Abstract
Localization of nanos (nos) mRNA to the posterior pole of the Drosophila oocyte is essential for abdominal segmentation and germline development during embryogenesis. Posterior localization is mediated by a complex cis-acting localization signal in the nos 3′ untranslated region that comprises multiple partially redundant elements. Genetic analysis suggests that this signal is recognized by RNA-binding proteins and associated factors that package nos mRNA into a localization competent ribonucleoprotein complex. However, functional redundancy among localization elements has made the identification of individual localization factors difficult. Indeed, only a single direct-acting nos localization factor, Rumpelstiltskin (Rump), has been identified thus far. Through a sensitized genetic screen, we have now identified the Argonaute family member Aubergine (Aub) as a nos localization factor. Aub interacts with nos mRNA in vivo and co-purifies with Rump in an RNA-dependent manner. Our results support a role for Aub, independent of its function in RNA silencing, as a component of a nos mRNA localization complex.
INTRODUCTION
mRNA localization is a widespread mechanism used to achieve intracellular polarity in diverse cellular and developmental contexts. Over 1500 transcripts are localized to multiple distinct subcellular locations in the early Drosophila embryo alone, representing approximately 70% of all mRNAs expressed at this time (Lecuyer et al., 2007). Thus, developmental processes require that localized mRNAs be distinguished not only from uniformly distributed transcripts, but also from mRNAs destined for other subcellular locations.
Four localized mRNAs involved in axial patterning within the Drosophila oocyte - gurken (grk), bicoid (bcd), oskar (osk) and nanos (nos) - have established a paradigm for studying the localization of different transcripts to distinct regions within a cell. All four are maternal mRNAs that are synthesized by the ovarian nurse cells, then transported into the oocyte through connecting cytoplasmic channels. During midoogenesis, grk mRNA is localized to the dorsal-anterior region of the oocyte, where it is translated to produce a TGFα-like ligand that signals to overlying somatic follicle cells to establish the dorsoventral (D-V) axis (Neuman-Silberberg and Schüpbach, 1993). Concurrently, osk mRNA localizes to the posterior pole, where its translation is activated. Production of Osk protein is required to maintain osk mRNA and protein localization and to initiate the assembly of the germ plasm, a specialized cytoplasm containing determinants for germ cell formation and, ultimately, nos mRNA (Ephrussi et al., 1991; Kim-Ha et al., 1991; Rongo et al., 1995; Vanzo and Ephrussi, 2002). Transport of grk and osk mRNAs to their destinations is mediated respectively by dynein and kinesin motors and relies on the polarization of the oocyte microtubule cytoskeleton that occurs earlier in oogenesis (reviewed in Becalska and Gavis, 2009).
Although some bcd mRNA is localized to the anterior margin of the oocyte during midoogenesis, the majority does not localize until late stages of oogenesis, after the nurse cells have initiated apoptosis and extruded or “dumped” their contents into the oocyte (Berleth et al., 1988; Weil et al., 2006). It is also during this late phase of oogenesis that nos accumulates within the germ plasm at the oocyte posterior (Forrest and Gavis, 2003). Whereas bcd transport is dynein-dependent, nos is localized passively, being dispersed throughout the oocyte by the concerted streaming of the ooplasm that follows nurse cell dumping, and trapped at the posterior by association with the germ plasm (Forrest and Gavis, 2003; Weil et al., 2006). Localized bcd and nos mRNAs function subsequently during embryogenesis to pattern the anterior-posterior (A-P) axis, through the production of protein gradients that emanate from the anterior and posterior poles respectively. Bcd specifies the development of head and thoracic structures, and mutations that compromise Bcd activity or localization disrupt anterior patterning (Driever and Nüsslein-Volhard, 1988). Nos function is required at the posterior of the embryo for the formation of the eight abdominal segments of the animal, and mutation of nos, or disruption of nos mRNA localization, results in decreased abdominal segmentation (Lehmann and Nusslein-Volhard, 1991; Wang et al., 1994).
The information regulating the specificity of mRNA localization is contained in cis-acting localization elements, typically found in the 3′ untranslated regions (3′UTRs) of localized transcripts (Gavis et al., 2007). These sequences are recognized by trans-acting localization factors that are thought to recruit additional proteins to package mRNAs into ribonucleoprotein (RNP) particles for transport to and anchoring at target destinations. A number of localization factors have been identified for bcd, osk and grk mRNAs (Kugler and Lasko, 2009) but much less is known about the trans-acting factors involved in nos mRNA localization. Traditional genetic screens have been unsuccessful in identifying nos mRNA localization factors due to the complex organization of the nos localization signal. The nos 3′UTR contains four localization elements that play partially redundant roles in nos localization (Gavis et al., 1996). The lack of sequence or structural similarity between the nos localization elements suggests that each is recognized by distinct localization factors that act in combination to mediate localization (Bergsten and Gavis, 1999). However, because no single element is necessary or sufficient for wild-type localization, eliminating a single localization factor is unlikely to result in a phenotypically detectable nos localization defect. Supporting this idea, mutations in the two identified nos localization factors, Rumpelstiltskin (Rump) and Hsp90 cause minimal defects in nos accumulation at the posterior pole (Jain and Gavis, 2008; Song et al., 2007). Rump was identified biochemically by its ability to bind directly to the nos localization signal; however, its requirement in nos mRNA localization is only revealed when the nos localization signal itself is compromised by deletion of two localization elements (Jain and Gavis, 2008). Hsp90 was identified in a dominant modifier screen for nos localization factors using a similar compromised localization signal and, like Rump, its requirement in nos localization is only apparent under these sensitized conditions (Song et al., 2007). How directly Hsp90 regulates nos localization remains unknown.
