Abstract
Necrosis has been thought to be an accidental or uncontrolled type of cell death rather than programmed. Recent studies from diverse organisms show that necrosis follows a stereotypical series of cellular and molecular events: swelling of organelles, increases in reactive oxygen species and cytoplasmic calcium, a decrease in ATP, activation of calpain and cathepsin proteases, and finally rupture of organelles and the plasma membrane. Genetic and chemical manipulations demonstrate that necrosis can be inhibited, indicating that necrosis can indeed be controlled and follows a specific “program.” This review highlights recent findings from C. elegans, yeast, Dictyostelium, Drosophila, and mammals that collectively provide evidence for conserved mechanisms of necrosis.
Introduction
The term necrosis has been used for hundreds, if not thousands, of years to describe the death of tissue in an organism. The word derives from the Greek, meaning death or dead body. With the coining of the term apoptosis by Kerr, Wyllie and Currie in 1972, necrosis became known as a distinct type of cell death [1]. Kerr defined two types of cell death occurring in the rat liver following disruption of the blood supply. Patches of swollen necrotic cells showing lysosomal leakage were found but cells showing a condensed morphology were also observed. Kerr and colleagues first called this form of cell death “shrinkage necrosis” and later termed it “apoptosis” (reviewed in [2]).
While others had previously described the cellular events of apoptotic cell death (reviewed in [3]), the papers from Kerr and colleagues have been highly influential in defining modes of cell death. These authors used morphological characteristics to distinguish apoptosis from necrosis. They also argued that necrosis occurred in response to gross cellular damage caused by environmental conditions and was less likely to be inherently controlled. The appreciation of the central role of apoptosis in development and disease has triggered prolific amounts of research, whereas progress towards understanding necrosis has lagged. In addition to apoptosis and necrosis, other types of cell death have been described, including autophagic cell death and cell death with mixed characteristics [4•].
Because historically necrosis has been the term used for all cell death, confusion has arisen as to the precise modern definition of necrosis. Several groups have tried to address this problem, devising alternative terms such as oncosis [3], oncotic necrosis [5], necroptosis [6•], programmed necrosis, necrotic cell death, and Type III cell death [4]. All of these terms refer to a form of cell death characterized by swelling which is morphologically distinct from apoptosis. Following the recommendations of the Nomenclature Committee on Cell Death [4], I will use the term necrosis to refer to this form of cell death.
As with apoptosis, cells undergoing necrosis show similar morphologies across a wide range of stimuli, cell types and organisms (reviewed in [4,7]). Not all necrotic cells show precisely the same features, but a general progression has emerged. Necrotic cells first show clumping of chromatin and swelling of organelles. This is followed by swelling of the cell, and the rupture of nuclei, mitochondria, and the plasma membrane [8,9]. Necrosis is characterized by a rise in cytosolic Ca2+, increased reactive oxygen species (ROS), intracellular acidification, and a depletion of ATP (reviewed in [7]). In mammals, rupture of necrotic cells leads to the discharge of “danger” molecules and stimulates immune cells to release pro-inflammatory cytokines. In contrast, apoptotic cells are engulfed by phagocytic cells which typically release anti-inflammatory molecules (reviewed in [10]). Thus, necrosis can trigger inflammation while apoptosis usually does not.
Necrosis is a major form of cell death occurring in multiple diseases including neurodegenerative disorders, heart disease, neuronal ischemia and toxicity, muscular dystrophy, diabetes, and infections [11–16•]. Additionally, apoptotic cells that fail to be engulfed by phagocytic cells can undergo “secondary” necrosis [17]. This review focuses on the emergence of non-mammalian models of necrosis, which collectively demonstrate that the events of necrosis may be programmed after all.
