uniform clonal expansions [108].
8. Emerging genome-wide technologies
The mutation-based methods described so far all rely upon targeted screening of a relatively modest number of uniquely mutable hotspots. Such loci are experimentally practical to work with using conventional technologies, but encompass only a small fraction of the total mutational lineage information encoded in the genome. Recent large-scale sequencing studies have illustrated that a diversity of tumor types carry tens of thousands of somatic alterations in non-hotspot DNA [6], most of which are likely to be neutral passengers [27, 29, 30]. Presumably the bulk of these arose well before the terminal cancer outgrowth and could serve as markers of early clones, but their identification would require screening hundreds of millions of base pairs. A new generation of genomic technologies is rapidly bringing such an approach into the realm of possibility for individualized diagnostics.
Early methods that may be considered “whole genome” mutation screens include measurement of ploidy imbalances by DNA content cytometry [109] and cytogenetic assessment by traditional G-banding, florescent in situ hybridization (FISH) [110] or spectral karyotyping (SKY) [111]. The granular resolution of these approaches necessitates that deletions, amplifications or structural rearrangements be on the order of 105-109 bases in size to be detectable. Another technique, known as DNA fingerprinting, can detect significantly smaller changes by using one or more short primers to randomly amplify a fraction of the genome during low-stringency PCR. The appearance, disappearance or change in size of product fragments during electrophoresis reflects clonal alterations that may be used as a marker of preneoplastic fields [112].
Newer technologies for identifying focal copy number changes by hybridization to solid-state probe arrays enable tens of thousands of sites across the genome to be systematically interrogated. Comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) arrays have been used in the last several years to investigate preneoplastic populations in Barrett’s Esophagus [113, 114] and ulcerative colitis [115], among a variety of other cancer-predisposing diseases. Navin et al recently used the profile of CGH-identified copy number changes to study the clonal architecture of different regions of advanced breast cancers through phylogenetic inference [116].
Direct genomic sequencing provides the most detailed means possible of identifying clonal mutant markers. In contrast to conventional capillary-based techniques where individual PCR products or bacterial clones must be sequenced individually, a powerful new class of “Next Generation” sequencing technologies allows for simultaneous genotyping of tens of billions of base pairs [117]. The rapidly decreasing costs associated with these platforms have recently made it feasible to sequence the entire aggregate genome of a tissue sample without any regional targeting. From a clone detection perspective this means that multiple types of mutations of all functional varieties (both likely passengers and suspected drivers) can be simultaneously assessed. While it only takes a single clonal mutation to identify an expanded population, the redundancy conferred by screening the entire genome provides a huge amount of additional lineage data with the potential to be used for subanalyses such as approximation of a clone’s mitotic age or the phylogenetic relationship between different clones.
The digital manner in which these novel sequencing technologies operate lend them a much greater dynamic range of sensitivity than conventional techniques, making it possible to resolve populations that are subclonal relative to a collected sample. Such an ability means that in situations where spatial coherence of an expanding clone is not maintained, for example in myelodysplasia preceding blood cancers, detection at an early stage can still be accomplished [118]. Similarly, a tolerance for clone mixing should allow for convenient, minimally invasive sampling techniques that disrupt cohesive growth patterns in epithelial tissues such as cell isolation from lavage, scrapings or body fluids rather than biopsy. The relatively high error rate of individual sequencing reads currently limits the average depth to which rare subclonal mutations can be accurately detected to about 2 orders of magnitude below pure clonality [26]. A variety of improvements at the level of chemistry, hardware and analysis are continuing to enhance this resolution for all current platforms [118, 119]. An even newer generation of exotic “Fourth Generation” sequencing technologies on the horizon promises ultra-long read lengths with the ability to continuously re-sequence the same molecule for extremely accurate detection of rare molecular populations [120–122]. The pace of innovation in this area is staggering. Perhaps the only thing that can be stated with certainty about the technologies that will be available five years in the future is that they will look nothing like those from five years in the past.
9. The complex interpretation of clonality: false negatives, positives and assumptions
The reasoning associated with clonality determination is complex and many potential confounders exist. In this section we highlight seven important ideas for consideration when interpreting experimental findings.