Here we describe the results of a new sensitized genetic screen for additional nos localization factors and demonstrate that the repeat-associated small interfering RNA (rasiRNA) pathway gene aubergine (aub) plays a role in spatial restriction of nos mRNA. While aub has previously been implicated in localization of osk mRNA (Cook et al., 2004; Wilson et al., 1996), we show that the effect of aub on nos mRNA localization is independent of its upstream effect on osk and the rasiRNA pathway. Furthermore, we demonstrate in vivo interactions between Aub, nos mRNA, and Rump, implicating Aub directly as a novel nos mRNA localization factor.
MATERIALS AND METHODS
Fly stocks and genetics
The y w67c23 strain (Lindsley and Zimm, 1992) was used for wild-type controls and generation of nos+1+3 transgenic lines. Deficiency stocks consisted of deficiency kits covering chromosomal arms 2L and 2R (Bloomington). The following mutants and transgenic lines were used: nosBN (Wang et al., 1994), aubQC42, aubHN2 (Schüpbach and Wieschaus, 1991), squHE47 (Pane et al., 2007), mnkp6 (Abdu et al., 2002), oskA87 (Jenny et al., 2006), nos-gal4-vp16; gfp-aub (Harris and Macdonald, 2001), FM7i GFP (Bloomington), hsp83-MCP-GFP (Forrest and Gavis, 2003), rump1 (Jain and Gavis, 2008). Unless otherwise indicated, aubQC/aubHN transheterozygotes were used as aub mutants and aubQC/+ as heterozygotes. For mnk, aub double mutants, mnkp6, aubQC42 and mnkp6, aubHN2 recombinant chromosomes were used in trans-heterozygous combination (Klattenhoff et al., 2007).
Generation of the nos+1+3 sensitized background
The nos+1+3 transgene includes the nos 5′UTR and coding region followed by the sensitized 3′UTR containing nucleotides 6-96 (the +1 element), 181-408 (the +3 element), and 548-786 of the nos 3′UTR (Gavis et al., 1996), inserted into pCaSpeR2. The transgene also contains MS2 stem-loops for in vivo mRNA visualization, inserted near the terminus of the 3′UTR; they do not affect the behavior of nos mRNA (Forrest and Gavis, 2003). Transgenic lines were generated by standard P element-mediated germline transformation (Spradling, 1986). Cloning details are available upon request.
Screening procedure
Each deficiency stock from the second chromosome kits was crossed to generate Df/+; nosBN, nos+1+3/nosBN females from which embryos were collected overnight on apple juice plates. Embryonic cuticle preparation was performed as previously described (Crucs et al., 2000). The average number of segments and the standard error (SEM) were calculated using standard histogram distribution analysis (Graphpad Prism).
In situ hybridization and immunostaining
Embryos were collected from 0–2 hours after egg laying and in situ hybridization was performed as described (Gavis and Lehmann, 1992). Immunofluorescent staining of embryos aged 3–5 hours was performed as previously described (Duchow et al., 2005) using rabbit anti-Vas (1:10000; gift from R. Lehmann), Alexa Fluor-568 goat anti-rabbit (1:1000; Molecular Probes) and DAPI (1:1000). For the analysis of pole cell number, a z-series of each embryo, at 1.5 μm intervals from the first focal plane with a visible pole cell to the last, was taken with a Zeiss LSM510 confocal microscope and 40x/1.3 oil objective, and projections were generated using ImageJ (NIH).
Northern blot analysis
Total RNA was extracted from dechorionated 0–1.5 hour old embryos using TRIzol reagent (Invitrogen) and northern blotting was carried out according to Bergsten and Gavis (1999). Quantitation of blots was performed by phosphorimaging (Molecular Dynamics).
Immunoblotting
Immunoblotting of extracts from 0–2 hour old embryos was performed as previously described (Forrest et al., 2004). Final antibody concentrations were: rabbit anti-Nos (1:1000; gift of A. Nakamura), rabbit anti-Osk (1:3000;Vanzo and Ephrussi, 2002), mouse anti-Snf (1:20000; gift of P. Schedl), and rabbit anti-Khc (1:30000; Cytoskeleton). Proteins were detected using the appropriate HRP conjugated secondary antibodies and Lumi-Light Western Blot Substrate (Roche). Quantitation was performed using an Alpha Innotech imager and AlphaEase software (Alpha Innotech) or using ImageJ (NIH).
Immunoprecipitation
RNA co-immunoprecipitation and RT-PCR analysis were performed as described (Jain and Gavis, 2008) except that extracts were prepared from freshly dissected ovaries and immunoprecipitation buffer contained 150 mM NaCl. RT-PCR analysis was performed with primers for nos (Jain and Gavis, 2008), osk (5′-AATGGATCCAGTGTGCAGAAAATC-3′ and 5′-AGCGAATGCTGTCACCTA-3′) and his3.3b (5′-GATTGATTCCGCATAAAGCGCG-3′ and 5′-AAGGAGCACGGCGCAACGTACA-3′). Protein co-immunoprecipitation using polyclonal anti-GFP antibody (Abcam 290) was performed according to Kalifa et al. (Kalifa et al., 2009) except that fresh ovaries were used and extracts were pre-incubated with Protein G Dynabeads (Invitrogen) for 1 hour at 4° C, then applied to anti-GFP-coated Protein G Dynabeads (Invitrogen) for 1 hour at 4° C. Samples were analyzed by SDS-PAGE and immunoblotting with monoclonal anti-GFP (Clontech JL-8) and anti-Rump (5G4) antibodies.
