Abstract
Multi-cellular animals have evolved a variety of mechanisms to respond to diverse apoptotic stimuli. In general these proceed through activation of apical caspases and culminate in executioner caspase activation and cell death. Due to breadth of possible initiators, various molecular platforms are used to trigger different apical caspases. Although some common protein domains are used to assemble the Apoptosome, the PIDDosome and death receptor complexes, an array of checks-and-balances are employed to ensure appropriate activation. Notwithstanding, these pathways share the underlying principle of proximity-dependent activation and post-translational modification. Here we will describe our current structural understanding of assembly and regulation of these signaling platforms.
Introduction
Timely cell death is an essential requirement for normal growth and development in multi-cellular organisms, and stimuli can arise from within the cell itself, or from a variety of extracellular sources. Therefore multiple signaling pathways have evolved to activate caspases and ultimately apoptosis. Due to the imperative nature of the process, much of the protein signaling machinery for programmed cell death preexists in the cell, poised in an inactive state. The platforms that activate this machinery generally consist of proteins bearing modular domains that form oligomeric complexes, and recent studies have strikingly clarified our structural understanding of how these complexes are assembled. Many of these interactions involve proteins that are members of the death domain superfamily. The domain consists of a conserved six-helical fold, and includes variants such as the death domain (DD), death effector domain (DED), and caspase recruitment domain (CARD) [1]. Here we will describe the molecular assembly mechanisms for three different cell death platforms involving these domains, the PIDDosome, Apoptosome and DISC complexes, and contrast this with diffuse platforms at the cytoplasmic face of TNF receptors nucleated by TNF receptor associated factors (TRAFs).
The Death domain superfamily
The benign six-helical bundle shared by proteins of the death domain superfamily performs two key functional roles in the formation of cell death platforms. Firstly, DDs are able to assemble into oligomeric complexes, which vary in stoichiometry depending on the particular DD and signaling pathway. This is essential for the second role carried out by CARD and DED modules, recruitment of downstream effector proteins (Figure 1). CARDs and DEDs homotypically interact with other CARD or DED domains that are present in many effector proteins including caspases and kinases, bringing them into sufficient proximity for activation [2–5].
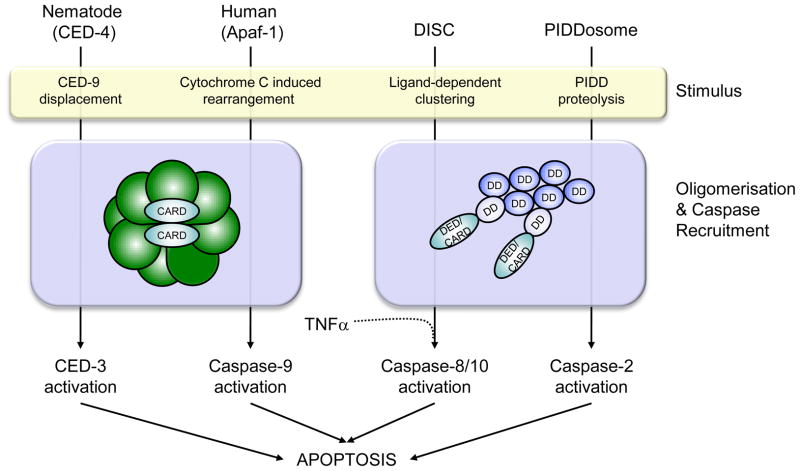
Figure 1. Regulation and assembly of cell death platforms.
Schematic summarizing formation of the various cell death platforms discussed. The nematode and human apoptosomes consist of heptameric and octameric rings respectively, which bring together CARD domains to recruit CED-3 and Caspase-9 for proximity-induced activation. The DISC and PIDDosome form clusters of DD, which incorporate adaptor proteins such as FADD and RAIDD that use their CARD and DED domains to recruit and activate Caspase-8/-10 and -2 respectively. TNFα is also able to activate Caspase-8 through a mechanism that remains to be clarified mechanistically.
The Death Domain as a Clustering Module
The PIDDosome: a soluble platform for cell death
Caspase-2 is an initiator caspase thought to participate in inducing apoptosis downstream of genotoxic insults such as DNA damage. Key to its activation is recruitment to a multisubunit assembly centered on the oligomerization of the DDs of RAIDD and PIDD (p53-induced protein with a death-domain), known as the PIDDosome [6]. PIDD contains a C-terminal DD, and an N-terminus incorporating seven leucine-rich repeats and two ZU-5 domains [7,8]. It undergoes proteolytic events to produce the C-terminal DD-containing fragment competent for nuclear translocation and classical PIDDosome assembly [9]. RAIDD is an adaptor molecule containing a C-terminal DD that is a core component of the PIDDosome, and an N-terminal CARD domain responsible for recruitment of Caspase-2 via CARD-CARD interaction [10]. CARD dependent recruitment of procaspase-2 to the PIDDosome is then thought to bring its catalytic regions into sufficient proximity for activation by induced dimerization, and initiate cell death pathways (Figure 2a).
