Abstract
When released from damaged erythrocytes free heme not only provides a source of iron for invading bacteria but is also highly toxic due to its ability to catalyze free radical formation. Hemopexin (Hx) binds free heme with very high affinity and thus protects against heme toxicity, sequesters heme from pathogens, and helps conserve valuable iron. Hx is also an acute-phase serum protein (APP), whose expression is induced by inflammation. To date Hx has been identified as far back in phylogeny as bony fish where it is called Warm-temperature Acclimation-related 65 kDa Protein (WAP65), as serum protein levels are increased at elevated environmental temperatures as well as by infection. During analysis of nurse shark (Ginglymostoma cirratum) plasma we isolated a Ni2+-binding serum glycoprotein and characterized it as the APP Hx. We subsequently cloned Hx from nurse shark and another cartilaginous fish species, the little skate Leucoraja erinacea. Functional analysis showed shark Hx, like that of mammals, binds heme but is found at unusually high levels in normal shark serum. As an Hx orthologue could not be found in the genomes of jawless vertebrates or lower deuterostomes it appears to have arisen just prior to the emergence of jawed vertebrates, coincident with the second round of genome wide duplication and the appearance of tetrameric haemoglobin (Hb).
Keywords: Hemopexin, WAP65, cartilaginous fish, shark, evolution
1. Introduction
Although hemoglobin (Hb) emerged over 1,500 million years ago (MYA) (Hardison, 1996), the tetrameric (α2-β2) form of Hb found in jawed vertebrates allows for a sophisticated regulation of oxygen binding. However this Hb form is much more harmful when released from damaged erythrocytes, having highly toxic oxidative and pro-inflammatory effects and providing a source of iron for invading bacteria (Lee, 1995). Therefore, several proteins have emerged in gnathostomes that bind and sequester free heme, thus neutralizing its detrimental features. Of these hemopexin (Hx), a ~60 kDa serum glycoprotein, binds heme with the highest affinity (Kd <10−12 M) (Hrkal et al., 1974). Hx is folded into 2 homologous domains of ~200 aa, joined by a highly flexible, ~20 aa linker. Each hx domain (N.B. herein hemopexin domains are denoted hx and the hemopexin protein Hx) folds into four β-sheets, which are arranged as the blades of a flat, four-bladed β-propeller, with the edge of the C-terminal domain locked at a 90° angle against the face of the N-terminal domain (Paoli et al., 1999). Hx binds a single heme molecule in a pocket formed between the two hx domains and bounded by the interdomain linker. The Fe(III) of the heme is coordinated by two histidine residues, His213 from the linker and His266 of the C-terminal hx domain and further stabilized by a host of non-covalent interactions provided by a large number of invariant aromatic and basic residues (Paoli et al., 1999). The Hx-heme complex in mammals is taken up by liver parenchymal cells through receptor-mediated endocytosis mediated by the scavenger receptor LRP1 (also known as CD91) (Hvidberg et al., 2005). The bound heme is released from Hx in the low pH environment of the endosome, likely by protonation of one or both of the coordinating histidine residues and movement of the hx domains and/or linker (Paoli et al., 1999). The heme is moved into the cell cytoplasm where it is used to build new hemoproteins or is safely catabolised (reviewed in Schaer and Alayash, 2010). Thus heme binding and transport by Hx protects against oxidative damage, limits access by pathogens to heme, and recycles valuable iron.
In mice and humans Hx is produced by the liver and found in serum at ~0.5–1.25 mg/ml (Tolosano et al., 2010). However as Hx is an acute-phase protein the concentration of Hx greatly increases during inflammation (Wagener et al., 2001; Graca-Souza et al., 2002), its production being upregulated by proinflammatory cytokines including IL-6, IL-11, IL-1β and TNF-α (Immenschuh et al., 1995). Phylogentically, hemopexin has been identified in the mammals (placental, marsupial and monotreme), amphibians, and birds. Additionally an Hx homologue, the 65 kDa protein WAP65, has been found in teleost fish, although orthology has yet to be confirmed. Most teleost species possess two WAP65 genes which are differentially expressed; whilst WAP65-1 is widely and constitutively expressed, WAP65-2 expression is limited to the liver and is upregulated upon warm acclimation (e.g. an environmental temperature shift from 10°C to 30°C), following bacterial infection, or administration of LPS (Kikuchi et al., 1995; Peatman et al., 2007); however, it is not yet known whether the two forms have different roles in Hb physiology. Herein we describe the cloning and functional characterization of Hx from the nurse shark (Ginglymostoma cirratum) and little skate (Leucoraja erinacea), members of the most ancient gnathostome lineage. We will discuss the emergence of this APP and potential modifications of its function in different vertebrate taxa.
2. Materials and methods
2.1. Preparation and SDS-PAGE of shark serum proteins
Animals were maintained in artificial seawater at approximately 28°C in large indoor tanks at the University of Maryland Biotechnology Institute, Center of Marine Biotechnology (COMB), Baltimore. Sharks were anaesthetized with MS222 prior to bleeding from the caudal sinus and the blood spun at 300g for 10 min to isolate serum. From this 1ul of serum was mixed with non-reducing Laemmli sample buffer containing 40 mg/ml SDS, loaded onto a 5% SDS-PAGE gel and electrophoresed in SGT buffer (1.5g Tris, 7.2g glycine, 0.5g SDS per litre dH2O). All gels were stained with Gelcode blue (Pierce) according to manufacturer’s instructions.
2.2. IMAC-purification and N-terminal sequencing of shark serum proteins
Ni2+-binding serum-proteins were purified by immobilized metal-affinity chromatography (IMAC). Serum (1 ml) was diluted 1/10 in PBS and passed over a Ni2+-sepharose column. The column was subsequently washed with 5 volumes of PBS, 2x 10 ml 20 mM imidazole followed by a further 5 volumes of PBS. Bound proteins were eluted with 5 ml of 200 mM imidazole. Eluted proteins were mixed with reducing Laemmli sample buffer containing 40 mg/ml SDS and characterized by SDS-PAGE on 12% gels, run as above. His-purified samples were sent to Emory University, Microchemical and Proteomics core facility for N-terminal sequencing by Edman degradation.