Models of necrosis
Caenorhabditis elegans
The nematode C. elegans was the organism in which genetic control over apoptosis was first demonstrated. Likewise, C. elegans has been a leading model for the genetic dissection of necrosis. Necrosis does not occur during normal C. elegans development, but certain mutant backgrounds lead to necrotic cell death, which has provided an excellent opportunity to determine the relevant cellular mechanisms (reviewed in [18,19•]). Gain-of-function mutations in ion channel subunit genes, including mec-4 and deg-1, or expression of a constitutively active G-protein Gα, lead to swelling and death of neurons [20,21]. The progression of degeneration in mec-4(d) mutants has been carefully characterized by electron microscopy; early stages of cell death are accompanied by membranous whorls and vacuole formation, while later stages show increased swelling with loss of organelles and the nucleus [20]. A screen for suppressors of mec-4(d) induced necrosis identified calreticulin, a calcium binding protein localized to the endoplasmic reticulum, and similar effects were obtained with other calcium modulators [22]. These findings led to the model that calcium influx is a common underlying cause for necrotic neurodegeneration in C. elegans. Furthermore, calpains, a family of calcium-regulated proteases, act to promote necrosis in mec-4(d), deg-1(d), deg-3(d) and Gα (gf) mutants, pointing to a significant role for calcium [23].
The lysosomal compartment also plays a critical role in C. elegans necrosis. Lysosomes cluster around the nuclei of necrotic cells and eventually rupture, acidifying the cytoplasm [24]. Mutations in vacuolar H+-ATPase subunits, disruption of lysosome biogenesis, or alkali treatment are sufficient to inhibit necrosis induced by distinct stimuli [24,25]. Further, lysosomal aspartyl proteases (cathepsins) are required for necrosis in C. elegans [23], and necrosis can be reduced by expression of serpin-6, an inhibitor of calpains and lysosomal cysteine proteases [26]. These findings in C. elegans have provided strong support for an evolutionarily conserved “calpain-cathepsin hypothesis,” first described as mediating necrosis in mammalian neurodegeneration [11]. Mutations in autophagy genes partially suppress necrosis, indicating that autophagy acts to facilitate necrosis in C. elegans [27,28• •]. Genetic analysis indicates that calpains and autophagy genes act within the same pathway, but cathepsins and acidification of the cytosol act synergistically with autophagy [28• •].
In addition to neurons, necrotic cell death can be induced in uterine vulval 1 (uv1) cells in C. elegans (Figure 1A). Mutations in pnc-1, which encodes an enzyme in the salvage pathway of NAD+ biosynthesis, cause specific necrotic cell death of uv1 cells [29,30•]. The necrotic phenotype appears to be caused primarily by a build up of nicotinamide (NAM). As with the examples of necrotic cell death in neurons, uv1 cell death is not blocked by mutations in the apoptosis gene ced-4, but is partially inhibited by RNAi of calpain and aspartyl protease genes [29]. How NAM induces necrotic cell death is unclear, but Hanna-Rose and colleagues suggest that it could be through NAM’s role as an inhibitor of NAD+ consumers such as sirtuins [30•]. Sirtuins are NAD+-dependent histone deacetylases linked to aging and neurodegeneration in diverse organisms (reviewed in [31]). In several models of mammalian necrosis, NAM treatment is neuroprotective. It is thought that this occurs by replenishing depleted NAD+ [32]. It is intriguing that the NAD+ biosynthesis pathway is linked to necrosis in both mammals and C. elegans, yet NAM treatment produces opposite outcomes in the two organisms. Whether these differences are due to truly distinct mechanisms or different feedback pathways in the two systems remains to be determined.
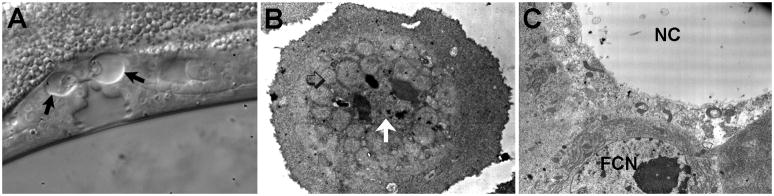
Figure 1.
Necrotic cells in different species. A) C. elegans pnc-1 mutant uv1 cells undergoing necrosis, visualized with Nomarski optics. Arrows indicate dramatic swelling of the cells. B) Dictyostelium atg1-deficient stalk cell undergoing necrosis, visualized by transmission electron microscopy. Mitochondria (open arrow) are clustered around the nucleus (white arrow) and the periphery of the cell is clear of organelles. C) Electron micrograph of a Drosophila wild-type nurse cell (NC) at the end of oogenesis; the region is largely devoid of organelles. Healthy follicle cell is nearby (FCN, follicle cell nucleus). Images kindly provided by Wendy Hanna-Rose (a, Reprinted from [29], Copyright (2006), with permission from Elsevier), Pierre Golstein (b, [35] © 2004 The American Society for Biochemistry and Molecular Biology) and Elizabeth Tanner (c, unpublished).