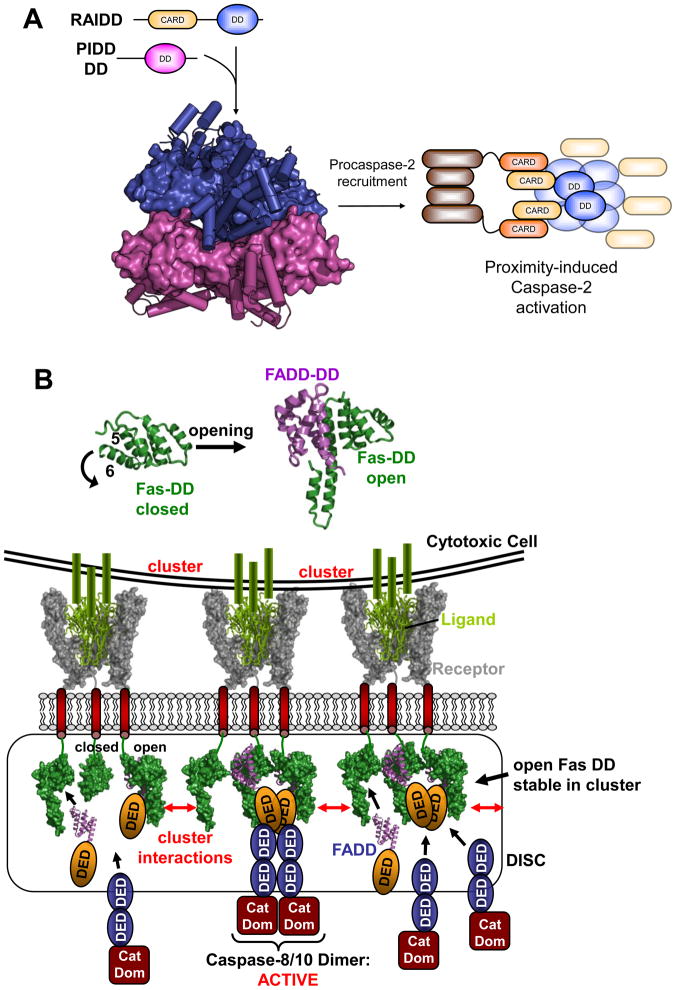
Figure 2. Death Domain assemblies: PIDDosome and DISC.
A. PIDDosome. Following processing to remove the N-terminal LRR and ZU5 domains of full-length PIDD, its C-terminal DD forms the PIDDosome along with RAIDD. Pentameric rings of PIDD (magenta) and RAIDD (blue) DD form extensive contacts and an additional two RAIDD DD bind atop the RAIDD ring (PDB entry 2of5). DD are shown alternately in surface and cartoon representation for clarity. The CARD domains of RAIDD then recruit procaspase-2 to facilitate proximity-induced dimerization and activation.
B. DISC formation: DD opening and illustrative rendering of Death Receptor clustering. Top: DD of the Death receptor Fas is adopting the classic globular shape comprised of six helices (left). In order to bind the DD of the adaptor protein FADD (magenta) the DD of Fas (green) has to undergo an opening mechanism where helix 6 moves outward and fuses with helix 5 (right). Bottom: Illustration of receptor clustering and DISC formation. The structure of a TRAIL (green)/TRAIL receptor (grey) complex [79] is used to illustrate a cell presenting clustered ligands leading to a clustering of receptor trimers in the target cell. On the insight of the affected cell DDs (from Fas DD structures) are brought into close proximity. Open Fas forms can interact and stabilize each other and are able to recruit FADD. FADD in turn recruits and activates target caspases via DED interaction (based on PDB entries: 1d0g, 1ddf, 3ezq).