2.3. Cloning of Hx from shark and skate
The SMART RACE cDNA amplification kit (Clonetech) was used as per the manufacturer’s instructions to generate 5’ and 3’ RACE-primed cDNA with 2.5 µg of liver RNA as the template per reaction. The degenerate primer GcP75-f1 was designed based upon a reverse translation of the nurse shark p75 N-terminal sequence and 5’ RACE yielded a single band of ~1200 bp. The sequence was completed using the primers GcP75-f2 and GcP75-r1 for nested 3’ and 5’ RACE (respectively). The full sequence was confirmed with the primers GcP75-f3 & GcP75-r3 (Genbank accession number HM347595).
The partial Hx sequence constructed from the little skate EST database was confirmed using the primers LeHx-f1 and LeHx-r1, then nested 3’ RACE performed with the primers LeHx-f1 for the first round and LeHx-f4 for the second round (Genbank accession number HM347594).
2.4. Northern blots
Total RNA was prepared for Northern blotting as detailed previously (Bartl et al., 1997) and 10 µg loaded per lane. The primers GcP75-f3 and GcP75-r3 were used to amplify the full length coding sequence of Hx. The PCR product was radiolabelled with 32P dCTP according to usual methods (Mertz and Rashtchian, 1994), cleaned of free nucleotides and used as the probe for Hx under high (final wash: 20 min agitation in 0.2X SSC + 01.% SDS at 65°C) stringency conditions. Expression of the housekeeping gene NDPK (nucleoside diphosphate kinase; Genbank accession number M63964.1) was also probed on the blot as a loading control (data not shown).
2.5. Hemoglobin- and IMAC-precipitation of plasma proteins
Shark Hb (sHb)-sepharose was prepared by lysing shark red blood cells in 10 volumes of PBS at a 1/10 dilution then the liquid spun at 13K rpm for 10 min to remove any debris or clots. The resultant supernatant was diluted 1/10 in binding buffer and conjugated to cyanogen bromide-sepharose (Sigma-Aldrich) according to manufacturer’s instructions. Prepared Hb-sepharose was washed extensively with PBS to remove any loosely bound Hb.
Plasma from shark was diluted 1/10 in PBS and 1 ml added to 100 µl Ni2+-sepharose (Qiagen) or 200 µl species matched Hb-sepharose and incubated for 1h with rotation. Ni2+-sepharose was washed x4 with 1ml PBST (PBS + 0.05% Tween20), 2 times with 1 ml 20 mM imidazole and 4 times with PBST. Hb-sepharose was washed 10 times with PBST. Following the final wash all supernatant was carefully aspirated, 200 µl of 2X reducing Laemmli buffer containing 40 mg/ml SDS added to the sepharose, and the resultant slurry boiled for 5 min to remove bound proteins. The boiled slurry was zip-spun and the supernatant loaded onto 12% SDS-PAGE gels and run as above.
2.6. Phylogenetic tree generation
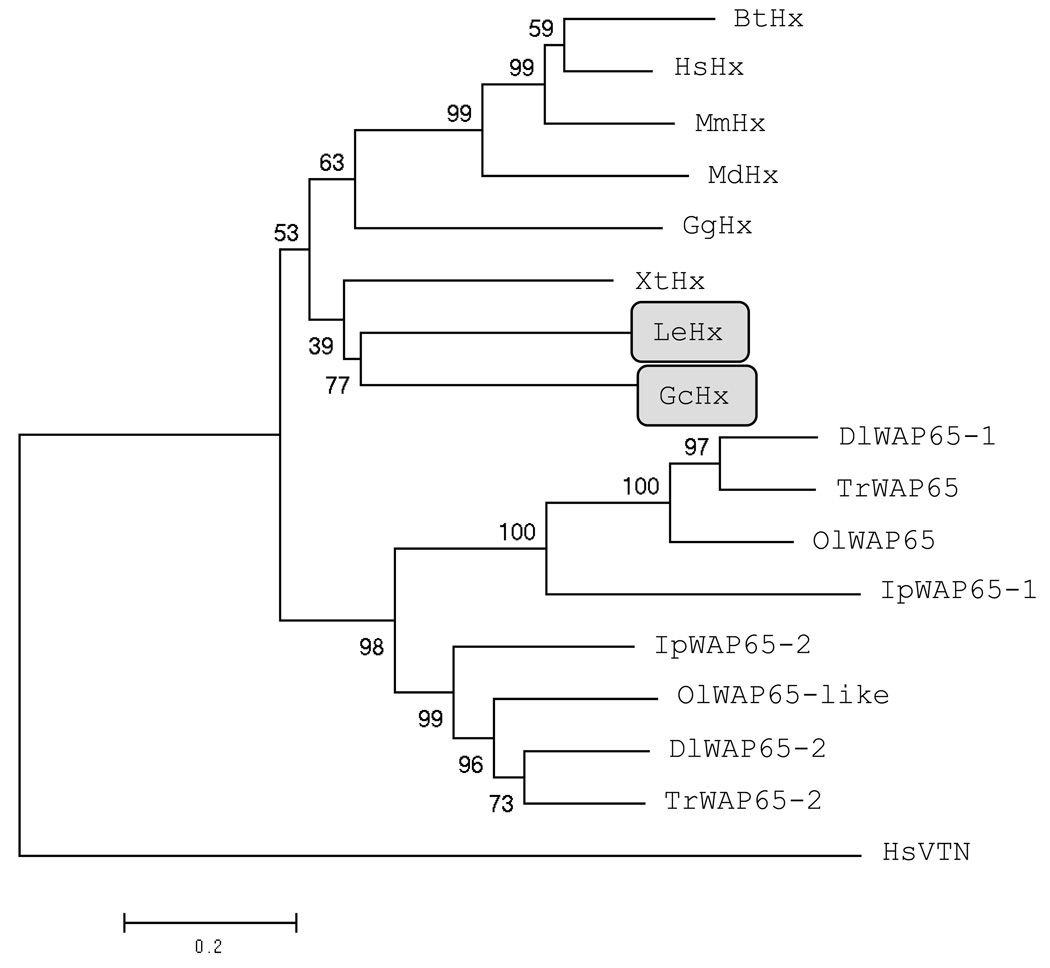
The evolutionary histories reported in Fig 5 were inferred using the Neighbour-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the sequences analyzed. The percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Dayhoff matrix based method (Schwarz and Dayhoff, 1979) and are in the units of the number of amino acid substitutions per site. All columns containing alignment gaps and missing data were eliminated in all comparisons (complete deletion option). There were a total of 235 positions in the final dataset. Genbank accession numbers for sequences used are as follows: Homo sapiens Hx (NP_000604.1), Mus musculus Hx (AAH19901.1), Monodelphis domestica Hx (predicted; XP_001380277), Bos taurus Hx (NP_001029784.1), Gallus gallus Hx (partial; AAL29887.1), Xenopus tropicalis Hx (putative, partial; jgi|Xentr4|435537|e_gw1.3417.2.1), Ictalurus punctatus WAP65-1 (ABW07853.1) and WAP65-2 (ABW07852.1), Takifugu rubripes WAP65 (BAD18109.1) and WAP65-2 (BAD18110.1), Dicentrarchus labrax WAP65-1 (DAA12503.1) and WAP65-2 (DAA12504.1), Oryzias latipes WAP65 (BAB97303.1) and WAP-65-like (BAB97304.1). The tree was rooted with Homo sapiens vitronectin (VTN; Genbank accession NP_000629.3). Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). Evolutionary histories were also inferred using the Maximum Parsimony method in MEGA and the trees obtained with this method did not differ significantly, and supported the conclusions drawn from the Neighbour-Joining trees.