RESULTS
Deficiency screen for nos mRNA localization factors
The nos localization signal is composed of four unique elements designated as +1, +2′, +3 and +4, no one of which is necessary or sufficient for wild-type nos mRNA localization (Gavis et al., 1996) (Fig. 1A). To sensitize nos mRNA to the loss of a candidate localization factor, we deleted the +2′ and +4 elements resulting in a transgene bearing only the +1 and +3 localization element sequences (nos+1+3; Fig. 1A). When this transgene is introduced into nosBN females, the transgenic nos+1+3 mRNA is the only maternal nos mRNA expressed. Because posterior localization of nos+1+3 mRNA is decreased relative to wild-type nos mRNA (Fig. 2A), embryos produced by these females exhibit reduced abdominal segmentation, forming an average of 6 of the 8 abdominal segments characteristic of wild-type embryos (Figs. 1A and B). For simplicity, we refer hereafter to this maternal genetic background as nos+1+3 and the resulting embryos as nos+1+3 embryos. To identify novel nos mRNA localization factors, we performed a deficiency screen of the second chromosome in the nos+1+3 background, reasoning that decreasing the dosage of a candidate localization factor would further compromise nos localization and, consequently, abdominal segmentation (Fig. 1A). A total of 18 of the 123 deficiencies tested produced a reduction of segment number to below a threshold of 3 abdominal segments (Table 1). These included deficiencies uncovering cappuccino (cap), chickadee (chic), staufen (stau), valois (vls) and vasa (vas), all of which have previously been implicated in patterning of the A-P axis (Hay et al., 1988; Lasko and Ashburner, 1988; Manseau and Schüpbach, 1989), validating the premise of the screen.
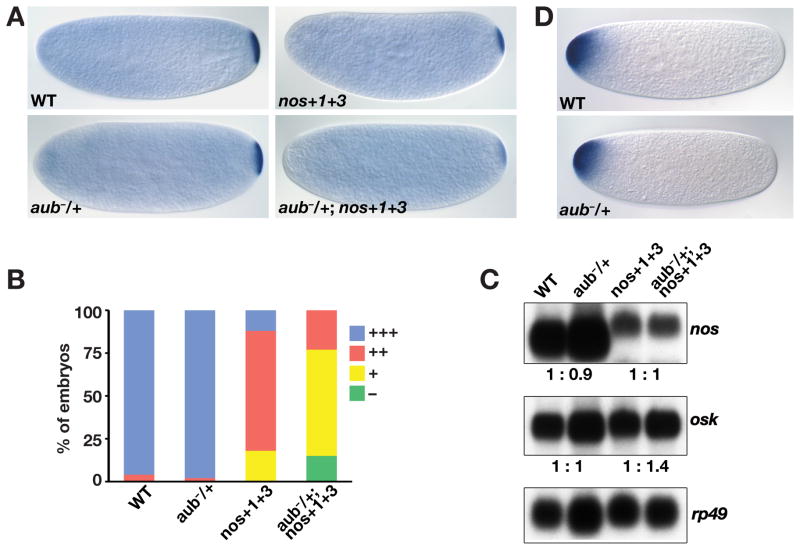
Figure 1.
Identification of aub as a nos mRNA localization factor. (A) Schematics of the 3′UTR of wild-type nos mRNA and the sensitized nos+1+3 transgene. The four previously defined localization signal elements are indicated, the black bar represents the remainder of the nos 3′UTR. (B) Bar chart showing the average number of abdominal segments in embryos from wild-type females (8 segments, n = 106), nos+1+3 females (5.8 segments, n = 332), and nos+1+3 females that are heterozygous for Df(2L)BSC32 (2.4 segments, n = 70), aubQC42 (n = 64), aubHN2 (n=145), or squHE47 (6.2 segments, n = 216). Df(2L)BSC32, aubQC42, and aubHN2 significantly reduce segment number in nos+1+3 derived embryos (*P < 0.0001). Error bars indicate SEM.
Figure 2.
aub affects localization of a sensitized nos mRNA. (A, B) Localization of wild-type nos or nos+1+3 mRNA was evaluated by in situ hybridization to embryos from wild-type females (WT), females heterozygous for aubQC42 (aub−/+) and nos+1+3 females heterozygous for aubQC42 (aub−/+; nos+1+3). A typical example of the nos RNA localization pattern in each genotype is shown (A). Localization was classified as wild-type (+++), moderate (++), weak (+), or undetectable (−). Localization of wild-type nos is unaffected by heterozygosity for aub whereas localization of the nos+1+3 transgene is further compromised by reducing aub (wild-type, n = 200; aub−/+, n = 97; nos+1+3, n = 305; aub−/+; nos+1+3, n = 341). (C) Northern blot of total mRNA isolated from embryos of the indicated genotypes, probed simultaneously for nos and for rp49 as a loading control. The blot was stripped and reprobed for osk. The relative nos and osk mRNA levels for each sample pair, after normalization to rp49, are indicated below. (D) In situ hybridization to bcd mRNA embryos from wild-type or aubQC/+ females. Heterozygosity for aub does not affect bcd mRNA localization.