The X-ray crystal structure of the “PIDDosome” revealed it to be formed by five-membered DD rings of PIDD and RAIDD respectively, stacked against one another. A further two RAIDD DD bind atop of the RAIDD ring, resulting in a 7 RAIDD, and 5 PIDD DD stoichiometry (Figure 2a) [11]. The pentameric rings do not possess rotational symmetry, but their stacked arrangement means that both PIDD and RAIDD DDs exist in a regular array and partake in qualitatively similar interactions with neighboring DD. These interfaces comprise three binding modes, which bear some resemblance to binary complexes formed by other death domain superfamily proteins [12,13]. More recently the structure of the Myd88-IRAK4-IRAK2 DD complex (or MyDosome) revealed a homologous array of DDs, albeit with a modified stoichiometry of six MyD88, four IRAK4 and four IRAK2 DDs [14]. For both the MyDosome and the PIDDosome a progressively assembled platform can be imagined where each new DD is accepted into a composite binding site that relies on the proper arrangement of preceding DD for continued assembly. This modular spiral provides an elegant model for the assembly of signaling nodes such as DD complexes, which could be easily adapted to various stoichiometries, symmetries and constituents.
DISCs – two dimensional membrane dependent clusters
The extrinsic apoptotic pathway is triggered by death inducing signaling complexes (DISCs), which are membrane dependent forms of DD assemblies [15–17]. DISCs are formed when a death ligand binds to a death receptor such as the prototypic Fas, or TRAIL receptors [18,19]. While it was initially believed that trimeric death ligands trimerise death receptors, it has now been shown that death receptors exist as preformed trimers in lipid rafts [20–23]. The actual activation stimulus is clustering of these trimers by death ligands, which are themselves anchored in clusters in the membrane of cytotoxic cells (Figure 2b) [24,25]. The key role of clustering is underlined by various pieces of evidence, most recently that soluble ligands fail to propagate DISC formation, and that O-glycosylation that promotes clustering of receptors is essential for DISC formation [26,27].
Structural studies of the Fas/FADD DD complex have revealed a mechanistic basis for this strong clustering dependency, showing that FADD utilizes a binding interface that is only accessible in an open form of the Fas DD [28]. This open form deviates from the classic DD fold in that helix six rotates away from the domain and fuses to form an extended helix five (Figure 2b) [28–30]. This contrasts with the previous model for receptor activation, which suggested oligomerisation by clustering of the classic globular form of the Fas DD [11,31]. The new structure-based model suggests that upon receptor clustering Fas DDs come into sufficient proximity for otherwise unstable open forms to interact and mutually stabilize each other. In this way Fas DD acts as a stringent signaling switch, whereby the FADD binding site is only stably presented when Fas is clustered in its open form [32]. Once recruited FADD is able to fulfill its role as a central DISC adaptor, where it utilizes its DED to recruit the DEDs of apical caspases -8 and -10, leading to their activation [33–35].
Apoptosomes – regulatory AAA+ assemblies
A soluble counterpart of the DISC is the apoptosome, which initiates the intrinsic apoptotic pathway. The human apoptosome is formed following cytochrome C release from the mitochondria and oligomerisation of Apaf-1, and subsequently recruits and activates caspase-9 [36–40]. Apaf-1 bears a complex domain structure evolved to facilitate its signaling role (Figure 3a). It possesses an N-terminal CARD domain followed by a nucleotide binding and oligomerisation domain (herein referred to as NOD). This domain consists of an α/β type nucleotide binding domain (NBD) and a helical domain (HD), and resembles an ATPase domain that places Apaf-1 in the AAA+ super family of ATPases [41]. The NOD is followed by a winged helix domain (WHD) and a super helical connector domain (HC) linking to two WD40 repeat regions responsible for cytochrome C binding. Recently two complementary studies have provided unprecedented structural insight into the apoptosome as an oligomeric signaling platform. One study resolved the human apoptosome using electron microscopy [42], and the other solved the crystal structure of the C. elegans apoptosome to 3.5 Å [43]. Both reports reveal exciting features with regards to apoptosome shape, formation and signaling.