Fig. 5. Neighbour-Joining tree showing the phylogenetic relationships between Hx and WAP65 from different species.
Hx from the cartilaginous fish are shaded. Two letter abbreviations for genus and species are used as follows: Human (Homo sapiens), mouse (Mus musculus), opossum (Monodelphis domestica), cow (Bos taurus), chicken (Gallus gallus) Hx, clawed frog (Xenopus tropicalis), channel catfish (Ictalurus punctatus), Fugu pufferfish (Takifugu rubripes), european seabass (Dicentrarchus labrax), Japanese medaka (Oryzias latipes), nurse shark (Ginglymostoma cirratum) and little skate (Leucoraja erinacea). The tree was rooted with the human hx domain-containing protein vitronectin (VTN).
3. Results
3.1. Cloning of Hx from the nurse shark and little skate
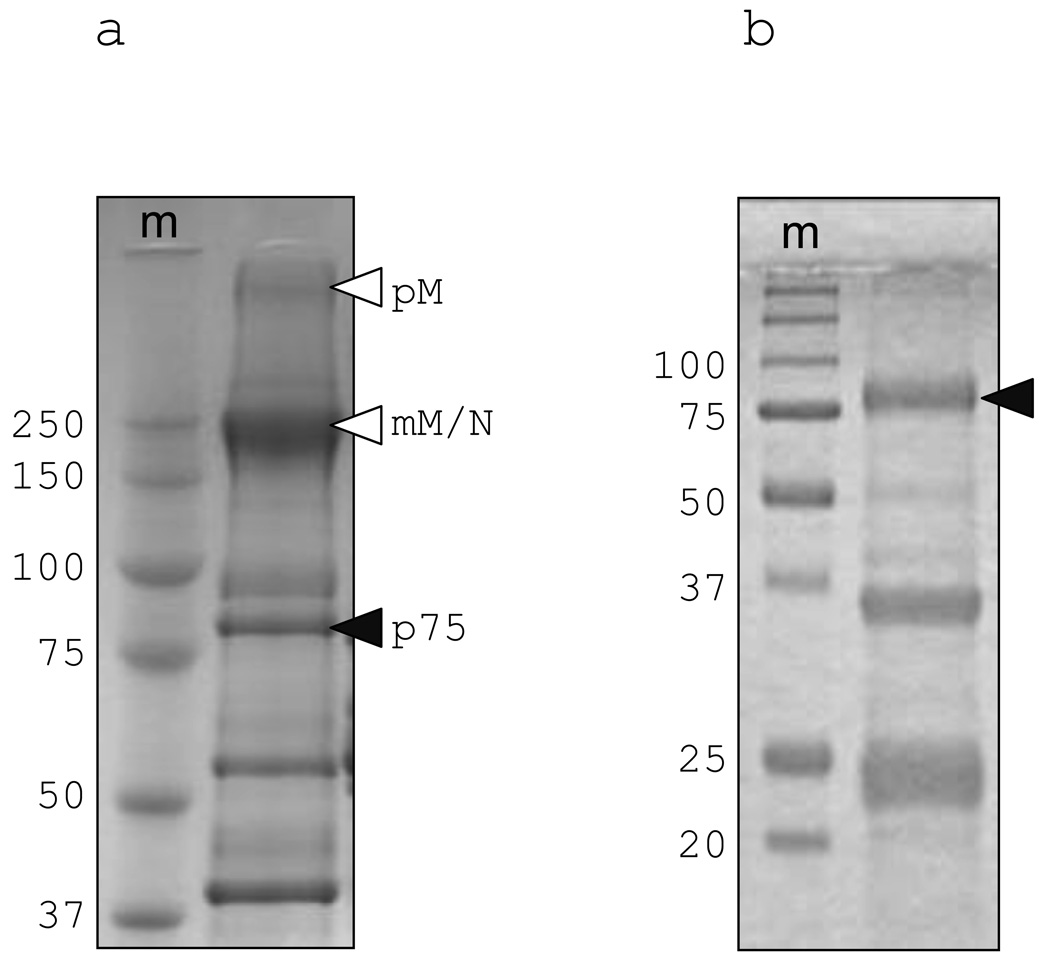
During routine size-exclusion fractionation and SDS-PAGE of nurse shark plasma we observed several unknown proteins that were present at very high levels. A ~75 kDa protein (p75) was of special interest as it was found to be enriched by immobilized metal-ion affinity chromatography (IMAC) on a Ni2+-sepharose column (Fig. 1). This protein ran at ~75 kDa on both reducing and non-reducing SDS-PAGE showing its monomeric nature. N-terminal sequence of p75 yielded 16 aa of sequence from which a degenerate primer could be designed (GcP75-f1; table 1) for 3’ RACE. The sequence was extended by 5’ and 3’ RACE and the final full-length sequence was confirmed with gene-specific primers. The full-length sequence encoded three methionine residues upstream of the N-terminal protein sequence but only one (−19) was predicted by SignalP (v3.0) to initiate a typical leader peptide. The best matches by BLAST analysis with the mature shark protein were the mammalian serum protein Hx and teleost WAP65.