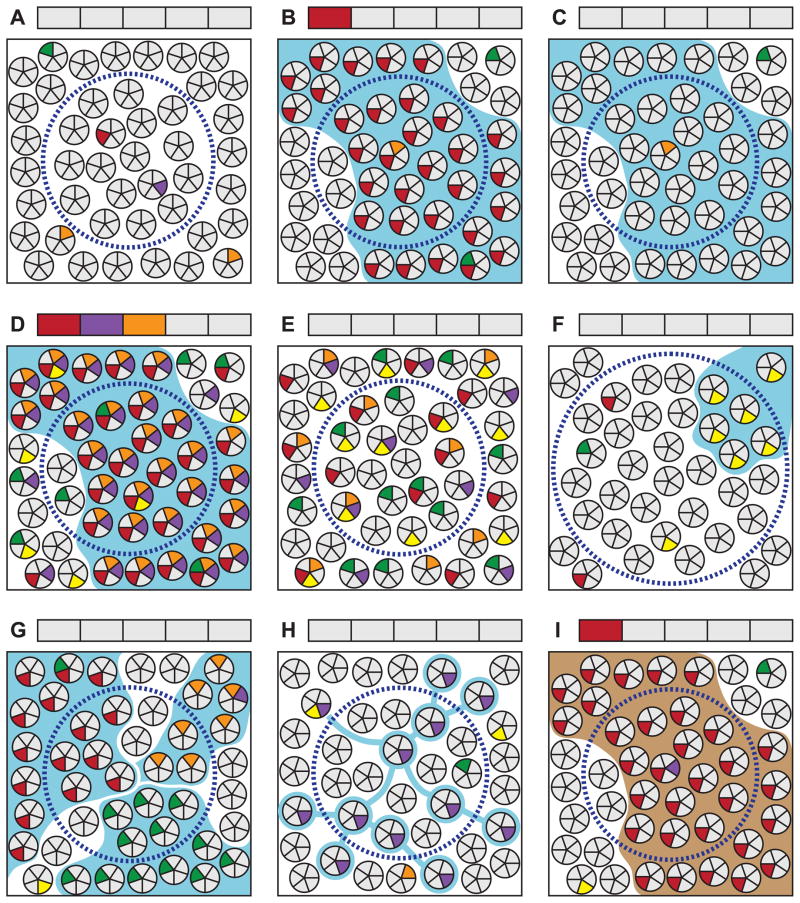
(i) Identification of a mutation in a clonal population does not indicate causality
Any mutation in a cell that undergoes clonal expansion will be passed onto progeny, irrespective of functional status (Fig. 3A,B). Given the size of the genome, more mutations carried by a neoplastic clone will be passengers than drivers [27, 29, 30]. To demonstrate driver status, supporting functional studies and/or repeated observations of a mutated loci in neoplastic clones from independent individuals are needed.
Figure 3.
Selected examples of important factors for consideration when interpreting clonality studies. See section 9 in the text for a detailed discussion of each scenario. Clusters of small circles in each panel represent a population of cells in a tissue. Each wedge represents a genomic site; gray and colored wedges respectively indicate wild-type and mutant genotypes. Blue boundaries indicate the true clonal relationship of cells derived from a neoplastic clonal expansion. Brown boundaries indicate an embryonically established clonal relationship. Dashed circles represent an area sampled for analysis. Boxes above each panel correspond to the genomic sites in the cell wedges and indicate the sample genotype as reported by a conventional method of sequencing that requires the majority of harvested cells to bear a mutation for it to be detectable. (A) Random mutations arising within individual cells in a tissue are not detectable by standard techniques. (B) If a cell bearing a unique mutation undergoes clonal expansion, it will become detectable when assessing this site. Note that new random mutations arise in some cells during clonal expansion which are not detected. (C) Clonal expansions will not be detectable if none of the genomic sites screened happen to be uniquely mutated in the founder cell. The probability of detecting a clone is a function of the number of sites screened and the frequency of random mutations in the founder cell. The latter, in turn, is dependent upon the per-cell-division mutation rate of each site and the number of cell divisions having occurred between the zygote and clone’s founding. (D) An elevated mutation frequency will facilitate a clone’s detection, although (E) in the absence of clonal expansion will have no effect on the detectable mutation load. (F) Clones that are small relative to the population sampled or that (G) become extensively mixed with other clones or (H) adjacent normal cells during expansion, are not detectable by conventional sequencing methods. (I) Based only on the presence of a detectable mutation, clonal patches arising during embryogenesis will be indistinguishable from those derived from neoplastic processes. Mutation spectrum, frequency and other contextual factors must be relied upon to infer the distinction.
(ii) Mutational marking of a clone is stochastic and not guaranteed
A clone may exist and not be detected if none of the genomic sites being screened carry a unique mutation permitting it to be distinguished from the germline (Fig. 3C). The more sites examined, the higher the probability of detection becomes. Restricting a marker panel to suspected driver sites precludes detection of pathological clones driven by unknown factors.