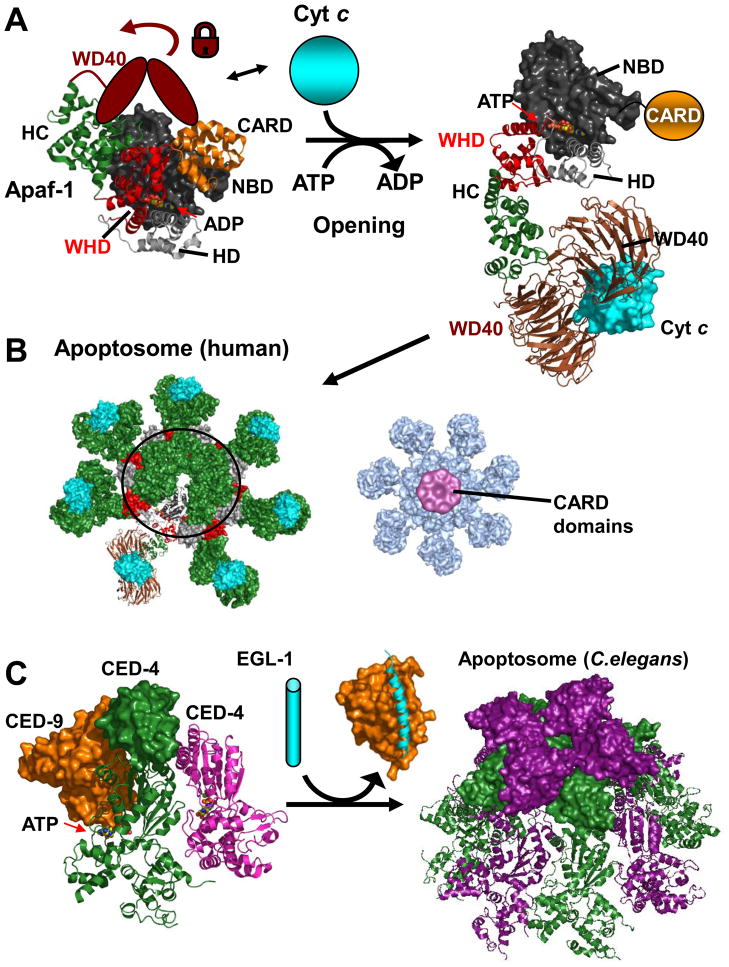
Figure 3. Apoptosomes.
A. Apaf-1 - opening. The structure of monomeric Apaf-1 shows a compact domain arrangement containing a central ADP molecule. Abbreviations are as in the main text and the WD40 region absent from the crystal structure is depicted as a cartoon locking the closed conformation. Cytochrome C releases the lock and ADP/(d)ATP exchange can occur along with a opening of Apaf-1 (inferred from the apoptosome structure under B). The CARD is not included in the electron microscopy structure since it becomes flexibly attached upon opening–illustrated in orange. B. Human Apoptosome. Open and cytochrome C bound Apaf-1 molecules can now interact to form the oligomeric apoptosome. Note that an outer ring is formed by alternating WHD/HD entities. Right: Map of Apoptosome with bound caspase-9 CARD domains. In this 3D map the CARD/CARD complexes hover in a ring-like manner above the apoptosome posed to dimerize and activate the catalytic domains of caspase-9 in the context of full length caspase-9. (PDB accession and structures: monomeric Apaf-1 1z6t, apoptosome based on 3iyt; cytochrome C bound model and density map for CARD bound apoptosome kindly provided by Drs. S. Yuan and C.W. Akey.) C. C. Elegans Apoptosome. CED-9 (orange) binds and inhibits an ATP bound CED-4 dimer (green, magenta; parts of the second CED-4 molecule are not defined in the structure). EGL-1 (cyan) binds and displaces CED-9 releasing the CED-4 dimers that form the octameric C. Elegans apoptosome. CARD domains are displayed in surface representation (PDB accession codes: 2a5y,1ty4, 3lqq)
In line with earlier reports the recent human apoptosome structure shows a heptameric arrangement of Apaf-1 molecules, but reveals a distinct mode of oligomerisation, and allows the movements of individual domains to be traced upon Apaf-1 activation [42,44]. In the absence of a sufficient stimulus Apaf-1 exists as a monomer adopting a compact shape (Figure 3a). In this form the HC, WHD and CARD closely interact with the NBD of the NOD to bind an ADP molecule. Although absent from the crystal structure the WD40 repeats are assumed to bind this compact form [45], locking monomeric Apaf-1 into this conformation. Upon an apoptotic signal cytochrome C is released from the mitochodria, binds to the WD40 regions, and releases the lock. Although Apaf-1 initially remains in the compact conformation the bound nucleotide can now be exchanged for (d)ATP, which triggers a dramatic rearrangement of Apaf-1 [38,46]. The rearrangement corresponds to an opening of Apaf-1 where the WHD flips outward and rotates ~120 degrees (Figure 3a). As a result the WD40 and HC point outwards, while on the opposite side the CARD domain is flexibly attached to the NBD [42]. This open form of Apaf-1 is now capable of oligomerisation in line with typical AAA ATPase-machines, with some distinct deviations [46–48]. The major adaptation distinguishing Apaf-1 as a signaling platform from typical AAA+ members is that the oligomer is comprised of an outer ring of alternating WHD and HDs (Figure 3b). With regards to catalysis, the Apaf-1 oligomer lacks the Arg/Lys finger often found in AAA+ machines even though the major catalytic residues are in principle present. However, the ATP is largely buried, in line with the fact that no- or low-level hydrolysis has been observed in several studies [41,44,49,50]. This may point to a regulatory timer function built into the apoptosome, but requires higher resolution investigations. The ultimate outcome of apoptosome formation is a clustering of CARD domains that can then recruit the CARDs of caspase-9. The new electron microscopy images of the apoptosome bound to procaspase-9 CARD show the CARDs hovering in a ring like structure above the main body of the apoptosome (Figure 3b) well poised to activate the caspase through proximity-induced dimerization.