Fig. 1. Characterization of the unknown 75 kDa shark serum protein.
(a) whole nurse shark plasma run on 6% non-reducing SDS-PAGE and (b) IMAC-purified plasma run on 12% reducing SDS-PAGE. The lane marked m is molecular weight marker, white arrows indicate pentameric IgM (pM) and monomeric IgM/IgNAR (mM/N), black arrows unknown protein p75 (characterization of the unlabelled bands in Fig 1b will be detailed elsewhere. Dooley et al., manuscript in preparation).
Table 1. a) N-terminal protein sequencing result for the unknown 75 kDa serum protein and b) sequence of primers used for cloning.
For protein sequencing lower case letters denote the probable amino acid at that position and×denotes the residue could not be determined. Residues underlined and in bold denote potential N-linked glycosylation site.
| Protein | N-terminal protein sequence |
|---|---|
| Gc p75 | FPwLKHQPNITEDELNxxh |
| Primer name | Sequence |
| GcP75-f1 | 5’-CAYCARCCiAAYATHACiGARG-3’ |
| GcP75-f2 | 5’-CACTCGCTTCTATCCTCACTGGG-3’ |
| GcP75-f3 | 5’-GAAGTTTCTGCTGGGAGGATTGTG-3’ |
| GcP75-r1 | 5’-CTGGGGTGGGAGTGATGCTG-3’ |
| GcP75-r3 | 5’-GTACACAAGACATGAAATCTGCGCG-3’ |
| LeHx-f1 | 5’-ATGAAGTCCCTGCTGGGAGGAGTG-3’ |
| LeHx-r1 | 5’-CGAGCCACCGCCTTGTAGATGAAC-3’ |
| LeHx-f4 | 5’-GTTCATCTACAAGGCGGTGGCTCG-3’ |
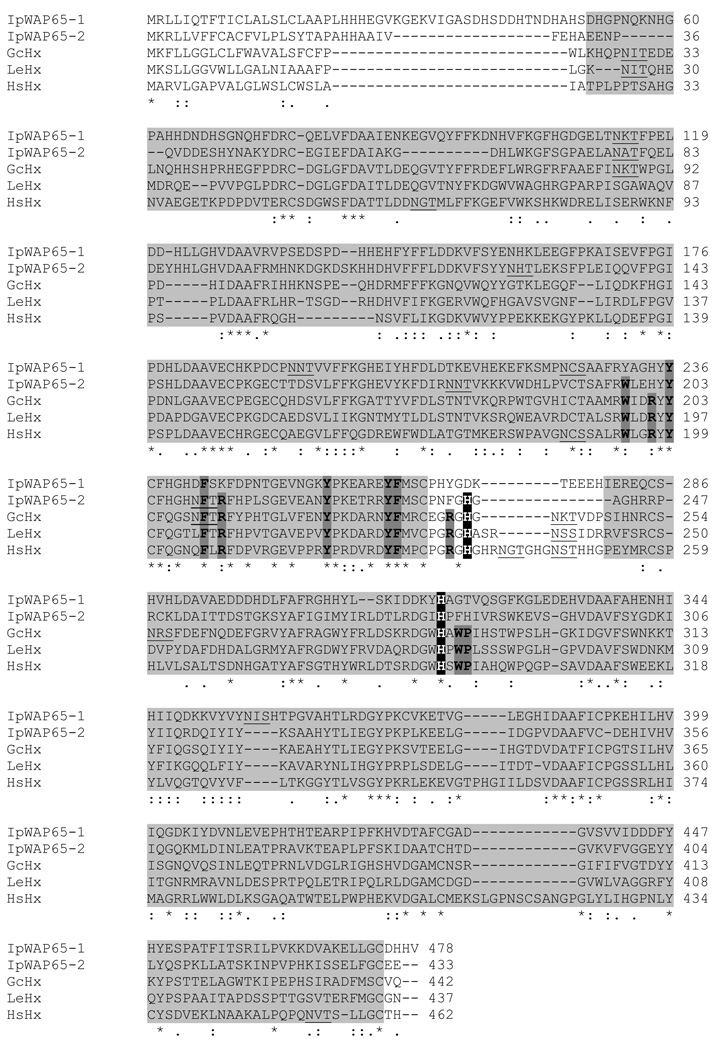
Using the nurse shark Hx sequence we searched the EST databases which are available for two other cartilaginous fish species; the little skate and the spiny dogfish (Squalus acanthias). No significant hits were obtained from spiny dogfish, but a partial (333 aa) Hx sequence was generated from 3 overlapping little skate ESTs. This was used to design primers and a complete Hx sequence confirmed from this second, evolutionary distant, cartilaginous fish species (Fig. 2); amino acid identity between nurse shark and little skate Hx is ~50%.
Fig. 2. Amino acid alignment of nurse shark (Gc) and little skate (Le) Hx with that of human Hx (Hs) shows conservation of residues important for function in these species. These residues are also conserved in fugu (Tr) WAP65-2 but not WAP65-1.
The two hx domains are shaded, the conserved histidine residues which co-ordinate Hb are highlighted by white text on black, conserved aromatic and basic residues in the Hb binding region are shown with darker shading and bolded. Potential N-linked glycosylation sites are underlined.