(iii) Elevated mutation rates facilitate clone detection but are not an absolute requirement
Screening of mutational hotspots makes it practical with conventional technologies to have a reasonable probability of detecting a clone by random passenger mutations. Clones derived from a mutator lineage or cells residing in a highly mutagenic environment should be more densely marked with identifiable passengers (Fig. 3D), potentially reducing the number of markers needing to be interrogated to identify them. Emerging high-throughput sequencing technologies will eventually obviate the need to restrict screening to a fraction of the genome.
(iv) Identification of one or more clonal mutations is not proof of genetic instability in the absence of collateral information
Detection of a mutation requires clonal expansion with most traditional methods of aggregate DNA analysis. The probability of identifying clonal mutations in an expanded population is a function of the number of sites screened, the mutability of these sites and the number of cell divisions having occurred in the lineage leading up to the final expansion. Particularly when considering hotspots, mutations will be occasionally detectable in any clonally-derived population at a statistically definable frequency. Because mutations are not routinely encountered in normal tissues, it is tempting to explain their presence in preneoplastic populations as the result of “genetic instability” (an elevated mutation rate). However, the phenomenon is more precisely explained by the fact that expansion does not routinely occur in most normal tissues. An elevated mutation rate itself will have no observable effect on clonal mutation frequency in the absence of expansion (Fig. 3E). To determine mutation rate from a clonal mutation frequency it is necessary to (A) know that the population being assessed is clonal and (B) have some metric of the number of cell divisions that occurred in the period between the zygote and the founding of the final clonal outgrowth. To infer that a rate is elevated, it is also necessary to know the normal in vivo mutation rate, which is, in turn, impossible to measure by this approach given that large clonal expansions do not routinely occur in normal adult tissue. Considering these complexities, the lack of resolution as to whether mutation rates are elevated in cancer is not surprising [1, 8–11].
(v) Detectability of a clone depends on spatial coherence of cells during expansion, sampling size relative to clone size and assay sensitivity
Clones which are small relative to the portion of tissue sampled (Fig. 3F) or which become mixed with other clones (Fig. 3G) or adjacent normal cells (Fig 3H) during expansion will not be detectable by methods that have a low dynamic range of sensitivity. Moderate-to-large sample sizes coupled with a low sensitivity assay limits detection to relatively large, coherently expanded clones, which in some situations may be of particular clinical relevance. For scenarios where mixing is anticipated, the most extreme example being in blood cell populations, a high sensitivity assay and/or enrichment prior to conventional detection methods is needed to identify early clones.
(vi) Not all clones that are identifiable in adults arose during adulthood
As humans we ourselves are a clonal entity derived from a single fertilized egg. Every cell that arises after the first zygotic cleavage forms the root of a new subclone that is propagated through development. When progeny derived from a common root remain spatially clustered during embryogenesis, a clonal patch is formed. Patches that happen to be marked by a founder mutation (Fig. 3I) will be indistinguishable in an adult from a similarly marked clone that arose post-zygotically as the result of a neoplastic process (Fig. 3B). Because Lyonization occurs very early in development, maximum patch size is represented by those defined by X-linked markers. In some tissues, X-linked patches appear to be relatively small [38], whereas in others they are quite large: on the order of a standard clinical biopsy [43–46]. In terms of non-X-linked lineage markers, however, it is only those embryonic patches which bear identifiable mutations that have the potential to be confused with adult-derived clones. Both the abundance of mutagenic insults, and the number of fallible cell divisions during very early embryogenesis, are small compared to those of later development and adult life. This is particularly true of the highly proliferative epithelial tissues most susceptible to field-associated cancers. The number of mutations marking embryonic patches should thus be relatively low compared to the quantity marking clones arising from adult-onset neoplastic processes. Determining the frequency and, possibly, spectrum of mutations in a detected clone may help distinguish embryonic from post-zygotic origins but the separation is unlikely to be absolute. In a diagnostic situation, it will always be necessary to establish the baseline mutational signature of “normal” for each type of tissue and marker being assessed.
vii) Not all clones arising during adulthood will develop into cancer
The clonal proliferation of B and T lymphocytes in response to antigen is part of a normal immune response. A few clonal processes including skewing of X-inactivation patterns in blood [123], and the development of patches of related crypts in the colon [79], appear to occur during normal aging, possibly as a result of drift coinciding with depletion of stem cell populations. For some clones that do represent early neoplastic processes, it may take years to accrue the other necessary alterations to progress to overt malignancy, and some may never advance in a patient’s lifetime. For example, chromosomal translocations pathognomonic for particular types of leukemia are not uncommonly found in leukocytes of healthy newborns and do not portend malignancy for the overwhelming number of individuals in whom they are present [124]. Correlating the detection of clones under different circumstances with future outcomes is essential for determining clinical relevance.