The breakthrough crystal structure of the C. elegans apoptosome reveals a similar oligomerisation mode for the Apaf-1 homologue CED-4, in this case forming an apoptosome comprising eight molecules of CED-4 [43]. As in the human apoptosome the WHD/HD are key oligomerisation elements, but in general apoptosome formation in C. elegans is regulated differently [51,52]. Prior to activation CED-4 dimers already adopt an open conformation approximately resembling the conformation they adopt in the oligomer, but are prevented from oligomerising by a Bcl-2 protein family member termed CED-9 (Figure 3c) [53]. Displacement of CED-9 by the BH3 only protein Egl-1, another Bcl-2 member, then frees the way for the apoptosome to form [53–55]. The structure of the C. elegans apoptosome further reveals that the CARD domains bind in a bimodal manner onto the apoptosome (Figure 3c) as opposed to the more flexibly detached mode observed in the human counterpart. However, this compact CARD arrangement is sterically challenging to recruitment of the caspase-9 CARD domain in the manner used by the isolated Apaf-1/caspase-9 CARD domains [13]. This region also serves as a scaffold mediating contacts in the protein crystal, leaving the possibility that they may be more flexible in solution. Interestingly the study also reports a unique recruitment of only two CED-3 molecules into the inner space of the apoptosome, as opposed to the observed disc-like arrangement in the Apaf-1 apoptosome. Continued use of complementary methods such as those used in these studies will further deconvolute these crucial details. Nevertheless, these studies undoubtledly represent hallmarks that provide for the first time a comprehensive understanding of these prototypical signaling platforms. They may also serve as basis to understand aspects of their relatives in the NLR family, which are key mediators that regulate innate immunity and other pathways [56].
Diffuse networks mediated by TRAFs
In contrast to modular assembly by death domain superfamily members, the network of protein-protein interactions that trigger signaling pathways at the cytoplasmic face of TNF receptor are comparatively diverse and woven upon a web of interactions [57]. The eventual signaling outcome is determined by the cellular context and the partners recruited by TNF receptor associated factors (TRAFs) [58], the best studied of which is TRAF2. Although not strictly a platform for cell-death, signals from the TNF receptor may bifurcate to either induce the NFκ-B pathway, or trigger apoptosis by activating caspase-8, and either outcome can be triggered by ligand-dependent trimerization of the receptor. The defining feature of TRAF proteins is their C-terminal TRAF domain, which is composed of an N-terminal coiled-coil preceding a C-terminal MATH domain [59,60]. In isolation a shallow groove on the MATH domain binds weakly to a spectrum of regulatory proteins, which can then be displaced by the trimeric intracellular regions of activated TNF receptors to stimulate signal propagation [61]. Herein we will briefly discuss signals mediated by TRAF2 and inhibitor of apoptosis (IAP) proteins cIAP1 and cIAP2.