Table 1.
Deficiencies enhancing the abdominal segmentation defects of nos+1+3
| Deficiency | Average segment number | Cytological breakpoints |
|---|---|---|
| control | 4.58 ± 1.79 | - |
| Df(2L)dp-79b | 2.48 ± 1.22 | 22A2-3;22D5-E1 |
| Df(2L)ed1 | 2.94 ± 2.22 | 24A2;24D4 |
| Df(2L)E110 | 2.75 ± 0.97 | 25F3-26A1;26D3-11 |
| Df(2L)Dwee1-W05 | 2.80 ± 1.41 | 27C2-3;27C4-5 |
| Df(2L)BSC32 | 2.41 ± 1.22 | 32A1-2;32C5-D1 |
| Df(2L)FCK-20 | 2.85 ± 1.22 | 32D1;32F1-3 |
| Df(2L)b87e25 | 1.67 ± 0.67 | 34B12-C1;35B10-C1 |
| Df(2L)r10 | 2.00 ± 0.49 | 35D1;36A6-7 |
| Df(2L)TW161 | 2.97 ± 1.08 | 38A6-B1;40A4-B1 |
| Df(2R)X1 | 2.70 ± 0.78 | 46C;47A1 |
| Df(2R)en-A | 2.39 ± 1.00 | 47D3;48B2 |
| Df(2R)ED2222 | 2.07 ± 0.65 | 47F13;48B6 |
| Df(2R)BSC39 | 2.71 ± 1.18 | 48C5-D1;48D5-E1 |
| Df(2R)BSC44 | 2.32 ± 1.09 | 54B1-2;54B7-10 |
| Df(2R)BSC161 | 2.73 ± 1.25 | 54B2;54B17 |
| Df(2R)robl-c | 2.71 ± 0.94 | 54B17-C4;54C1-4 |
| Df(2R)14H10W-35 | 2.90 ± 1.05 | 54E5-7;55B5-7 |
| Df(2R)or-BR6 | 2.00 ± 1.38 | 59D5-10;60B3-8 |
The cytological breakpoints of each deficiency are provided according to FlyBase. The average number of abdominal segments and the standard deviation are given for each deficiency. +/CyO; nos+1+3 females were used as a baseline control. Only deficiencies leading to an average abdominal segment number less than 3 are listed.
One deficiency that reduced abdominal segmentation to an average of 2.4 segments, Df(2L)BSC32 (Fig. 1B), is the subject of this report. Of the 78 genes contained within this deficiency, we focused on aubergine (aub), which had been previously identified in a screen for heterozygous suppressors of the bicaudal phenotype produced by misexpression of osk, and the consequent mislocalization of nos mRNA, at the anterior of the embryo. Although Osk protein is still produced at the anterior when osk is misexpressed in embryos heterozygous for aub mutations, nos mRNA is no longer ectopically localized. To explain the requirement for aub, it was proposed that Aub might regulate translation of factors acting downstream of Osk to mediate nos localization (Wilson et al., 1996). The identification of a deficiency uncovering aub by our screen suggests that aub could play a more direct role in nos mRNA localization. We therefore examined whether the reduction in abdominal segmentation observed in nos+1+3 embryos heterozygous for Df(2L)BSC32 is due to the elimination of aub. Maternal heterozygosity for a strong aub mutation (aubQC42/+; hereafter referred to as aub−/+) produced similar results to Df(2L)BSC32 while heterozygosity for a weaker aub allele (aubHN2/+) had an intermediate effect (Fig. 1B), suggesting that elimination of aub function results in the effect of Df(2L)BSC32 on abdominal segmentation.
More recently, Aub has been shown to be a member of the Piwi-related Argonaute family of proteins involved in the repeat-associated small interfering RNA (rasiRNA) pathway that silences retrotransposons in the germline (Kennerdell et al., 2002; Klattenhoff et al., 2007; Savitsky et al., 2006; Vagin et al., 2004). Aub interacts physically with Squash (Squ), a protein with nuclease homology that is involved in rasiRNA biogenesis (Pane et al., 2007). In contrast to aub, heterozygosity for a strong loss of function squ mutation does not reduce abdominal segmentation of nos+1+3 embryos (Fig. 1B). This result indicates that the observed decrease in nos+1+3 activity is specific to aub and not a general effect of mutations in rasiRNA pathway components.
aub affects nos mRNA localization
To determine if the effect of aub on abdominal segmentation in the sensitized nos background is due to a defect in nos localization, we investigated the distribution of nos mRNA in aub−/+ embryos. Wild-type endogenous nos mRNA is localized to the posterior pole in these embryos, indistinguishably from wild-type embryos (Figs. 2A and B). However, localization of nos+1+3 mRNA, which is initially weaker than wild-type nos, is further decreased when nos+1+3 embryos are also aub−/+ (Figs. 2A and B), consistent with observed decrease in abdominal segmentation. Northern blot analysis confirmed that the total amount of nos+1+3 mRNA is comparable between the two genetic backgrounds (Fig. 2C). Additionally, localization of bcd mRNA to the anterior of the embryo is not disrupted, suggesting that reducing aub activity does not generally affect A-P patterning (Fig. 2D). Thus, these results suggest that aub plays a role in nos mRNA localization, possibly acting redundantly with other nos localization factors.