Dictyostelium discoideum
Dictyostelium is a protist that undergoes differentiation into a multicellular spore-producing organism under starvation conditions. Dictyostelium cells do not undergo apoptosis and lack genes encoding Bcl-2 family proteins, caspases and metacaspases. However, differentiating stalk cells undergo autophagic programmed cell death [33]. This form of cell death can also be induced in monolayers of cells subjected to starvation and treated with differentiation-inducing factor DIF-1 [34]. Genetic disruption of the autophagy gene atg-1 does not prevent cell death, but cells die with a distinct non-vacuolar morphology with characteristics of necrosis (Figure 1B) [34,35]. This system has become a very useful model for the dissection of events in necrotic cell death.
Dictyostelium atg-1 mutant cells stimulated to undergo necrosis show perinuclear clustering of mitochondria, an increase in ROS, and a depletion of ATP [34]. These changes occur within minutes of adding DIF-1 to the cultures, but interestingly, the effects on mitochondria and ROS are reversible if DIF-1 is removed, even after several hours of treatment [36• •]. The effects on ATP depletion are not reversible but cells in these early stages of necrosis do not progress further when DIF-1 is removed. Later steps in the progression of DIF-1-induced necrotic cell death include lysosome permeabilization and ultimately plasma membrane rupture. Lysosome permeabilization is not reversible upon removal of DIF-1, suggesting that this step is the “point of no return” for necrotic cell death in Dictyostelium.
Saccharomyces cerevisiae
The yeast Saccharomyces cerevisiae has recently emerged as a model for necrotic cell death. Yeast have been shown to undergo necrosis at old age, when exposed to certain chemicals, or when ectopically expressing genes associated with human neurodegeneration [37,38•]. Twenty day-old yeast cells show hallmarks of necrosis such as plasma membrane rupture and swollen organelles, and this death can be inhibited by treatment with spermidine [39• •]. Additionally, spermidine treatment of aged yeast inhibited nuclear release of the chromatin-associated high mobility group Box 1 protein (HMGB1) [39• •], a phenomenon characteristic of mammalian necrosis. Interestingly, spermidine can extend lifespan in multiple organisms, suggesting a link between increased necrosis and aging. Spermidine was also found to increase autophagy, suggesting that this could be a mechanism for the suppression of necrosis. Further support for a protective role for autophagy against necrosis was shown in yeast cells treated with acetic acid. Treatment of wild-type yeast with acetic acid leads to cells dying by both apoptosis and necrosis whereas mutants defective in yeast vacuole formation (analogous to autophagy in higher organisms) become exclusively necrotic, as revealed by propidium iodide uptake [40]. This role for autophagy in protecting against necrosis is at odds with the findings in C. elegans.
Drosophila melanogaster
Drosophila has been remarkably under-developed as a model for necrosis. Numerous models for neurodegeneration have been reported, but the type of cell death in many cases is not clear (reviewed in [41–43]). Most fly examples of neurodegeneration fall into three broad classes of mutants: those that affect the phototransduction cascade, those that produce protein aggregates, and those that affect ion homeostasis. In several mutants with disruption in the phototransduction cascade, cell death is not blocked with caspase inhibitors, although the morphology may resemble apoptosis [44,45]. Some Drosophila models of human neurodegeneration also fail to be inhibited by caspase inhibitors [41], although Ark, which acts upstream of caspases in the apoptosis cascade, has been shown to inhibit polyglutamine toxicity [46]. In several models, autophagy acts as a protective mechanism against neurodegeneration [47,48•], and blocking autophagy alone is sufficient to lead to light-dependent retinal degeneration [48•]. Thus, several examples of neurodegeneration do not appear to occur by typical apoptosis or autophagic cell death, but a necrotic morphology has not been described either. In some cases, events common to necrosis have been reported, such as an increase in oxidative stress or calcium levels [42,44,49], or large tissue vacuolization [44,50,51]. However, no specific descriptions of necrosis have been reported in Drosophila neurodegeneration models.