10. Conclusions and future directions
Most adult cancers appear to develop over a period of years, but exist for the majority of this time as clinically unrecognizable entities [125]. While tumorigenesis is strongly associated with age, environmental exposures and certain predisposing conditions, many unknown factors and randomly occurring replication errors contribute to the unpredictable clinical emergence of the disease. The underlying process of clonal evolution, itself, leaves a molecular signature in the genome of neoplastically transforming cell populations with the potential to be identified prior to the appearance of overt malignancy.
In this article we have reviewed different techniques for using neutral passenger mutations as lineage markers of preneoplastic clones and discussed important considerations for, and limitations of, their use. While the presence of certain driver mutations is of unquestionable clinical utility for partially predicting cancer risk in some predisposing conditions, passenger-based analysis has the benefit of identifying anomalous clonal proliferation in a mechanism-independent manner which remains operative in scenarios were growth results from unknown molecular drivers. Clinically, the mutational signature of evolving clones offers a means of precisely defining the margins of a disease process so that affected tissues may best be treated, resected or monitored for recurrence. With sufficient mutational information it should be possible, not only to detect neoclones, but to characterize their specific features such as size [126], spatial arrangement [23, 127], diversity among foci [65, 116] and lineage age [60, 107, 128], all of which are important metrics of evolution and evolvability in traditional ecological populations.
Powerful new tools for genetic analysis are rapidly making their way into the mainstream and promise to significantly alter the way in which cancer is diagnosed and managed over the next decade. The unprecedented amount of data generated by these technologies will soon afford complete access to the vast amount of somatic evolutionary history encrypted in the genome of early neoplasia, leaving only the daunting task of meaningfully deciphering it. Early detection has historically been among the most effective means of preventing cancer deaths [129] and we hope that the concepts and past approaches discussed herein will help stimulate creative thinking about ways in which this wealth of information can be translated to a new generation of clinical diagnostics.
Acknowledgments
JJS was supported by NIH grant F30AG033485 and the University of Washington Medical Scientist Training Program under NIH grant T32GM007266. MSH was supported by NIH grants R01DK078340 and DP1OD003278.
Abbreviations
- CGH
comparative genomic hybridization
- COX1
cytochrome c oxidase gene
- FISH
fluorescent in situ hybridization
- G6PD
glucose-6-phosphate dehydrogenase gene
- HUMARA
human androgen receptor gene
- LOH
loss-of-heterozygosity
- mtDNA
mitochondrial DNA
- PCR
polymerase chain reaction
- PGK
phosphoglycerate kinase gene
- poly(dG)
polydeoxyguanosine
- SKY
spectral karyotyping
- SNP
single nucleotide polymorphism
- UC
ulcerative colitis
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–21. [PubMed] [Google Scholar]
- 2.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 4.Pepper JW, Findlay CS, Kassen R, Spencer SL, Maley CC. Cancer research meets evolutionary biology. Evolutionary Applications. 2009;2:62–70. doi: 10.1111/j.1752-4571.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Fox EJ, Salk JJ, Loeb LA. Cancer genome sequencing--an interim analysis. Cancer Res. 2009;69:4948–50. doi: 10.1158/0008-5472.CAN-09-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepper JW, Sprouffske K, Maley CC. Animal cell differentiation patterns suppress somatic evolution. PLoS Computational Biology. 2007;3:e250. doi: 10.1371/journal.pcbi.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 9.Bodmer W. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68:3558–60. doi: 10.1158/0008-5472.CAN-07-6544. discussion 60–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–7. doi: 10.1158/0008-5472.CAN-07-5835. discussion 7. [DOI] [PubMed] [Google Scholar]
- 11.Shibata D, Lieber MR. Is there any genetic instability in human cancer? DNA Repair (Amst) 2010;9:858. doi: 10.1016/j.dnarep.2010.04.011. discussion 9–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–8. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 13.Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–30. [PubMed] [Google Scholar]
- 14.Bedi GC, Westra WH, Gabrielson E, Koch W, Sidransky D. Multiple head and neck tumors: evidence for a common clonal origin. Cancer Res. 1996;56:2484–7. [PubMed] [Google Scholar]