aub affects nos mRNA localization independent of osk and the Chk2 pathway
nos mRNA localization requires the prior localization and translation of osk at the posterior pole. In aub mutant ovaries, osk mRNA is prematurely translated, suggesting that Aub plays a role in osk mRNA silencing at early stages of oogenesis (Cook et al., 2004). Additionally, during mid-oogenesis, localization of osk mRNA and accumulation of Osk protein at the posterior of the oocyte is greatly diminished in aub mutant ovaries (Cook et al., 2004; Wilson et al., 1996). This effect is likely to be indirect, due to an earlier requirement for aub in the A-P polarization of the oocyte microtubule cytoskeleton that is in turn required for osk mRNA localization (Cook et al., 2004). However, because Osk levels are altered in aub mutants, we investigated whether the decreased localization of nos+1+3 mRNA in aub−/+ embryos could be also be due to an upstream defect in osk regulation. In aub−/+ embryos, osk localization appears wild-type (Fig. 3A) and these embryos express wild-type levels of osk mRNA and Osk protein (Figs. 2C and 3B). Osk is a critical component of the germ plasm that is necessary to generate the germ cell precursors, or pole cells, at the posterior of the embryo (Ephrussi et al., 1991; Kim-Ha et al., 1991; Lehmann and Nusslein-Volhard, 1986). Pole cell formation is highly sensitive to osk and a decrease in Osk protein levels of as little as 15% results in a significant reduction in pole cell number (Riechmann et al., 2002). In aub−/+ embryos, the average number of pole cells is equivalent to the wild-type number (30.2 ± 6.7 versus 29.2 ± 5.0; Figs. 3C and D). Similarly, nos+1+3 and aub−/+; nos+1+3 embryos have comparable numbers of pole cells (33.0 ± 6.5 versus 29.8 ± 5.7; data not shown). Together, these results indicate that the loss of a single copy of aub affects nos mRNA localization without affecting osk activity.
Figure 3.
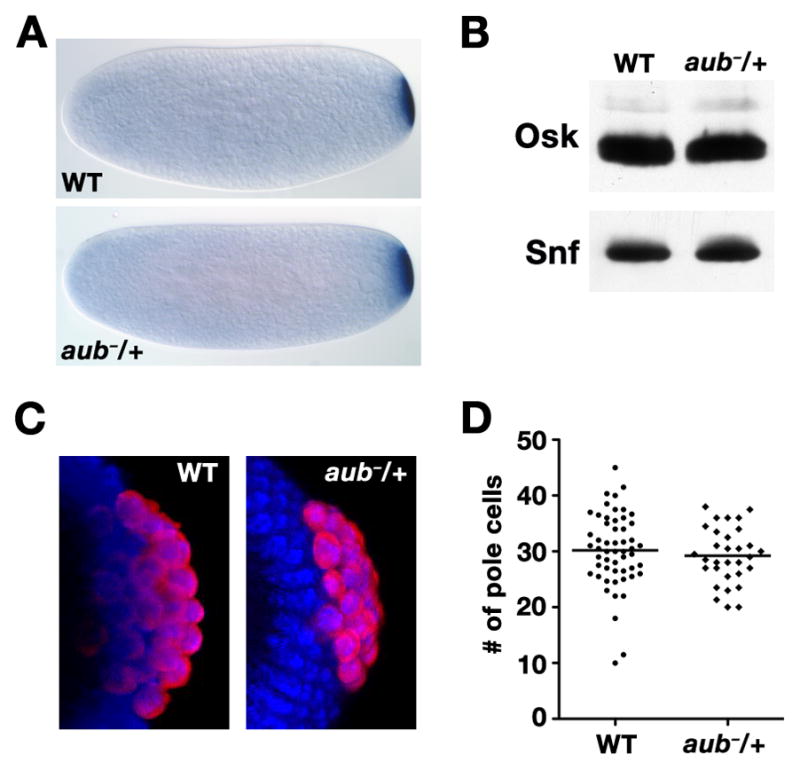
osk regulation is unaffected in aub heterozygotes. (A) In situ hybridization to osk mRNA in embryos from wild-type or aubQC/+ females. (B) Immunoblot of total protein from wild-type and aubQC/+ embryos. After transfer, the membrane was cut at the 37 kDa marker. The top portion was blotted with anti-Osk, which recognizes both the short and long Osk isoforms, and the bottom with anti-Snf as a loading control. Quantitation of the blot showed that Osk expression is not affected by heterozygosity for aub. (C) Confocal projections of anti-Vas immunofluorescence (red) in wild-type or aubQC/+ embryos. Nuclei are labeled with DAPI (blue). (D) Scatterplot of the number of pole cells per embryo in wild-type (n = 54) or aubQC/+ (n = 31) embryos, with the mean indicated by a horizontal bar. The average number of pole cells is not significantly different between wild-type (30.2 ± 6.7) and aubQC/+ (n = 29.2 ± 5.0) as determined by the Student’s t-test.