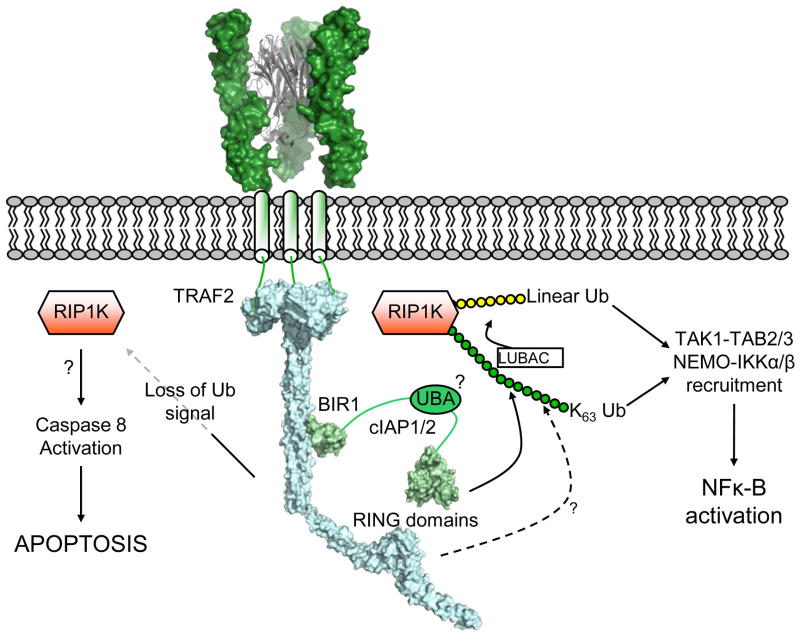
TRAF2 uses its coiled-coil region to bind BIR1 from cIAP1/2 with a 3:1 stoichiometry [62–64], and from this juncture a web of ubiquitin chains is woven (Figure 4) [57,65]. Both TRAF2 and cIAP1/2 contain RING domains that are ubiquitin E3 ligases, and various substrates, most notably RIP1K, NIK, but also TRAF2 and cIAP1/2 themselves are ubiquitinated with either K63- or K48-linked ubiquitin chains. K63-linked ubiquitin chains on RIP1K in particular appear to be central, as they serve as a platform for recruitment of effectors such as TAK1/TAB2/TAB3 and NEMO/IKKα/IKKβ [57,66], and further ubiquitin ligases such as LUBAC [67]. IAP proteins themselves contain a UBA-type ubiquitin binding domain that is required for robust IAP function [68,69], but its mechanistic role remains undetermined. Although it has recently been reported that TRAF2 itself can ubiquitinate RIP1K [70], it is clear that loss of cIAP-1 and -2 quickly leads to loss of appropriate ubiquitin homeostasis at the receptor complex [71,72]. In particular, loss of K63 chains on RIP1K redirects it from associating with the prosurvival kinase TAK1, to activating caspase-8 and downstream apoptosis [73]. Structurally this signaling network evolves around a combination of complexes: homotrimers such as the trimeric TRAF domain, homodimers such as dimeric RING domains [74–76] and ubiquitin binding by various ubiquitin binding domains [57]. Additionally, there is the suggestion that BIR1 from cIAP1 may dimerize in a homologous manner to BIR1 from XIAP [62,77,78]. For XIAP, BIR1 dimerization has been proposed bring together TAB1 molecules, which in turn activate TAK1 and NFκ-B signaling [78]. In combination these interactions result in a potentially extensive heterogeneous platform to initiate either cell death or proliferation.
Figure 4. Signaling at the cytoplasmic face of the TNF receptor.
Illustrative representation of the complex signaling network assembled upon receptor ligation by TNF ligands. Trimerization of the TNF receptor (based on [80]) recruits TRAF2 to the cytoplasmic tail of the receptor. cIAP1 and cIAP2 are subsequently recruited through the coiled-coil domain of TRAF2, and as E3 ligases cIAP1/2 (and possibly TRAF2) facilitate ubiquitination of various substrates using their RING domains. K63-linked ubiquitin chains on RIP1K serve as a platform for recruitment of proteins containing ubiquitin-binding domains such as NEMO, TAB2 and IAP proteins themselves. Loss of this ubiquitin signal by degradation of cIAPs or other means leaves RIP1K free to activate Caspase-8, in a manner that is currently not clear at a structural level. (Based upon PDB entries: 1tnr, 1ca9, 3m0a, 3hcs, 3eb6).
Conclusions
Our understanding of the structural geography of signaling at the cytoplasmic face of the TNF receptor is obviously still a work in progress. Its extended scaffold built upon ubiquitin chains and coiled-coil proteins such as TRAF2 and NEMO represents a stark contrast to the modular brickwork of the more defined signaling platforms that operate in a more concerted and processive manner. Examples are the DDs that assemble the PIDDosome, and the conserved modus operandi of CARD-CARD dimerization to activate apical caspases. This culminates in regulation by conformational switches such as clustering dependent DISCs and the AAA+ motif based apoptosome. However, all of these cell death platforms share a mutual reinforcement of interactions that serves to govern the delicate balance between survival and the most irreversible decision of all, to commit to apoptosis.