As in C. elegans, it is not clear that there is any developmental necrosis in Drosophila. One possible exception is an unusual form of cell death that occurs in the fly ovary at the completion of oogenesis (reviewed in [52]). Each developing Drosophila oocyte is connected to fifteen support cells called nurse cells. Near the end of oogenesis, the nurse cell cytoplasm is rapidly transferred to the oocyte through intercellular bridges. At this point, the nurse cell nuclei are excluded from the oocyte, retaining only a small amount of cytoplasm and few organelles. The nurse cell nuclei disappear over a few hours, displaying chromatin condensation and becoming TUNEL-positive [53–55]. Caspases appear to be weakly activated during this process, and autophagosomes can be detected; however, inhibition of caspases or autophagy causes only a partial inhibition of nurse cell death [56–60]. Importantly, caspase- or autophagy-inhibited nurse cells die with the same gross morphology as wild-type nurse cells, suggesting that they are not shifted to a different mode of cell death, although this has not been examined by electron microscopy. These findings suggest that nurse cell death does not normally occur by apoptosis or autophagic cell death.
Developmental nurse cell death displays several of the common features of necrotic cell death seen in Dictyostelium and C. elegans. Early stages of nurse cell death show condensed chromatin, but in later stages the chromatin becomes largely diffuse, and eventually only an empty space remains (Figure 1C) [59•]. The disappearance of the nuclei is accompanied by acidification of the nurse cell remnants, and ruptured organelles are detectable by electron microscopy [59•]. Mutations in lysosomal fusion proteins and enzymes can suppress nurse cell death. Another common feature is the involvement of calcium which rises in the cytosol of nurse cells prior to cytoplasmic transfer [61]. The timing is appropriate for calcium-dependent signaling or proteases to play a role in nurse cell death, although this remains to be shown.
Mammalian pathways to necrosis
Necrosis is induced in response to diverse stimuli in mammals, and the topic has been covered in several excellent recent reviews [6•,14, 62]. I will briefly highlight two key effectors of mammalian necrosis. The kinase RIP1 has been shown to play an essential role in promoting necrosis in response to TNFα, TRAIL or FasL stimulation in mammals. RIP1 also acts to promote NF-kB activation and cell survival, and caspase-8 dependent apoptosis in certain contexts [6•,14, 62]. However, when caspases are inhibited, RIP1 interacts with a second kinase, RIP3, and switches to a necrotic pathway [6•,14, 62]. Necrotic effectors acting downstream of RIP1 still remain to be elucidated, but a recent siRNA screen for effectors of RIP1-mediated necrosis has identified a number of intriguing possibilities [63• •]. Cyclophilin-D, a mitochondrial peptidyl prolyl cis-trans isomerase that regulates the mitochondrial permeability transition, has been shown to control necrosis activated by diverse stimuli [64,65]. Cyclophilin-D-deficient mice have proven to be a powerful tool for dissecting cell death mechanisms in several mouse disease models [12,15,16]. There are no reports of RIP1, RIP3 or Cyclophilin-D orthologs in invertebrates [66], suggesting that these proteins direct vertebrate-specific pathways to activate necrosis, or divergent molecules carry out these functions in invertebrates. Despite these differences, there are many common events among different organisms, including the involvement of calcium, ROS, organelle rupture, acidification of the cytoplasm, and a requirement for specific proteases (Figure 2).
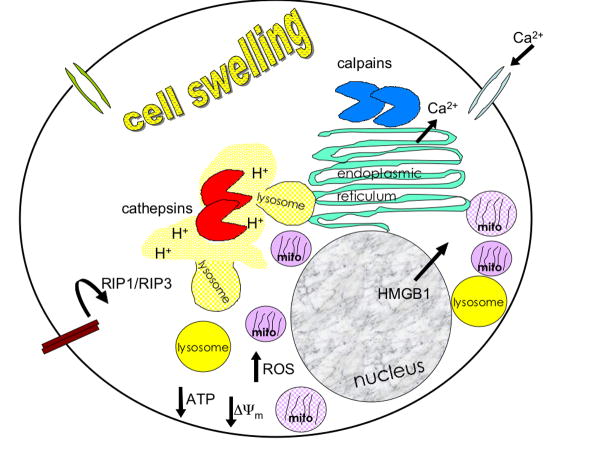
Figure 2.