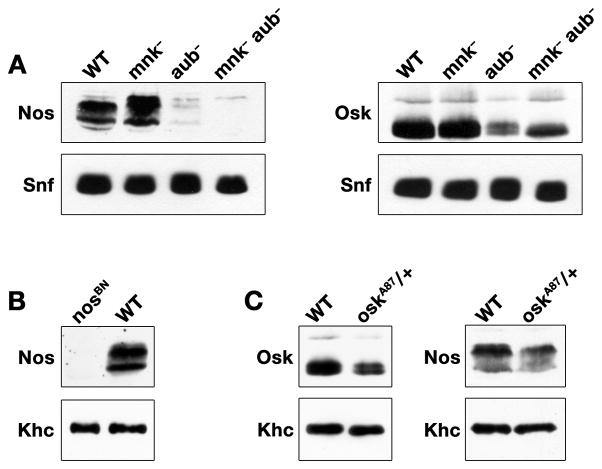
aub mutants affect microtubule polarization and osk mRNA regulation by activating a DNA damage signaling pathway within the germline mediated by mnk and mei-41, the Drosophila homologs of Chk2 and ATR kinase respectively. Double mutants between aub and either mnk or mei-41 bypass the DNA damage response and suppress the effects of aub on osk translation leading to wild-type posterior accumulation of Osk protein (Klattenhoff et al., 2007). To address whether aub similarly affects nos mRNA localization by triggering the DNA damage response, we assessed Nos expression as a quantitative measure of nos mRNA localization in embryos from aub, mnk double mutant females. Embryos from aub mutant females express minimal Nos protein, consistent with their diminished Osk expression (Figs. 4A and B). While mutation of mnk alone has no effect on Nos or Osk levels, Osk expression is partially restored in aub, mnk double mutant females (Fig. 4A). By contrast, Nos protein remains largely undetectable in the aub, mnk double mutant extract (Fig. 4A).
Figure 4.
The effect of aub on nos is independent of the DNA damage response pathway. (AC) Immunoblots of total protein extracted from 0–2 hour embryos of the indicated genotypes, probed with either anti-Nos or anti-Osk antibodies. Each membrane was also probed with anti-Snf or anti-Khc to control for loading. (A) Extracts from wild-type, mnk, aub, and mnk, aub double mutant embryos. Nos levels are reduced in aub and mnk, aub mutant embryos, however the reduction in Osk levels observed in aub mutants is partially suppressed by eliminating mnk. (B) Extracts from wild-type or nosBN embryos, which lack nos mRNA, confirming specificity of the anti-Nos antibody as previously shown (Gavis et al., 2008). (C) Extracts from wild-type or oskA87 heterozygous embryos, which contain half the wild-type level of osk mRNA. Quantitation of the blot shows that Nos protein levels are reduced to a lesser extent than Osk (70% versus 50% of wild-type levels, respectively).
To provide calibration for this assay, we determined the expected relationship between Osk and Nos levels by monitoring nos and osk mRNA localization and protein accumulation in embryos heterozygous for oskA87, an RNA null mutation (Jenny et al., 2006). In oskA87/+ embryos, osk mRNA is present at 60% of the wild-type level resulting in less osk localized at the posterior of the embryo (Supplemental Fig. 1). Consequently, Osk protein accumulates to approximately 50% of the wild-type level (Fig. 4D). The effect on osk is paralleled by a decrease in posteriorly localized nos (Supplemental Fig. 1), with Nos protein present at approximately 70% of wild-type levels (Fig. 4D). This response of Nos, which is largely correlated with the behavior of Osk, contrasts sharply with the complete failure of Nos to recover when Osk levels are partially restored in aub−, mnk− embryos. Together, these results demonstrate that aub affects Nos expression in a manner independent of the DNA damage response and its effect on osk localization. Moreover, they indicate that the nos localization defect in aub mutant embryos cannot be accounted for solely by the defect in osk localization, consistent with an osk-independent role for aub in nos localization.
Aub forms a complex with nos mRNA and the nos localization factor Rump in vivo
To determine whether Aub might directly act as a nos localization factor, we performed co-immunoprecipitation experiments from ovarian extracts to test the interaction of Aub with nos mRNA in vivo. To facilitate Aub immunoprecipitation, we took advantage of a transgenic line expressing a functional GFP-Aub fusion protein (Harris and Macdonald, 2001). Following immunoprecipitation with anti-GFP antibody, RNA was isolated and analyzed by RT-PCR using primers for nos mRNA or for a control RNA (his3.3b). nos mRNA is detected in immunoprecipitates from GFP-Aub ovary extract but not from control extract of ovaries expressing GFP alone (Fig. 5A). Moreover, the control his3.3b RNA is not detected in either of the immunoprecipitates, indicating specificity of the interaction between Aub and nos. Because Aub is implicated in silencing of osk mRNA early in oogenesis (Cook et al., 2004), we also tested whether osk mRNA is co-immunoprecipitated with Aub. RT-PCR with primers for osk mRNA showed that this is indeed the case (Fig. 5A).
Figure 5.
Aub associates with nos mRNA and Rump in vivo. (A) RT-PCR to detect nos or osk mRNA co-immunoprecipitated from ovaries expressing either GFP alone (G) or GFP-Aub (A) using anti-GFP antibody. his3.3b serves as a negative control. Total RNA from the extracts used for immunoprecipitation was used as a positive control for the RT-PCR reaction. Reactions were performed in the presence (+RT) or absence (−RT) of reverse transcriptase. (B) Immunoblot of anti-GFP immunoprecipitates from ovaries expressing either MCP-GFP or GFP-Aub in the absence (−) or presence (+) of RNase. Duplicate immunoblots were prepared for detection with either anti-Rump or anti-GFP antibodies. Lanes with extract used for immunoprecipitation (E) contain 1/20 volume equivalent of the immunoprecipitate sample (IP). (C) Bar graph with results from a genetic interaction assay for aub and rump. The percentage of embryos developing with 8 or fewer than 8 (≤8) abdominal segments in rump1 embryos (10%, n = 185), rump1 embryos heterozygous for aubQC (31%, n = 326), rump1 embryos heterozygous for nosBN (27%, n = 391), or rump1 embryos heterozygous for both aubQC and nosBN (56%, n = 268) was determined from embryonic cuticle preparations. ***P<0.0001 as determined by the Chi squared test.