Acknowledgments
We thank Drs. S. Yuan and C.W. Akey for kindly providing the cytochrome C bound pdb model of the apoptosome and density map for CARD bound apoptosome. The work on cell death platforms is supported by R01 AA017238 to SJR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Reed JC, Doctor KS, Godzik A. The domains of apoptosis: a genomics perspective. Sci STKE. 2004;2004:re9. doi: 10.1126/stke.2392004re9. This review provides a comprehensive overview of the common structural domains that regulate apoptosis. [DOI] [PubMed] [Google Scholar]
- 2.Yin Q, Park HH, Chung JY, Lin S-C, Lo Y-C, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baliga BC, Read SH, Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–1241. doi: 10.1038/sj.cdd.4401492. [DOI] [PubMed] [Google Scholar]
- 4.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 5.Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 7.Telliez JB, Bean KM, Lin LL. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta. 2000;1478:280–288. doi: 10.1016/s0167-4838(00)00029-7. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- 9.Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. Embo J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 11**.Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. The crystal structure of the PIDDosome revealing basis of its assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao T, Towb P, Wasserman SA, Sprang SR. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 14.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 16.Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 17.Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Current Opinion in Cell Biology. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 18.Chaigne-Delalande B, Moreau J-F, Legembre P. Rewinding the DISC. Arch Immunol Ther Exp (Warsz) 2008;56:9–14. doi: 10.1007/s00005-008-0002-9. [DOI] [PubMed] [Google Scholar]
- 19.Pennarun B, Meijer A, de Vries EGE, Kleibeuker JH, Kruyt F, de Jong S. Playing the DISC: turning on TRAIL death receptor-mediated apoptosis in cancer. Biochim Biophys Acta. 2010;1805:123–140. doi: 10.1016/j.bbcan.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Siegel RM, Muppidi JR, Sarker M, Lobito A, Jen M, Martin D, Straus SE, Lenardo MJ. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K-H, Feig C, Tchikov V, Schickel R, Hallas C, Schütze S, Peter ME, Chan AC. The role of receptor internalization in CD95 signaling. Embo J. 2006;25:1009–1023. doi: 10.1038/sj.emboj.7601016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hueber A-O, Bernard A-M, Herincs Z, Couzinet A, He H-T. An essential role for membrane rafts in the initiation of Fas/CD95-triggered cell death in mouse thymocytes. EMBO reports. 2002;3:190–196. doi: 10.1093/embo-reports/kvf022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gajate C, Gonzalez-Camacho F, Mollinedo F. Lipid raft connection between extrinsic and intrinsic apoptotic pathways. Biochem Biophys Res Commun. 2009;380:780–784. doi: 10.1016/j.bbrc.2009.01.147. [DOI] [PubMed] [Google Scholar]
- 24.Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 25*.Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010 doi: 10.1038/onc.2010.221. Review describing current models of death receptor signalling from Apo2L/TRAIL. [DOI] [PubMed] [Google Scholar]
- 26.O’ Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, Haynes NM, Tarlinton DM, Zhang J-G, Belz GT, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461:659–663. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF, Totpal K, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 28**.Scott F, Stec B, Pop C, Dobaczewska M, Lee J, Monosov E, Robinson H, Salvesen G, Schwarzenbacher R, Riedl S. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2008 doi: 10.1038/nature07606. The crystal structure of the Fas-FADD DD complex revealing conformational changes that regulate DISC formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Park HH, Lo Y-C, Lin S-C, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. Comprehensive review summarizing death domains and their regualtion and signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 31.Weber CH, Vincenz C. A docking model of key components of the DISC complex: death domain superfamily interactions redefined. FEBS Lett. 2001;492:171–176. doi: 10.1016/s0014-5793(01)02162-7. [DOI] [PubMed] [Google Scholar]
- 32.Salvesen GS, Riedl SJ. Structure of the Fas/FADD complex: a conditional death domain complex mediating signaling by receptor clustering. Cell Cycle. 2009;8:2723–2727. doi: 10.4161/cc.8.17.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 35.Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T, Gavathiotis E, Wei Y, Werner MH. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 36.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 38.Kim H-E, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colin J, Gaumer S, Guenal I, Mignotte B. Mitochondria, Bcl-2 family proteins and apoptosomes: of worms, flies and men. Front Biosci. 2009;14:4127–4137. doi: 10.2741/3517. [DOI] [PubMed] [Google Scholar]
- 40.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 41**.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. Crystal structure of monomeric Apaf-1 revealing basis for locked inactive conformation. [DOI] [PubMed] [Google Scholar]
- 42**.Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. To date the highest resolution cryoelectron microscopy structure of the human apoptosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. Crystal structure of the C. elegans apoptosome to 3.5 Å resolution. [DOI] [PubMed] [Google Scholar]