Common cellular events during necrosis. In some examples of necrosis, an increase in cytoplasmic Ca2+ is an early event, leading to the activation of calcium-dependent proteases such as calpains. Influx of Ca2+ may be from intracellular stores or extracellular sources. Mitochondria (pink) and lysosomes (yellow) are seen to cluster around the nucleus (gray). Disruptions of mitochondrial permeability transition lead to a loss of membrane potential (ΔΨm), mitochondrial swelling and rupture. Lysosomal rupture leads to acidification of the cytoplasm (H+) and release of cathepsins. Nuclear export of the chromatin protein HMGB1 has been reported in yeast and mammals. TNFR-activation of RIP1 is an important initiator of necrosis in mammals but has not been shown to be conserved in other species. Many necrotic cells will eventually show rupture of the plasma membrane.
Conclusions
It is now apparent from work in multiple organisms that necrosis is not an uncontrolled process, but that it is under genetic control. Necrosis can be suppressed by treatment with specific inhibitors, genetic mutation of downstream effectors, or alkalinization. Many of the molecules thus far identified in necrosis are evolutionarily conserved, suggesting that necrosis is an ancient process. However, some aspects are puzzling, such as the opposing effects of autophagy, acting to promote necrosis in C. elegans but protecting against necrosis in yeast and Dictyostelium. This difference may depend on specific requirements for subcellular events, such as ROS generation (expected to be inhibited by autophagy) versus lysosomal activity (which could be promoted by autophagy).
One remaining enigma is that there are few examples of necrosis occurring during development. However, necrosis appears to be widespread during infection and disease. It has been suggested that a key role of necrosis is to allow for cell death to occur when apoptosis is specifically inhibited, as may occur during viral infection or tumorigenesis [62• •]. Others have suggested that necrosis may be the more primitive form of cell death, with apoptosis and autophagic cell death having evolved later [67]. Some stimuli such as ROS can promote either apoptosis or necrosis, with necrosis being activated after a massive insult. Necrosis may be the major type of cell death triggered by a significant ion imbalance. Many details of the mechanisms of necrosis have yet to be uncovered, but are of high clinical relevance. The initial studies highlighted here show that necrosis can be genetically programmed rather than accidental, and provide a strong foundation for further dissection of the molecular pathways.
Acknowledgments
I thank Wendy Hanna-Rose, Pierre Golstein and Elizabeth Tanner for providing images, Wendy Hanna-Rose and Patrick Dolph for helpful discussions, and members of my laboratory for comments on the manuscript. KM is supported by National Institutes of Health grant R01 GM060574.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review are highlighted as:
•Of special interest
• • Of outstanding interest
- 1.Kerr JFR, Wyllie AH, Currie AR. Apoptosis, a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerr JF. History of the events leading to the formulation of the apoptosis concept. Toxicology. 2002;181–182:471–474. doi: 10.1016/s0300-483x(02)00457-2. [DOI] [PubMed] [Google Scholar]
- 3.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 4•.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. The paper makes important recommendations for nomenclature, methods and criteria used to describe different cell death modes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin S, Bucci TJ, Cohen SM, Fix AS, Hardisty JF, LeGrand EK, Maronpot RR, Trump BF. The nomenclature of cell death: recommendations of an ad hoc Committee of the Society of Toxicologic Pathologists. Toxicol Pathol. 1999;27:484–490. doi: 10.1177/019262339902700419. [DOI] [PubMed] [Google Scholar]
- 6•.Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol. 2010;22:263–268. doi: 10.1016/j.ceb.2009.12.003. This article is a recent review on RIP1-mediated necrosis in mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem Sci. 2007;32:37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Wyllie AH, Kerr JF, Currie AR. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 9.Searle J, Kerr JF, Bishop CJ. Necrosis and apoptosis: distinct modes of cell death with fundamentally different significance. Pathol Annu. 1982;17(Pt 2):229–259. [PubMed] [Google Scholar]