3.2. Sequence features and expression profile of Hx from cartilaginous fish
Despite the fact that shark and skate Hx have low amino acid identity with the human orthologue (shark v human 37%, skate v human 38%), those residues defining the structure and function of the heme binding site (Paoli et al., 1999) are conserved in these species: the heme-coordinating residues His213 and His266 are both conserved, as are all of the aromatic and basic residues that line the heme-binding pocket (Fig. 2). Whilst these residues are also highly conserved in WAP65-2 of teleosts this is not true for WAP65-1; in WAP65-1 approximately half of the ‘conserved’ aromatic and basic residues in the pocket have been lost, as have one or both, of the heme-coordinating His (illustrated by the example of Fugu (Tr)WAP65-1 and -2 in Fig. 2).
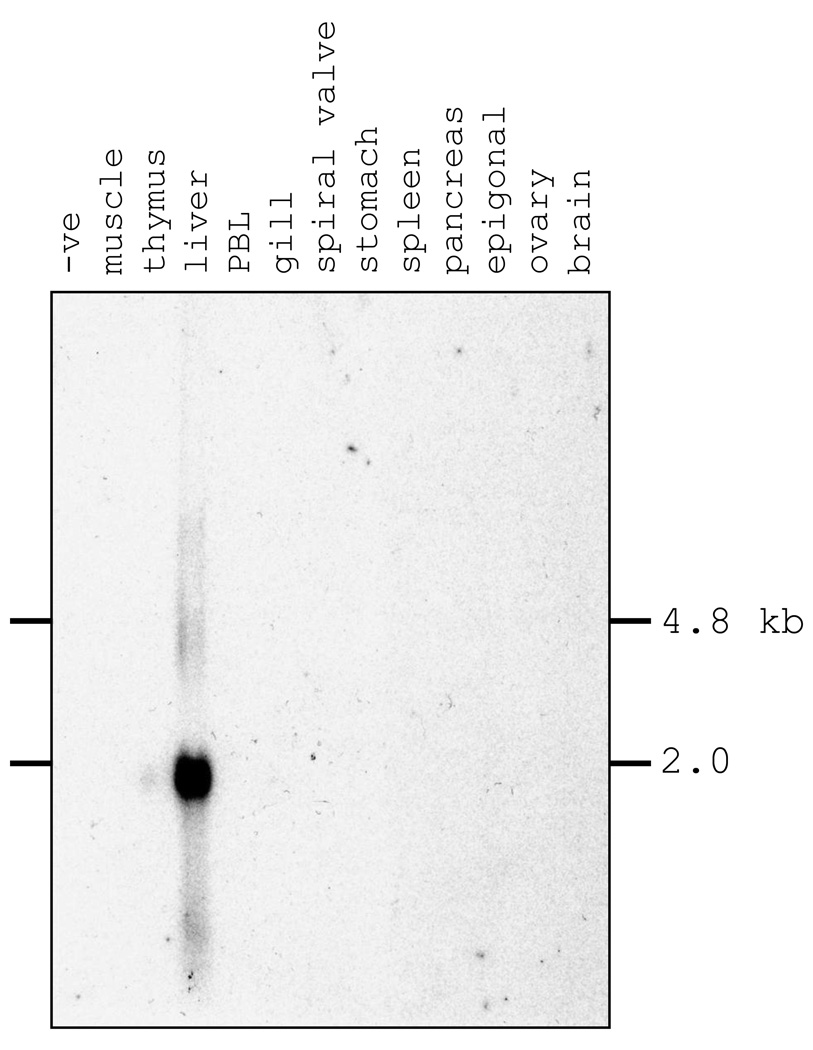
Glycosylation of human Hx is important for high-affinity binding to heme (Satoh et al., 1994), and the elasmobranch Hx also has a number of potential N-linked glycosylation sites (5 in nurse shark; 2 in little skate), most of which are apparently used in the nurse shark protein as demonstrated by PNGaseF deglycosylation experiments (data not shown). Interestingly the N-linked glycosylation site in the interdomain linker is conserved in all lineages except the teleost fishes indicating it may play some special role, perhaps acting to protect the highly flexible linker (Paoli et al., 1999) from proteolysis. As in mammals, shark Hx mRNA expression was restricted to the liver in a northern blotting experiment, giving a single band of < 2.0 kb that correlated well with the cDNA size of ~1.7 kb (Fig. 3).
Fig. 3. Northern blot results indicate that shark Hx is expressed only from liver.
Full length Hx cDNA was used to probe total RNA from multiple nurse shark tissues.
3.3. Heme-binding of nurse shark Hx
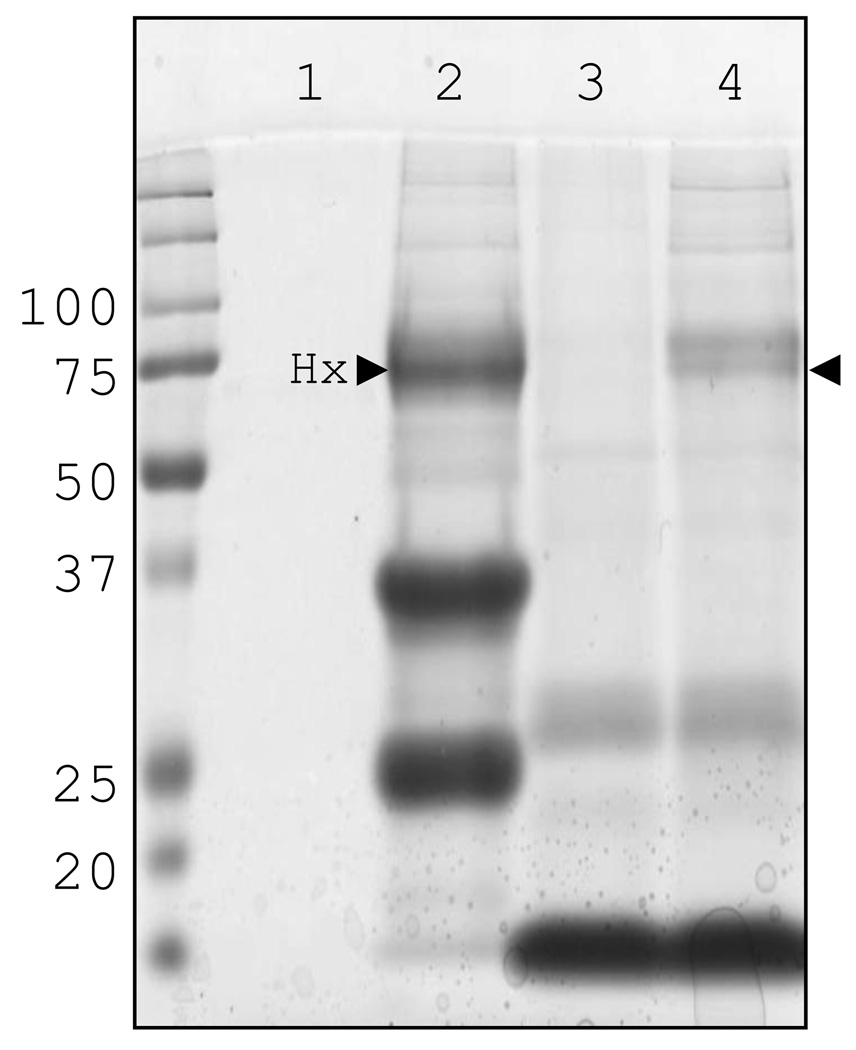
As detailed previously the primary role of Hx in mammals is the sequestration and neutralization of heme, we therefore decided to test the capability of nurse shark Hx to bind shark heme (Fig. 4). Following IMAC-purification of nurse shark plasma the same three bands as described earlier were observed (lane 2), Hx being the one at 75 kDa.