- 15.Garcia SB, Park HS, Novelli M, Wright NA. Field cancerization, clonality, and epithelial stem cells: the spread of mutated clones in epithelial sheets. J Pathol. 1999;187:61–81. doi: 10.1002/(SICI)1096-9896(199901)187:1<61::AID-PATH247>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Prevo LJ, Sanchez CA, Galipeau PC, Reid BJ. p53-mutant clones and field effects in Barrett’s esophagus. Cancer Res. 1999;59:4784–7. [PubMed] [Google Scholar]
- 17.Wong DJ, Paulson TG, Prevo LJ, Galipeau PC, Longton G, Blount PL, et al. p16(INK4a) lesions are common, early abnormalities that undergo clonal expansion in Barrett’s metaplastic epithelium. Cancer Res. 2001;61:8284–9. [PubMed] [Google Scholar]
- 18.Mao L, Lee JS, Kurie JM, Fan YH, Lippman SM, Lee JJ, et al. Clonal genetic alterations in the lungs of current and former smokers. J Natl Cancer Inst. 1997;89:857–62. doi: 10.1093/jnci/89.12.857. [DOI] [PubMed] [Google Scholar]
- 19.Franklin WA, Gazdar AF, Haney J, Wistuba II, La Rosa FG, Kennedy T, et al. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–7. doi: 10.1172/JCI119748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denzinger S, Mohren K, Knuechel R, Wild PJ, Burger M, Wieland WF, et al. Improved clonality analysis of multifocal bladder tumors by combination of histopathologic organ mapping, loss of heterozygosity, fluorescence in situ hybridization, and p53 analyses. Hum Pathol. 2006;37:143–51. doi: 10.1016/j.humpath.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–9. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 22.Brentnall TA, Crispin DA, Rabinovitch PS, Haggitt RC, Rubin CE, Stevens AC, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107:369–78. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 23.Salk J, Salipante S, Risques R, Crispin D, Li L, Bronner M, et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc Natl Acad Sci USA. 2009;106:20871–6. doi: 10.1073/pnas.0909428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leedham S, Graham T, Oukrif D, Mcdonald S, Rodriguez-Justo M, Harrison R, et al. Clonality, Founder Mutations, and Field Cancerization in Human Ulcerative Colitis-Associated Neoplasia. Gastroenterology. 2009;136:542–50. doi: 10.1053/j.gastro.2008.10.086. [DOI] [PubMed] [Google Scholar]
- 25.Galipeau PC, Li X, Blount PL, Maley CC, Sanchez CA, Odze RD, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salk J, Fox E, Loeb L. Mutational heterogeneity in human cancers: origin and consequences. Annual Review of Pathology. 2009;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 28.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubin AF, Green P. Mutation patterns in cancer genomes. Proc Natl Acad Sci USA. 2009;106:21766–70. doi: 10.1073/pnas.0912499106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 32.Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein C, Bernstein H, Payne CM, Dvorak K, Garewal H. Field defects in progression to gastrointestinal tract cancers. Cancer Letters. 2008;260:1–10. doi: 10.1016/j.canlet.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heaphy CM, Griffith JK, Bisoffi M. Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res Treat. 2009;118:229–39. doi: 10.1007/s10549-009-0504-0. [DOI] [PubMed] [Google Scholar]
- 35.Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci. 2009;39:331–7. [PubMed] [Google Scholar]
- 36.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–67. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol. 2010;84:8308–15. doi: 10.1128/JVI.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder D, Gartler SM. Glucose-6-phosphate dehydrogenase mosaicism: utilization as a cell marker in the study of leiomyomas. Science. 1965;150:67–9. doi: 10.1126/science.150.3692.67. [DOI] [PubMed] [Google Scholar]
- 39.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 40.Vogelstein B, Fearon ER, Hamilton SR, Feinberg AP. Use of restriction fragment length polymorphisms to determine the clonal origin of human tumors. Science. 1985;227:642–5. doi: 10.1126/science.2982210. [DOI] [PubMed] [Google Scholar]
- 41.Uchida T, Ohashi H, Aoki E, Nakahara Y, Hotta T, Murate T, et al. Clonality analysis by methylation-specific PCR for the human androgen-receptor gene (HUMARA-MSP) Leukemia. 2000;14:207–12. doi: 10.1038/sj.leu.2401631. [DOI] [PubMed] [Google Scholar]
- 42.Prchal JT, Guan YL, Prchal JF, Barany F. Transcriptional analysis of the active X-chromosome in normal and clonal hematopoiesis. Blood. 1993;81:269–71. [PubMed] [Google Scholar]
- 43.Linder D, Gartler SM. Distribution of glucose-6-phosphate dehydrogenase electrophoretic variants in different tissues of heterozygotes. Am J Hum Genet. 1965;17:212–20. [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai YC, Simoneau AR, Spruck CH, Nichols PW, Steven K, Buckley JD, et al. Mosaicism in human epithelium: macroscopic monoclonal patches cover the urothelium. J Urol. 1995;153:1697–700. doi: 10.1016/s0022-5347(01)67507-4. [DOI] [PubMed] [Google Scholar]