- 10.Kepp O, Tesniere A, Schlemmer F, Michaud M, Senovilla L, Zitvogel L, Kroemer G. Immunogenic cell death modalities and their impact on cancer treatment. Apoptosis. 2009;14:364–375. doi: 10.1007/s10495-008-0303-9. [DOI] [PubMed] [Google Scholar]
- 11.Yamashima T. Ca2+-dependent proteases in ischemic neuronal death: a conserved 'calpain-cathepsin cascade' from nematodes to primates. Cell Calcium. 2004;36:285–293. doi: 10.1016/j.ceca.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Millay DP, Sargent MA, Osinska H, Baines CP, Barton ER, Vuagniaux G, Sweeney HL, Robbins J, Molkentin JD. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat Med. 2008;14:442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy CL, Smith DJ, Lyras D, Chakravorty A, Rood JI. Programmed cellular necrosis mediated by the pore-forming alpha-toxin from Clostridium septicum. PLoS Pathog. 2009;5:e1000516. doi: 10.1371/journal.ppat.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challa S, Chan FK. Going up in flames: necrotic cell injury and inflammatory diseases. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0413-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto K, Chen Y, Polonsky KS, Dorn GW., 2nd Targeting cyclophilin D and the mitochondrial permeability transition enhances beta-cell survival and prevents diabetes in Pdx1 deficiency. Proc Natl Acad Sci U S A. 2010;107:10214–10219. doi: 10.1073/pnas.0914209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111. This article provides a detailed review of mechanisms of apoptosis, necrosis and autophagy, and their contributions to heart disease. [DOI] [PubMed] [Google Scholar]
- 17.Schulze C, Munoz LE, Franz S, Sarter K, Chaurio RA, Gaipl US, Herrmann M. Clearance deficiency--a potential link between infections and autoimmunity. Autoimmun Rev. 2008;8:5–8. doi: 10.1016/j.autrev.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 18.Kourtis N, Tavernarakis N. Non-developmentally programmed cell death in Caenorhabditis elegans. Semin Cancer Biol. 2007;17:122–133. doi: 10.1016/j.semcancer.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 19•.Blum ES, Driscoll M, Shaham S. Noncanonical cell death programs in the nematode Caenorhabditis elegans. Cell Death Differ. 2008;15:1124–1131. doi: 10.1038/cdd.2008.56. This is a recent review describing alternative cell death pathways in C. elegans. [DOI] [PubMed] [Google Scholar]
- 20.Hall DH, Gu G, Garcia-Anoveros J, Gong L, Chalfie M, Driscoll M. Neuropathology of degenerative cell death in Caenorhabditis elegans. J Neurosci. 1997;17:1033–1045. doi: 10.1523/JNEUROSCI.17-03-01033.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger AJ, Hart AC, Kaplan JM. Gαs-induced neurodegeneration in Caenorhabditis elegans. J Neurosci. 1998;18:2871–2880. doi: 10.1523/JNEUROSCI.18-08-02871.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K, Tavernarakis N, Driscoll M. Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca(2+) release from the endoplasmic reticulum. Neuron. 2001;31:957–971. doi: 10.1016/s0896-6273(01)00432-9. [DOI] [PubMed] [Google Scholar]
- 23.Syntichaki P, Xu K, Driscoll M, Tavernarakis N. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 24.Artal-Sanz M, Samara C, Syntichaki P, Tavernarakis N. Lysosomal biogenesis and function is critical for necrotic cell death in Caenorhabditis elegans. J Cell Biol. 2006;173:231–239. doi: 10.1083/jcb.200511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syntichaki P, Samara C, Tavernarakis N. The vacuolar H+-ATPase mediates intracellular acidification required for neurodegeneration in C. elegans. Curr Biol. 2005;15:1249–1254. doi: 10.1016/j.cub.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 26.Luke CJ, Pak SC, Askew YS, Naviglia TL, Askew DJ, Nobar SM, Vetica AC, Long OS, Watkins SC, Stolz DB, et al. An intracellular serpin regulates necrosis by inhibiting the induction and sequelae of lysosomal injury. Cell. 2007;130:1108–1119. doi: 10.1016/j.cell.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toth ML, Simon P, Kovacs AL, Vellai T. Influence of autophagy genes on ion-channel-dependent neuronal degeneration in Caenorhabditis elegans. J Cell Sci. 2007;120:1134–1141. doi: 10.1242/jcs.03401. [DOI] [PubMed] [Google Scholar]