Fig. 4. SDS-PAGE of Hb- and Ni2+-immunoprecipitated proteins shows shark Hx binds heme.
Immunoprecipitated nurse shark plasma, run under reducing conditions. Proteins eluted from Ni2+-sepharose incubated without (lane 1) or with plasma (lane 2) and Hb-sepharose without (lane 3) or with plasma (lane 4).
When shark plasma was incubated with Hb-sepharose a doublet at ~75 kDa was precipitated (lane 4). The presence of a tightly-migrating doublet suggests either we are observing the heme-bound and -free forms of Hx or slight differences in glycosylation. The other bands in this sample were also observed in the sample of heme-sepharose incubated with PBS in place of plasma (lane 3). In order to confirm this result we incubated Ni2+-purified shark plasma with a limited amount of heme and passed the mixture over an S300 size-exclusion column; fractions with a red coloration were analyzed by SDS-PAGE to assess their protein content. Shark Hx was found in the same fractions as heme (data not shown) thus confirming the immunoprecipitation result that shark Hx indeed binds heme.
4. Discussion
In mammals the primary role of the APP Hx is to bind free heme and prevent oxidative damage whilst sequestering iron away from bacteria. Hx is expressed by the liver and is present at 0.5–1.25 mg/ml levels in serum; however its expression is greatly upregulated (50-fold within 24 h) by the inflammation that accompanies injury or infection (Wang et al., 2001; Poli and Cortese, 1989). During the present study we identified one of the major serum proteins in nurse sharks as the cartilaginous fish orthologue of Hx. Cartilaginous fish diverged from a common ancestor with other vertebrates ~500 MYA and are members of the most ancient extant gnathostome lineage. However mammalian and elasmobranch Hx both show the ability to bind their respective Hbs and comparison of their sequences shows the residues important for function are highly conserved in these two lineages. The situation in the teleost fish is more complex with a number of species now being shown to express two forms of WAP65, the teleost Hx homologue. The two WAP65 genes are divergent in both their sequence and pattern of tissue expression; WAP65-1 shows mutation of many of the ‘conserved’ Hx residues and shows constitutive, multi-tissue expression whilst WAP65-2 is more conserved in both sequence and expression characteristics (Sha et al., 2008; Sarropoulou et al., 2009; Kikuchi et al., 1995). Phylogenetic trees (Fig. 5) show the teleost WAP65s cluster separately from Hx of all other species and the WAP65-1 and -2 genes group separately within the cluster. As the ancestor of the majority of ray-finned fish is thought to have experienced an additional, lineage-specific genome-wide duplication (so-called 3R) ~320 MYA (Vandepoele et al., 2004) the separation of the two WAP65 genes strongly suggests they are paralogues resulting from this additional round of duplication. Initial studies also suggest the two forms have different physiological functions (Sha et al., 2008; Hirayama et al., 2004; Sarropoulou et al., 2009) indicating duplication has been followed by diversification and neofuctionalization.