- 45.Jovanovic L, Delahunt B, McIver B, Eberhardt NL, Grebe SKG. Thyroid gland clonality revisited: the embryonal patch size of the normal human thyroid gland is very large, suggesting X-chromosome inactivation tumor clonality studies of thyroid tumors have to be interpreted with caution. J Clin Endocrinol Metab. 2003;88:3284–91. doi: 10.1210/jc.2002-021552. [DOI] [PubMed] [Google Scholar]
- 46.Novelli M, Cossu A, Oukrif D, Quaglia A, Lakhani S, Poulsom R, et al. X-inactivation patch size in human female tissue confounds the assessment of tumor clonality. Proc Natl Acad Sci USA. 2003;100:3311–4. doi: 10.1073/pnas.0437825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt GH, Mead R. On the clonal origin of tumours--lessons from studies of intestinal epithelium. Bioessays. 1990;12:37–40. doi: 10.1002/bies.950120109. [DOI] [PubMed] [Google Scholar]
- 48.Halberg RB, Dove WF. Polyclonal tumors in the mammalian intestine: are interactions among multiple initiated clones necessary for tumor initiation, growth, and progression? Cell Cycle. 2007;6:44–51. doi: 10.4161/cc.6.1.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons BL. Many different tumor types have polyclonal tumor origin: evidence and implications. Mutat Res. 2008;659:232–47. doi: 10.1016/j.mrrev.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kornberg A, Bertsch LL, Jackson JF, Khorana HG. Enzymatic synthesis of deoxyribonucleic acid, XVI. Oligonucleotides as templates and mechanism of their replication. Proc Natl Acad Sci USA. 1964;51:315–23. doi: 10.1073/pnas.51.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.La Spada AR, Paulson HL, Fischbeck KH. Trinucleotide repeat expansion in neurological disease. Ann Neurol. 1994;36:814–22. doi: 10.1002/ana.410360604. [DOI] [PubMed] [Google Scholar]
- 53.Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, et al. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 54.Budowle B, Moretti TR, Niezgoda SJ, Brown BL. Publication 98–06. Laboratory Division of the Federal Bureau of Investigation; Washington DC: 1998. CODIS and PCR-based short tandem repeat loci: law enforcement tools. [Google Scholar]
- 55.Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc Natl Acad Sci USA. 1995;92:6723–7. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paulson TG, Galipeau PC, Reid BJ. Loss of heterozygosity analysis using whole genome amplification, cell sorting, and fluorescence-based PCR. Genome Res. 1999;9:482–91. [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch HT, Shaw MW, Magnuson CW, Larsen AL, Krush AJ. Hereditary factors in cancer. Study of two large midwestern kindreds. Arch Intern Med. 1966;117:206–12. [PubMed] [Google Scholar]
- 58.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharyya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotypes in human colorectal carcinoma cell lines. Proc Natl Acad Sci USA. 1994;91:6319–23. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsao JL, Yatabe Y, Salovaara R, Järvinen HJ, Mecklin JP, Aaltonen LA, et al. Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci USA. 2000;97:1236–41. doi: 10.1073/pnas.97.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frumkin D, Wasserstrom A, Itzkovitz S, Stern T, Harmelin A, Eilam R, et al. Cell lineage analysis of a mouse tumor. Cancer Res. 2008;68:5924–31. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 62.Boyer JC, Yamada NA, Roques CN, Hatch SB, Riess K, Farber RA. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum Mol Genet. 2002;11:707–13. doi: 10.1093/hmg/11.6.707. [DOI] [PubMed] [Google Scholar]
- 63.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog. 2009;48:379–88. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao L, Lee DJ, Tockman MS, Erozan YS, Askin F, Sidransky D. Microsatellite alterations as clonal markers for the detection of human cancer. Proc Natl Acad Sci USA. 1994;91:9871–5. doi: 10.1073/pnas.91.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 66.Brentnall TA, Crispin DA, Bronner MP, Cherian SP, Hueffed M, Rabinovitch PS, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res. 1996;56:1237–40. [PubMed] [Google Scholar]
- 67.Meltzer SJ, Yin J, Manin B, Rhyu MG, Cottrell J, Hudson E, et al. Microsatellite instability occurs frequently and in both diploid and aneuploid cell populations of Barrett’s-associated esophageal adenocarcinomas. Cancer Res. 1994;54:3379–82. [PubMed] [Google Scholar]
- 68.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 69.Graham T, Halford S, Page KM, Tomlinson IPM. Most low-level microsatellite instability in colorectal cancers can be explained without an elevated slippage rate. J Pathol. 2008;215:204–10. doi: 10.1002/path.2351. [DOI] [PubMed] [Google Scholar]
- 70.Lai Y, Sun F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol Biol Evol. 2003;20:2123–31. doi: 10.1093/molbev/msg228. [DOI] [PubMed] [Google Scholar]