- 28••.Samara C, Syntichaki P, Tavernarakis N. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 2008;15:105–112. doi: 10.1038/sj.cdd.4402231. The authors show that autophagy acts to promote necrosis in C. elegans and provide genetic evidence that the autophagy pathway functions synergistically with lysosomal enzymes. [DOI] [PubMed] [Google Scholar]
- 29.Huang L, Hanna-Rose W. EGF signaling overcomes a uterine cell death associated with temporal mis-coordination of organogenesis within the C. elegans egg-laying apparatus. Dev Biol. 2006;300:599–611. doi: 10.1016/j.ydbio.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 30•.Vrablik TL, Huang L, Lange SE, Hanna-Rose W. Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development. 2009;136:3637–3646. doi: 10.1242/dev.028431. This paper further characterizes a non-neuronal model for necrosis in C. elegans first described in [28]. The authors demonstrate that excessive NAM can promote necrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang BL. Sirt1's complex roles in neuroprotection. Cell Mol Neurobiol. 2009;29:1093–1103. doi: 10.1007/s10571-009-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, Gharavi R, Pitta M, Gleichmann M, Mattson MP. Nicotinamide prevents NAD+ depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD+ consumption by SIRT1 may endanger energetically compromised neurons. Neuromolecular Med. 2009;11:28–42. doi: 10.1007/s12017-009-8058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornillon S, Foa C, Davoust J, Buonavista N, Gross JD, Golstein P. Programmed cell death in Dictyostelium. J Cell Sci. 1994;107 (Pt 10):2691–2704. doi: 10.1242/jcs.107.10.2691. [DOI] [PubMed] [Google Scholar]
- 34.Laporte C, Kosta A, Klein G, Aubry L, Lam D, Tresse E, Luciani MF, Golstein P. A necrotic cell death model in a protist. Cell Death Differ. 2007;14:266–274. doi: 10.1038/sj.cdd.4401994. [DOI] [PubMed] [Google Scholar]
- 35.Kosta A, Roisin-Bouffay C, Luciani MF, Otto GP, Kessin RH, Golstein P. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J Biol Chem. 2004;279:48404–48409. doi: 10.1074/jbc.M408924200. [DOI] [PubMed] [Google Scholar]
- 36••.Giusti C, Luciani MF, Klein G, Aubry L, Tresse E, Kosta A, Golstein P. Necrotic cell death: From reversible mitochondrial uncoupling to irreversible lysosomal permeabilization. Exp Cell Res. 2009;315:26–38. doi: 10.1016/j.yexcr.2008.09.028. The authors characterize the progression of necrosis in Dictyostelium, and present the remarkable finding that necrosis can be halted by removing the inducer, even after mitochondrial uncoupling has occurred. [DOI] [PubMed] [Google Scholar]
- 37.Braun RJ, Buttner S, Ring J, Kroemer G, Madeo F. Nervous yeast: modeling neurotoxic cell death. Trends Biochem Sci. 2010;35:135–144. doi: 10.1016/j.tibs.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 38•.Eisenberg T, Carmona-Gutierrez D, Buttner S, Tavernarakis N, Madeo F. Necrosis in yeast. Apoptosis. 2010;15:257–268. doi: 10.1007/s10495-009-0453-4. This article provides a recent review on necrosis in yeast, and describes methods for distinguishing necrosis from apoptosis. [DOI] [PubMed] [Google Scholar]
- 39••.Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. This paper shows that spermidine protects against necrosis and induces autophagy in yeast. [DOI] [PubMed] [Google Scholar]
- 40.Schauer A, Knauer H, Ruckenstuhl C, Fussi H, Durchschlag M, Potocnik U, Frohlich KU. Vacuolar functions determine the mode of cell death. Biochim Biophys Acta. 2009;1793:540–545. doi: 10.1016/j.bbamcr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 42.Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 43.Lu B. Recent advances in using Drosophila to model neurodegenerative diseases. Apoptosis. 2009;14:1008–1020. doi: 10.1007/s10495-009-0347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alloway PG, Howard L, Dolph PJ. The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28:129–138. doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 45.Hsu CD, Whaley MA, Frazer K, Miller DA, Mitchell KA, Adams SM, O'Tousa JE. Limited role of developmental programmed cell death pathways in Drosophila norpA retinal degeneration. J Neurosci. 2004;24:500–507. doi: 10.1523/JNEUROSCI.3328-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sang TK, Li C, Liu W, Rodriguez A, Abrams JM, Zipursky SL, Jackson GR. Inactivation of Drosophila Apaf-1 related killer suppresses formation of polyglutamine aggregates and blocks polyglutamine pathogenesis. Hum Mol Genet. 2005;14:357–372. doi: 10.1093/hmg/ddi032. [DOI] [PubMed] [Google Scholar]