Co-immunoprecipitation of Aub with nos mRNA suggests that Aub may be part of a nos mRNA localization complex. To further test this hypothesis, we investigated whether Aub also interacts with Rump, the only known direct-acting nos localization factor (Jain and Gavis, 2008). GFP-Aub or a control RNA-binding protein, MCP-GFP, that does not bind to nos mRNA (Forrest and Gavis, 2003), was immunoprecipitated from ovary extracts using an anti-GFP antibody and the immunoprecipitates were analyzed by immunoblotting with an anti-Rump antibody. Rump is detected specifically in GFP-Aub immunoprecipitates, but co-immunoprecipitation with GFP-Aub is abolished if the extract is treated with RNase (Fig. 5B). This behavior indicates that Aub and Rump do not interact directly; rather, they may be incorporated into an RNP complex together via their interactions with the same target mRNA.
To determine whether aub and rump function together in nos regulation, we took advantage of a genetic assay previously used to characterize Rump as a nos localization factor (Jain and Gavis, 2008). Consistent with previous data (Jain and Gavis, 2008), embryos lacking rump exhibit a weak abdominal segmentation defect, with 10% developing fewer than 8 abdominal segments (Fig. 5C). Lowering nos mRNA levels by reducing the nos gene dosage to one copy raises the sensitivity to localization factor loss, increasing both the frequency and severity of abdominal segmentation defects. Comparable to previous results (Jain and Gavis, 2008), 31% of rump mutant embryos that are also heterozygous for nosBN develop with fewer than 8 segments (Fig. 5C). Heterozygosity for aub similarly exacerbates the segmentation defect of rump mutant embryos, resulting in 27% of embryos with fewer than 8 segments. Moreover, reducing both nos and aub gene dosage in rump mutant embryos leads to a further deficit, with 58% of embryos showing loss of abdominal segments (Fig. 5C). Together, these genetic and biochemical results, support a role for Aub as a component of a nos localization RNP complex.
DISCUSSION
Localization of nos mRNA to the posterior of the Drosophila embryo is critical for patterning of the A-P body axis. Although a cis-acting nos mRNA localization signal has been identified, the complement of trans-acting factors required for assembly of a nos RNP complex competent for posterior localization has remained elusive. From a sensitized genetic screen, we have identified Aub as a novel nos mRNA localization factor and show that Aub interacts with nos mRNA in vivo. Importantly, we show that nos localization is affected by aub acting downstream of osk mRNA localization, implying independent roles for aub in regulating these two transcripts. Although the role of aub in osk localization appears to be indirect, through a requirement in oocyte microtubule organization (Cook et al., 2004; Klattenhoff et al., 2007), our results suggest that aub plays a more direct role in regulating nos mRNA.
A decrease in aub activity leads to defects in nos mRNA localization and, consequently, in patterning of the A-P axis when nos localization signal redundancy is reduced by removing two localization elements. A similar behavior is observed in rump mutants, which exhibit only weak segmentation defects unless redundantly acting elements are removed from the nos localization signal (Jain and Gavis, 2008). Presumably, elimination of individual localization signal elements compromises localization by stripping away the contributions of nos localization factors with overlapping functions in nos RNP assembly. Conversely, elimination of multiple nos localization factors should lead to a more severe defect than elimination of an individual factor. Consistent with this prediction, decreasing aub gene dosage in rump mutants also leads to more severe loss of abdominal segments.
In addition to allowing us to uncover a requirement for aub, the sensitized nos+1+3 background has allowed us to separate the indirect requirement for aub in osk localization from a more direct requirement in nos localization. Defects in osk regulation and abdominal segmentation are only observed when females are homozygous mutant for aub and not when they are heterozygous. By contrast, defects in nos+1+3 localization are observed when females are heterozygotes for aub mutations. Our results are further supported by previous data showing that the ability of ectopically expressed Osk to recruit nos mRNA is compromised in aub−/+ embryos (Wilson et al., 1996).
Aub has been implicated in the rasiRNA pathway that silences retrotransposons in the germline (Kennerdell et al., 2002; Klattenhoff et al., 2007; Savitsky et al., 2006; Vagin et al., 2004). However, mutation of squ, which encodes a rasiRNA pathway component that interacts with Aub (Pane et al., 2007), has no effect on the nos+1+3 transgene. Interestingly, another rasiRNA pathway component, piwi (Cook et al., 2004), has the opposite effect of aub on the nos+1+3 transgene, as heterozygosity for a piwi mutation results in increased segmentation in the sensitized background (data not shown). Mutations that inactivate the rasiRNA pathway, including aub mutations, activate the DNA damage checkpoint, presumably due to unsuppressed transposon activity (Chen et al., 2007; Klattenhoff et al., 2007). Checkpoint activation disrupts microtubule organization and grk translation, resulting in a failure of axis specification that is thought to lead to subsequent defects in osk mRNA localization (Klattenhoff et al., 2007). However, the effect of aub mutation on nos+1+3 mRNA localization is independent of the DNA damage pathway, providing further evidence that Aub regulates nos independently of osk. Moreover, these results indicate that Aub function in nos localization is distinct from its function in RNA silencing.