- 71.Salipante SJ, Horwitz MS. Phylogenetic fate mapping. Proc Natl Acad Sci USA. 2006;103:5448–53. doi: 10.1073/pnas.0601265103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salipante SJ, Kas A, McMonagle E, Horwitz MS. Phylogenetic analysis of developmental and postnatal mouse cell lineages. Evol Dev. 2010;12:84–94. doi: 10.1111/j.1525-142X.2009.00393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, et al. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–4. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 75.Kulawiec M, Salk JJ, Ericson NG, Wanagat J, Bielas JH. Generation, function, and prognostic utility of somatic mitochondrial DNA mutations in cancer. Environ Mol Mutagen. 2010;51:427–39. doi: 10.1002/em.20582. [DOI] [PubMed] [Google Scholar]
- 76.Dasgupta S, Yung RC, Westra WH, Rini DA, Brandes J, Sidransky D. Following mitochondrial footprints through a long mucosal path to lung cancer. PLoS ONE. 2009;4:e6533. doi: 10.1371/journal.pone.0006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dasgupta S, Koch R, Westra WH, Califano JA, Ha PK, Sidransky D, et al. Mitochondrial DNA mutation in normal margins and tumors of recurrent head and neck squamous cell carcinoma patients. Cancer prevention research. 2010;3:1205–11. doi: 10.1158/1940-6207.CAPR-10-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fellous T, Mcdonald S, Burkert J, Humphries A, Islam S, De-Alwis N, et al. A methodological approach to tracing cell lineage in human epithelial tissues. Stem Cells. 2009;27:1410–20. doi: 10.1002/stem.67. [DOI] [PubMed] [Google Scholar]
- 79.Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci USA. 2006;103:714–9. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald SAC, Greaves LC, Gutierrez-Gonzalez L, Rodriguez-Justo M, Deheragoda M, Leedham SJ, et al. Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology. 2008;134:500–10. doi: 10.1053/j.gastro.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 81.Gutierrez-Gonzalez L, Deheragoda M, Elia G, Leedham S, Shankar A, Imber C, et al. Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J Pathol. 2008 doi: 10.1002/path.2502. [DOI] [PubMed] [Google Scholar]
- 82.Fellous TG, Islam S, Tadrous PJ, Elia G, Kocher HM, Bhattacharya S, et al. Locating the stem cell niche and tracing hepatocyte lineages in human liver. Hepatology. 2009;49:1655–63. doi: 10.1002/hep.22791. [DOI] [PubMed] [Google Scholar]
- 83.Lin W-R, Lim S-N, McDonald SAC, Graham T, Wright VL, Peplow CL, et al. The histogenesis of regenerative nodules in human liver cirrhosis. Hepatology. 2010;51:1017–26. doi: 10.1002/hep.23483. [DOI] [PubMed] [Google Scholar]
- 84.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–9. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–4. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 86.Nooteboom M, Johnson R, Taylor RW, Wright NA, Lightowlers RN, Kirkwood TBL, et al. Age-associated mitochondrial DNA mutations lead to small but significant changes in cell proliferation and apoptosis in human colonic crypts. Aging Cell. 2010;9:96–9. doi: 10.1111/j.1474-9726.2009.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–4. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet. 2001;28:147–50. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 89.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–6. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yatabe Y, Tavaré S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci USA. 2001;98:10839–44. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cervantes RB, Stringer JR, Shao C, Tischfield JA, Stambrook PJ. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci USA. 2002;99:3586–90. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 93.Chu MW, Siegmund KD, Eckstam CL, Kim JY, Yang AS, Kanel GC, et al. Lack of increases in methylation at three CpG-rich genomic loci in non-mitotic adult tissues during aging. BMC Med Genet. 2007;8:50. doi: 10.1186/1471-2350-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Issa J-P. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 95.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–8. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 96.Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–74. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 97.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–9. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 98.Ishii T, Murakami J, Notohara K, Cullings HM, Sasamoto H, Kambara T, et al. Oesophageal squamous cell carcinoma may develop within a background of accumulating DNA methylation in normal and dysplastic mucosa. Gut. 2007;56:13–9. doi: 10.1136/gut.2005.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, et al. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–6. [PubMed] [Google Scholar]