- 47.Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Wang T, Lao U, Edgar BA. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J Cell Biol. 2009;186:703–711. doi: 10.1083/jcb.200904090. This paper shows that induction of autophagy can protect against multiple models of retinal degeneration in flies but does not protect against a caspase-dependent form of degeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias–Santagata D, Fulga TA, Duttaroy A, Feany MB. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J Clin Invest. 2007;117:236–245. doi: 10.1172/JCI28769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fergestad T, Ganetzky B, Palladino MJ. Neuropathology in Drosophila membrane excitability mutants. Genetics. 2006;172:1031–1042. doi: 10.1534/genetics.105.050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gnerer JP, Kreber RA, Ganetzky B. wasted away, a Drosophila mutation in triosephosphate isomerase, causes paralysis, neurodegeneration, and early death. Proc Natl Acad Sci U S A. 2006;103:14987–14993. doi: 10.1073/pnas.0606887103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pritchett TL, Tanner EA, McCall K. Cracking open cell death in the Drosophila ovary. Apoptosis. 2009;14:969–979. doi: 10.1007/s10495-009-0369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavaliere V, Taddei C, Gargiulo G. Apoptosis of nurse cells at the late stages of oogenesis. Dev Genes Evol. 1998;208:106–112. doi: 10.1007/s004270050160. [DOI] [PubMed] [Google Scholar]
- 54.Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- 55.McCall K, Steller H. Requirement for DCP-1 caspase during Drosophila oogenesis. Science. 1998;279:230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- 56.Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in Drosophila oogenesis. Dev Biol. 2003;260:113–123. doi: 10.1016/s0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 57.Mazzalupo S, Cooley L. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 2006;13:1950–1959. doi: 10.1038/sj.cdd.4401892. [DOI] [PubMed] [Google Scholar]
- 58.Baum JS, Arama E, Steller H, McCall K. The Drosophila caspases Strica and Dronc function redundantly in programmed cell death during oogenesis. Cell Death Differ. 2007;14:1508–1517. doi: 10.1038/sj.cdd.4402155. [DOI] [PubMed] [Google Scholar]
- 59•.Bass BP, Tanner EA, Mateos San Martin D, Blute T, Kinser RD, Dolph PJ, McCall K. Cell-autonomous requirement for DNaseII in nonapoptotic cell death. Cell Death Differ. 2009;16:1362–1371. doi: 10.1038/cdd.2009.79. This paper provides evidence that developmental nurse cell death in Drosophila has characteristics of necrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 190:523–31. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matova N, Mahanjan-Miklos S, Mooseker MS, Cooley L. Drosophila Quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development. 1999;126:5645–5657. doi: 10.1242/dev.126.24.5645. [DOI] [PubMed] [Google Scholar]
- 62.Moquin D, Chan FK. The molecular regulation of programmed necrotic cell injury. Trends Biochem Sci. 2010;5:434–41. doi: 10.1016/j.tibs.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63• •.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. This article presents the findings from a genome-wide siRNA screen for effectors of mammalian necrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 65.Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 66.Pemberton TJ, Kay JE. Identification and Comparative Analysis of the Peptidyl-Prolyl cis/trans Isomerase Repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe. Comp Funct Genomics. 2005;6:277–300. doi: 10.1002/cfg.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golstein P, Kroemer G. Redundant cell death mechanisms as relics and backups. Cell Death Differ. 2005;12 (Suppl 2):1490–1496. doi: 10.1038/sj.cdd.4401607. [DOI] [PubMed] [Google Scholar]