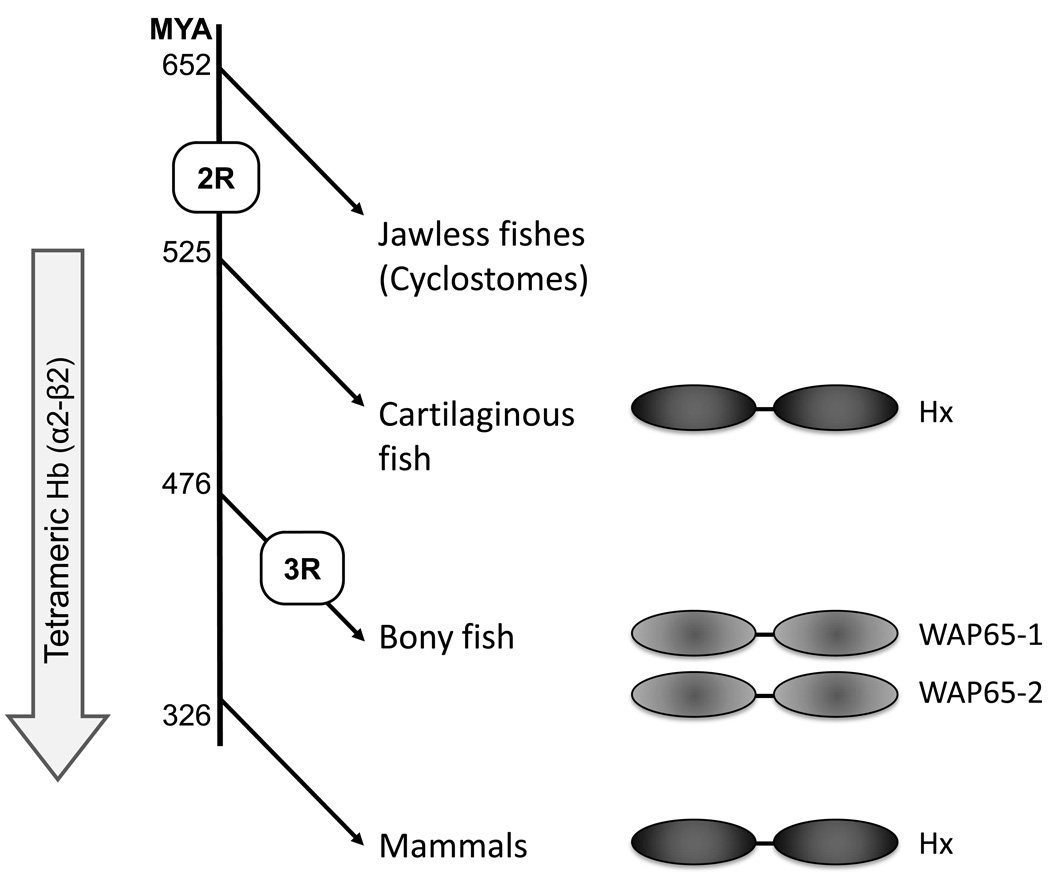
Searches for Hx in the genomes of more ancient species (lamprey, hagfish, sea urchin and amphioxus) have been conducted by us and others (Wicher and Fries, 2010) however no orthologue has yet been found. This strongly suggests that Hx evolved after the emergence of the cyclostomes (jawless fishes) but prior to that of the jawed vertebrates, coincident with the 2nd round of genome wide duplication (2R; Fig. 6). Additional searches with individual hx domains in these species returned only matrix metalloproteases (MMPs) or MMP-like molecules, proteins which also use hx domains as mediators of protein:protein interaction. Thus, we have not identified an unambiguous candidate related to the Hx ancestor.
Fig. 6. Schematic illustrating the putative evolution of Hx.
The second round of genome wide duplication in the jawed vertebrate ancestor (2R) and teleost specific genome wide-duplication (3R) are shown. The divergence times of the lineages (shown as millions of years ago) are based upon those of Blair and Hedges (2005).
Despite the crystal structure of Hx clearly showing that the heme-binding site lies between the two hx domains, bounded by the interdomain linker, when subject to proteolysis the isolated N-terminal hx domain is also able to bind heme (Morgan and Smith, 1984). This finding, combined with the high sequence identity between the two hx domains, has lead to the suggestion that Hx evolved from another protein containing a single Hx domain (Baker et al., 2003). We propose that this molecule was an MMP-like protein, similar to those we found in the genomes of the jawless vertebrates and invertebrates, which arose at approximately the same time as the second round of genome wide duplication (2R) in the ancestor of the jawed vertebrates. Freed from selective pressure the duplicate MMP-like gene could lose its pro-peptide and catalytic domains and duplicate its hx domain, thus generating the proto-Hx gene. The concomitant appearance of tetrameric (α2-β2) Hb in jawed vertebrates enabled more sophisticated regulation of oxygen binding (as compared to the monomeric/dimeric Hbs of the jawless fish) (Coates, 1975) but also proved much more harmful when released from erythrocytes; as the interdomain binding site gives much greater control of heme binding and release (Morgan and Smith, 1984) this improved mode of binding by Hx, once evolved, would be under very strong positive selection.
One feature that remains difficult to explain is the very high (>10 mg/ml) level of Hx in the serum of outwardly healthy sharks (10–20X the level of serum Hx in mammals). Studies in mammals have shown that released heme is removed by Hx, however when Hb is released from erythrocytes it is cleared by another acute-phase serum protein, haptoglobin (Nielsen et al., 2010 and references therein). Whilst we have found high levels of haptoglobin in nurse shark plasma, in our hands it does not bind Hb-coupled sepharose (Dooley et al., manuscript in preparation). Thus it seems that nurse shark Hx could bind either heme or Hb, or maybe even binds both, perhaps explaining why such high levels of serum Hx are required. This would be even more likely if there is a need to conserve valuable iron in these species and/or sequester it away from whatever bacteria routinely infect these animals? Additionally, recent work in mice has shown that Hx can suppress the production of TNF and IL6 by macrophages in response to LPS (Liang et al., 2009). As bacteria can be routinely cultured from the organs and blood of healthy sharks (Mylniczenko et al., 2007) perhaps high levels of Hx act to ‘dampen’ the immune response to their commensal microflora? Although this important question remains unresolved, with the cloning of shark Hx we have finally identified the 5S ‘pink protein’ which was first described during chromatography of shark serum over 45 years ago (Marchalonis and Edelman, 1965).