- 100.Guo M, House MG, Hooker C, Han Y, Heath E, Gabrielson E, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131–6. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- 101.Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, et al. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12:6626–36. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 102.Arai E, Kanai Y, Ushijima S, Fujimoto H, Mukai K, Hirohashi S. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int J Cancer. 2006;119:288–96. doi: 10.1002/ijc.21807. [DOI] [PubMed] [Google Scholar]
- 103.Habuchi T, Takahashi T, Kakinuma H, Wang L, Tsuchiya N, Satoh S, et al. Hypermethylation at 9q32–33 tumour suppressor region is age-related in normal urothelium and an early and frequent alteration in bladder cancer. Oncogene. 2001;20:531–7. doi: 10.1038/sj.onc.1204122. [DOI] [PubMed] [Google Scholar]
- 104.Shibata D. Inferring human stem cell behaviour from epigenetic drift. J Pathol. 2009;217:199–205. doi: 10.1002/path.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nicolas P, Kim K-M, Shibata D, Tavaré S. The stem cell population of the human colon crypt: analysis via methylation patterns. PLoS Computational Biology. 2007;3:e28. doi: 10.1371/journal.pcbi.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim JY, Siegmund KD, Tavaré S, Shibata D. Age-related human small intestine methylation: evidence for stem cell niches. BMC Med. 2005;3:10. doi: 10.1186/1741-7015-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim JY, Tavaré S, Shibata D. Counting human somatic cell replications: methylation mirrors endometrial stem cell divisions. Proc Natl Acad Sci USA. 2005;102:17739–44. doi: 10.1073/pnas.0503976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Siegmund K, Marjoram P, Tavaré S, Shibata D. Many colorectal cancers are “flat” clonal expansions. Cell Cycle. 2009:8. doi: 10.4161/cc.8.14.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rubin CE, Haggitt RC, Burmer GC, Brentnall TA, Stevens AC, Levine DS, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology. 1992;103:1611–20. doi: 10.1016/0016-5085(92)91185-7. [DOI] [PubMed] [Google Scholar]
- 110.Rabinovitch PS, Dziadon S, Brentnall TA, Emond MJ, Crispin DA, Haggitt RC, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59:5148–53. [PubMed] [Google Scholar]
- 111.Varella-Garcia M, Chen L, Powell RL, Hirsch FR, Kennedy TC, Keith R, et al. Spectral karyotyping detects chromosome damage in bronchial cells of smokers and patients with cancer. Am J Respir Crit Care Med. 2007;176:505–12. doi: 10.1164/rccm.200609-1329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen R, Rabinovitch PS, Crispin DA, Emond MJ, Koprowicz KM, Bronner MP, et al. DNA fingerprinting abnormalities can distinguish ulcerative colitis patients with dysplasia and cancer from those who are dysplasia/cancer-free. Am J Pathol. 2003;162:665–72. doi: 10.1016/S0002-9440(10)63860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li X, Galipeau PC, Sanchez CA, Blount PL, Maley CC, Arnaudo J, et al. Single nucleotide polymorphism-based genome-wide chromosome copy change, loss of heterozygosity, and aneuploidy in Barrett’s esophagus neoplastic progression. Cancer Prevention Research. 2008;1:413–23. doi: 10.1158/1940-6207.CAPR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lai LA, Kostadinov R, Barrett MT, Peiffer DA, Pokholok D, Odze R, et al. Deletion at fragile sites is a common and early event in Barrett’s Esophagus. Mol Cancer Res. 2010;8:1084–94. doi: 10.1158/1541-7786.MCR-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bronner MP, Skacel M, Crispin DA, Hoff PD, Emond MJ, Lai LA, et al. Array-based comparative genomic hybridization in ulcerative colitis neoplasia: single non-dysplastic biopsies distinguish progressors from non-progressors. Modern Pathology. 2010 doi: 10.1038/modpathol.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Research. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 118.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat Methods. 2010;7:119–22. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 121.Travers KJ, Chin C-S, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Research. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nature nanotechnology. 2009;4:265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 123.Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88:59–65. [PubMed] [Google Scholar]
- 124.Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA. 2002;99:8242–7. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maley CC, Galipeau PC, Li X, Sanchez CA, Paulson TG, Blount PL, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–33. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 127.Leedham SJ, Preston SL, McDonald SAC, Elia G, Bhandari P, Poller D, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008;57:1041–8. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wasserstrom A, Frumkin D, Adar R, Itzkovitz S, Stern T, Kaplan S, et al. Estimating cell depth from somatic mutations. PLoS Comp Biol. 2008;4:e1000058. doi: 10.1371/journal.pcbi.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]