Biochemical experiments indicate nos mRNA forms a complex with Aub in vivo, although whether Aub interacts directly with nos mRNA, or is recruited to the complex by other proteins that bind directly to nos, is not yet clear. We have been unable to obtain soluble recombinant Aub necessary to distinguish between these possibilities. However, the RNA-dependent co-purification of Aub and Rump, combined with evidence for genetic interactions between aub and rump further supports the contribution of Aub to the formation and/or function of a nos localization RNP complex. Whereas Rump is not concentrated at the posterior of the oocyte, Aub-GFP is localized to the posterior during midoogenesis and continues to accumulate at the posterior pole throughout the later stages of oogenesis when nos becomes localized (Harris and Macdonald, 2001)(data not shown). Thus, the contributions of Rump and Aub to nos RNP complexes may be dynamic, with both proteins accompanying nos as it is dispersed throughout the oocyte during ooplasmic streaming, but only Aub remaining associated with the nos RNP upon its entrapment at the posterior. Isolation and characterization of the full complement of nos localization factors will be essential to dissect the assembly pathway for nos localization complexes. The isolation of aub in a sensitized genetic screen validates the use of such an approach, in addition to biochemical purification strategies that proved successful for isolation of Rump, for achieving this goal.
Supplementary Material
(A) Northern blot of total mRNA isolated from either wild-type or oskA87/+ embryos, probed simultaneously for nos and for rp49 as a loading control. The blot was then stripped and reprobed for osk. Relative nos and osk mRNA levels, after normalization to rp49, are indicated below each sample set. (D) In situ hybridization to nos and osk in embryos from wild-type or oskA87/+ females.
Acknowledgments
We thank A. Pane, T. Schüpbach, and P. Macdonald for fly stocks and A. Nakamura, A. Ephrussi, R. Lehmann, and P. Schedl for antibodies. We also thank J. Goodhouse for assistance with confocal microscopy and S. Chatterjee for technical assistance. This work was supported by a grant from the National Institutes of Health (GM067758) to E.R.G. and by an NSERC postgraduate fellowship (A.N.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdu U, Brodsky M, Schupbach T. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr Biol. 2002;12:1645–1651. doi: 10.1016/s0960-9822(02)01165-x. [DOI] [PubMed] [Google Scholar]
- Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126:659–669. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Crucs S, Chatterjee S, Gavis ER. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol Cell. 2000;5:457–467. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Duchow HK, Brechbiel JL, Chatterjee S, Gavis ER. The nanos translational control element represses translation in somatic cells by a Bearded box-like motif. Dev Biol. 2005;282:207–217. doi: 10.1016/j.ydbio.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Clark IE, Jain RA, Gavis ER. Temporal complexity within a translational control element in the nanos mRNA. Development. 2004;131:5849–5857. doi: 10.1242/dev.01460. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Live imaging of endogenous mRNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Chatterjee S, Ford NR, Wolff LJ. Dispensability of nanos mRNA localization for abdominal patterning but not for germ cell development. Mech Dev. 2008;125:81–90. doi: 10.1016/j.mod.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Singer RH, Hüttelmaier S. Localized translation through messenger RNA localization. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 687–717. [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Jain R, Gavis ER. The Drosophila hnRNP M homolog, Rumpelstiltskin, regulates nanos mRNA localization. Development. 2008;135:973–982. doi: 10.1242/dev.015438. [DOI] [PubMed] [Google Scholar]
- Jenny A, Hachet O, Zavorszky P, Cyrklaff A, Weston MD, Johnston DS, Erdelyi M, Ephrussi A. A translation-independent role of oskar RNA in early oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- Kalifa Y, Armenti ST, Gavis ER. Glorund interactions in the regulation of gurken and oskar mRNAs. Dev Biol. 2009;326:68–74. doi: 10.1016/j.ydbio.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:1–14. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986;47:141–152. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. Academic Press, Inc; San Diego: 1992. [Google Scholar]
- Manseau LJ, Schüpbach T. cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes and Development. 1989;3:1437–1452. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, Gutierrez GJ, Filardo P, Nebreda AR, Ephrussi A. Par-1 regulates stability of the posterior determinant Oskar by phosphorylation. Nat Cell Biol. 2002;4:337–342. doi: 10.1038/ncb782. [DOI] [PubMed] [Google Scholar]
- Rongo C, Gavis ER, Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Fee L, Lee TH, Wharton RP. The molecular chaperone Hsp90 is required for mRNA localization in Drosophila melanogaster embryos. Genetics. 2007 doi: 10.1534/genetics.107.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. P element-mediated transformation. In: Roberts DB, editor. Drosophila: A Practical Approach. IRL Press; Oxford: 1986. pp. 175–197. [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- Vanzo NF, Ephrussi A. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development. 2002;129:3705–3714. doi: 10.1242/dev.129.15.3705. [DOI] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dynam. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Northern blot of total mRNA isolated from either wild-type or oskA87/+ embryos, probed simultaneously for nos and for rp49 as a loading control. The blot was then stripped and reprobed for osk. Relative nos and osk mRNA levels, after normalization to rp49, are indicated below each sample set. (D) In situ hybridization to nos and osk in embryos from wild-type or oskA87/+ females.