Acknowledgements
Our thanks to Dr Jan Pohl at the Microchemical and Proteomics core facility, Emory University, for advice concerning protein sequencing. This work was funded by NIH grant RR06603 awarded to MFF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker HM, Anderson BF, Baker EN. Dealing with iron: common structural principles in proteins that transport iron and heme. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3579–3583. doi: 10.1073/pnas.0637295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartl S, Baish MA, Flajnik MF, Ohta Y. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J. Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Coates ML. Hemoglobin function in the vertebrates: an evolutionary model. J. Mol. Evol. 1975;6:285–307. doi: 10.1007/BF01794636. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Graca-Souza AV, Arruda MA, de Freitas MS, Barja-Fidalgo C, Oliveira PL. Neutrophil activation by heme: implications for inflammatory processes. Blood. 2002;99:4160–4165. doi: 10.1182/blood.v99.11.4160. [DOI] [PubMed] [Google Scholar]
- Hardison RC. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama M, Kobiyama A, Kinoshita S, Watabe S. The occurrence of two types of hemopexin-like protein in medaka and differences in their affinity to heme. J. Exp. Biol. 2004;207:1387–1398. doi: 10.1242/jeb.00897. [DOI] [PubMed] [Google Scholar]
- Hrkal Z, Vodrazka Z, Kalousek I. Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur. J. Biochem. 1974;43:73–78. doi: 10.1111/j.1432-1033.1974.tb03386.x. [DOI] [PubMed] [Google Scholar]
- Hvidberg V, Maniecki MB, Jacobsen C, Hojrup P, Moller HJ, Moestrup SK. Identification of the receptor scavenging hemopexin-heme complexes. Blood. 2005;106:2572–2579. doi: 10.1182/blood-2005-03-1185. [DOI] [PubMed] [Google Scholar]
- Immenschuh S, Song DX, Satoh H, Muller-Eberhard U. The type II hemopexin interleukin-6 response element predominates the transcriptional regulation of the hemopexin acute phase responsiveness. Biochem. Biophys. Res. Commun. 1995;207:202–208. doi: 10.1006/bbrc.1995.1173. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Yamashita M, Watabe S, Aida K. The warm temperature acclimation-related 65-kDa protein, Wap65, in goldfish and its gene expression. J. Biol. Chem. 1995;270:17087–17092. doi: 10.1074/jbc.270.29.17087. [DOI] [PubMed] [Google Scholar]
- Lee BC. Quelling the red menace: haem capture by bacteria. Mol. Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- Liang X, Lin T, Sun G, Beasley-Topliffe L, Cavaillon JM, Warren HS. Hemopexin down-regulates LPS-induced proinflammatory cytokines from macrophages. J. Leukoc. Biol. 2009 doi: 10.1189/jlb.1208742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchalonis J, Edelman GM. Phylogenetic origins of antibody structure. I. Multichain structure of immunoglobulins in the smooth dogfish (Mustelus canis) J. Exp. Med. 1965;122:601–618. doi: 10.1084/jem.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz LM, Rashtchian A. Nucleotide imbalance and polymerase chain reaction: effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal. Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- Morgan WT, Smith A. Domain structure of rabbit hemopexin. Isolation and characterization of a heme-binding glycopeptide. J. Biol. Chem. 1984;259:12001–12006. [PubMed] [Google Scholar]
- Mylniczenko ND, Harris B, Wilborn RE, Young FA. Blood culture results from healthy captive and free-ranging elasmobranchs. J. Aquat. Anim Health. 2007;19:159–167. doi: 10.1577/H06-039.1. [DOI] [PubMed] [Google Scholar]
- Nielsen MJ, Moller HJ, Moestrup SK. Hemoglobin and heme scavenger receptors. Antioxid. Redox. Signal. 2010;12:261–273. doi: 10.1089/ars.2009.2792. [DOI] [PubMed] [Google Scholar]
- Paoli M, Anderson BF, Baker HM, Morgan WT, Smith A, Baker EN. Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains. Nat. Struct. Biol. 1999;6:926–931. doi: 10.1038/13294. [DOI] [PubMed] [Google Scholar]
- Peatman E, Baoprasertkul P, Terhune J, Xu P, Nandi S, Kucuktas H, Li P, Wang S, Somridhivej B, Dunham R, Liu Z. Expression analysis of the acute phase response in channel catfish (Ictalurus punctatus) after infection with a Gram-negative bacterium. Dev. Comp Immunol. 2007;31:1183–1196. doi: 10.1016/j.dci.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Poli V, Cortese R. Interleukin 6 induces a liver-specific nuclear protein that binds to the promoter of acute-phase genes. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8202–8206. doi: 10.1073/pnas.86.21.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sarropoulou E, Fernandes JM, Mitter K, Magoulas A, Mulero V, Sepulcre MP, Figueras A, Novoa B, Kotoulas G. Evolution of a multifunctional gene: The warm temperature acclimation protein Wap65 in the European seabass Dicentrarchus labrax. Mol. Phylogenet. Evol. 2009 doi: 10.1016/j.ympev.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Satoh T, Satoh H, Iwahara S, Hrkal Z, Peyton DH, Muller-Eberhard U. Roles of heme iron-coordinating histidine residues of human hemopexin expressed in baculovirus-infected insect cells. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8423–8427. doi: 10.1073/pnas.91.18.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaer DJ, Alayash AI. Clearance and control mechanisms of hemoglobin from cradle to grave. Antioxid. Redox. Signal. 2010;12:181–184. doi: 10.1089/ars.2009.2923. [DOI] [PubMed] [Google Scholar]
- Schwarz R, Dayhoff M. Matricies for detecting distant relationships. In: Dayhoff M, editor. Atlas of Protein Sequences. National Biomedical Research Foundation; 1979. pp. 353–358. [Google Scholar]
- Sha Z, Xu P, Takano T, Liu H, Terhune J, Liu Z. The warm temperature acclimation protein Wap65 as an immune response gene: its duplicates are differentially regulated by temperature and bacterial infections. Mol. Immunol. 2008;45:1458–1469. doi: 10.1016/j.molimm.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tolosano E, Fagoonee S, Morello N, Vinchi F, Fiorito V. Heme scavenging and the other facets of hemopexin. Antioxid. Redox. Signal. 2010;12:305–320. doi: 10.1089/ars.2009.2787. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, De VW, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener FA, Eggert A, Boerman OC, Oyen WJ, Verhofstad A, Abraham NG, Adema G, van KY, de WT, Figdor CG. Heme is a potent inducer of inflammation in mice and is counteracted by heme oxygenase. Blood. 2001;98:1802–1811. doi: 10.1182/blood.v98.6.1802. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kinzie E, Berger FG, Lim SK, Baumann H. Haptoglobin, an inflammation-inducible plasma protein. Redox. Rep. 2001;6:379–385. doi: 10.1179/135100001101536580. [DOI] [PubMed] [Google Scholar]
- Wicher KB, Fries E. Evolutionary aspects of hemoglobin scavengers. Antioxid. Redox. Signal. 2010;12:249–259. doi: 10.1089/ars.2009.2760. [DOI] [PubMed] [Google Scholar]