- 44.Yu X, Acehan D, Ménétret J-F, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure (London, England: 1993) 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 46.Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of Life, Wheel of Death: A Mechanistic Insight into Signaling by STAND Proteins. Structure (London, England: 1993) 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 48.Truscott KN, Lowth BR, Strack PR, Dougan DA. Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem Cell Biol. 2010;88:97–108. doi: 10.1139/o09-167. [DOI] [PubMed] [Google Scholar]
- 49.Reubold TF, Wohlgemuth S, Eschenburg S. A new model for the transition of APAF-1 from inactive monomer to caspase-activating apoptosome. J Biol Chem. 2009;284:32717–32724. doi: 10.1074/jbc.M109.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Benedict MA, Ding L, Núñez G. Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. Embo J. 1999;18:3586–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 52.Peden E, Killian DJ, Xue D. Cell death specification in C. elegans. Cell Cycle. 2008;7:2479–2484. doi: 10.4161/cc.7.16.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu J-W, Kokel D, Li H, Hao Q, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. Crystal structure revealing the basis of regulation of CED-9 by CED-4. [DOI] [PubMed] [Google Scholar]
- 54.Yan N, Gu L, Kokel D, Chai J, Li W, Han A, Chen L, Xue D, Shi Y. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 55.Woo J-S, Jung J-S, Ha N-C, Shin J, Kim K-H, Lee W, Oh B-H. Unique structural features of a BCL-2 family protein CED-9 and biophysical characterization of CED-9/EGL-1 interactions. Cell Death Differ. 2003;10:1310–1319. doi: 10.1038/sj.cdd.4401303. [DOI] [PubMed] [Google Scholar]
- 56.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 57*.Bianchi K, Meier P. A Tangled Web of Ubiquitin Chains: Breaking News in TNF-R1 Signaling. Mol Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. Review summarizing current models of ubiquitin-based regulation of TNF receptor signaling. [DOI] [PubMed] [Google Scholar]
- 58.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 59.McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, Alber T. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc Natl Acad Sci USA. 1999;96:8408–8413. doi: 10.1073/pnas.96.15.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 61.Sanjo H, Zajonc DM, Braden R, Norris PS, Ware CF. Allosteric regulation of the ubiquitin:NIK and ubiquitin:TRAF3 E3 ligases by the lymphotoxin-beta receptor. J Biol Chem. 2010;285:17148–17155. doi: 10.1074/jbc.M110.105874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mace PD, Smits C, Vaux DL, Silke J, Day CL. Asymmetric Recruitment of cIAPs by TRAF2. Journal of Molecular Biology. 2010 doi: 10.1016/j.jmb.2010.04.055. [DOI] [PubMed] [Google Scholar]
- 63**.Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell. 2010;38:101–113. doi: 10.1016/j.molcel.2010.03.009. Crystal structure showing basis of recruitment of cIAPs to the coiled-coil domain of TRAF2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vince J, Pantaki D, Feltham R, Mace P, Cordier S, Schmukle A, Davidson A, Callus B, Wong W, Gentle I, et al. TRAF2 must bind to cIAPs for TNF to efficiently activate NF-{kappa}B and to prevent TNF-induced apoptosis. J Biol Chem. 2009 doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends in Cell Biology. 2009 doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 68.Blankenship JW, Varfolomeev E, Goncharov T, Fedorova AV, Kirkpatrick DS, Izrael-Tomasevic A, Phu L, Arnott D, Aghajan M, Zobel K, et al. Ubiquitin binding modulates IAP antagonist-stimulated proteasomal degradation of c-IAP1 and c-IAP2(1) Biochem J. 2009;417:149–160. doi: 10.1042/BJ20081885. [DOI] [PubMed] [Google Scholar]
- 69*.Gyrd-Hansen M, Darding M, Miasari M, Santoro MM, Zender L, Xue W, Tenev T, da Fonseca PCA, Zvelebil M, Bujnicki JM, et al. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol. 2008;10:1309–1317. doi: 10.1038/ncb1789. First report describing characterization of a ubiquitin binding domain in IAP proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJA, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 72.Vince JE, Wong WW-L, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 73.Bertrand MJM, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 74.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75*.Yin Q, Lin S, Lamothe B, Lu M, Lo Y, Hura G, Zheng L, Rich R, Campos A, Myszka D, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1605. Structure of the Ubc13-bound RING domain of TRAF6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76*.Mace PD, Linke K, Feltham R, Schumacher F-R, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. Structure of the UbcH5b-bound RING domain of cIAP2. [DOI] [PubMed] [Google Scholar]
- 77.Lin Huang, Lo Lu, Wu H. Crystal Structure of the BIR1 Domain of XIAP in Two Crystal Forms. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu M, Lin S-C, Huang Y, Kang YJ, Rich R, Lo Y-C, Myszka D, Han J, Wu H. XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization. Mol Cell. 2007;26:689–702. doi: 10.1016/j.molcel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O’Connell M, Kelley RF, Ashkenazi A, de Vos AM. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell. 1999;4:563–571. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- 80.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]