Abstract
Survival and replication inside host cells by Brucella spp. requires a type IV secretion system (T4SS), encoded by the virB locus. However, the identity of the molecules secreted by the T4SS has remained elusive. We hypothesized that proteins translocated by the T4SS would be co-regulated with the virB operon. The LuxR family regulator VjbR, known to regulate virB, bound a fragment of the virB promoter containing an 18-bp palindromic motif (virB promoter box), showing that VjbR regulated the virB operon directly. To identify virB-coregulated genes, we searched the B. suis 1330 and B. abortus 2308 genomes for genes with an upstream virB promoter box. 144 promoters in the two genomes contained the virB promoter box, including those of fliC encoding flagellin and cgs encoding cyclic β-glucan synthetase. Thirteen of these proteins were tested for VirB dependent translocation into macrophages using a β-lactamase reporter assay. This analysis resulted in the identification of the proteins encoded by BAB1_1652 (VceA) and BR1038/BAB1_1058 (VceC) as novel protein substrates of the Brucella T4SS. VceC could also be translocated by the L. pneumophila Dot/Icm T4SS into host cells. Our results suggest that VjbR coordinates expression of the T4SS and at least two of its secreted substrates.
Introduction
In order to cause persistent infection of the reticuloendothelial system, all human pathogenic Brucella species require a type IV secretion system (T4SS) known as VirB (Hong et al., 2000, O'Callaghan et al., 1999). T4SS are multi-component protein structures in the bacterial envelope used by many Gram-negative bacterial pathogens of animals and plants for the translocation of virulence factors into eukaryotic host cells (Backert & Meyer, 2006). Examples of well-studied T4SS are the VirB system of the plant pathogen Agrobacterium tumefaciens, which facilitates export of T-DNA together with at least 3 effector proteins into plant cells (Berger & Christie, 1994, Fullner, 1998, Vergunst et al., 2000), and the Dot/Icm system of the accidental human pathogen Legionella pneumophila, which is used for secretion of more than 40 effector proteins into host cells (Nagai & Roy, 2001, Chen et al., 2004, Conover et al., 2003, Shohdy et al., 2005, Luo & Isberg, 2004). Together with Agrobacterium tumefaciens, Brucella spp. are classified in the α-2 group of the α-proteobacteria (Moreno et al., 1990), and correspondingly, their T4SS are closely related. However, the Brucella genome does not encode any homologs of A. tumefaciens effectors, probably because these two pathogens have different lifestyles. Agrobacterium is a soil dwelling bacterium that infects root cells of plants, and while remaining extracellular, uses its T4SS to induce tumorigenesis (Christie, 2004).
Brucella utilizes a similar T4SS, VirB, for a different purpose. Brucella spp. are facultative intracellular pathogens of many wild and domestic animals and can cause zoonotic disease in humans. Brucella spp. can survive within the phagocytic cells of the host and are able to evade normal mechanisms of bacterial killing by altering the intracellular trafficking of their vacuole (Arenas et al., 2000, Pizarro-Cerda et al., 1998a, Pizarro-Cerda et al., 1998b). The ability of phagocytosed Brucella to evade fusion of their endosomal vesicles with lysosomes requires the VirB T4SS (Comerci et al., 2001, Delrue et al., 2001, Sieira et al., 2000, Celli et al., 2003, Celli et al., 2005). Secretion of effector proteins by the Brucella T4SS likely alters this pathway, allowing the bacteria to reside in vacuoles with properties of rough endoplasmic reticulum, thereby promoting survival and replication (Celli et al., 2003, Celli et al., 2005). Mutants of Brucella lacking a functional T4SS are highly attenuated in vitro in macrophages and in vivo in the mouse model of infection (O'Callaghan et al., 1999, Delrue et al., 2001, den Hartigh et al., 2004, Sieira et al., 2000, Hong et al., 2000).
It has been shown that for both Type III secretion systems (T3SS) and T4SS, genes encoding the secretion apparatus are often co-regulated with the secreted substrates. For example, the same regulators control expression of genes encoding the structural components of Salmonella enterica serotype Typhimurium T3SS-1 and T3SS-2 and their effectors (Thijs et al., 2007, Worley et al., 2000). Further, in L. pneumophila the two-component system regulators PmrA and CpxR were found to regulate genes of the Dot/Icm T4SS as well as several Dot/Icm effector proteins (Zusman et al., 2007, Altman & Segal, 2008). Based on these findings, we hypothesized that secreted substrates of the B. abortus T4SS would be co-regulated with the structural components of the secretion apparatus by the same transcriptional regulator.
The Brucella T4SS is induced during intracellular infection (Sieira et al., 2004, Sieira et al., 2000, Boschiroli et al., 2002). The only direct regulator of the virB genes shown to date is integration host factor (IHF), which was shown to bind to the virB promoter (PvirB) of B. abortus (Sieira et al., 2004). IHF was found to be necessary for activity of PvirB inside the host cell and during vegetative growth, likely by its well characterized DNA bending activity, which is thought to provide the correct promoter structure for the action of additional transcriptional regulators. Recently, a transcriptional activator (VjbR) of the B. melitensis virB genes was described (Delrue et al., 2005, Delrue et al., 2001). In a B. melitensis vjbR mutant expression of the T4SS is greatly reduced (Delrue et al., 2005). Furthermore, the B. melitensis vjbR mutant is highly attenuated in both cellular and mouse models of infection. The VjbR protein belongs to the family of LuxR quorum-sensing (QS) regulators and contains a conserved motif required for binding to acylhomoserine lactone (AHL) pheromones. It was shown that the QS pheromone N-dodecanoylhomoserine lactone (C12-HSL) inhibits the activation of virB genes via VjbR (Delrue et al., 2005). The expression of VirB is downregulated by C12-HSL in B. melitensis (Delrue et al., 2005, Taminiau et al., 2002, Uzureau et al., 2007). However, it remains unknown whether VjbR binds directly at the virB promoter to activate its transcription, or whether intermediate regulators are involved.
In this report we present evidence for direct activation of PvirB by VjbR in Brucella abortus. Furthermore, by determining a conserved motif in PvirB required for activation by VjbR, we identified 143 additional promoter regions in the B. abortus 2308 and B. suis 1330 genomes containing this motif, including the promoters of vceA and vceC, encoding substrates of the VirB T4SS.
Results
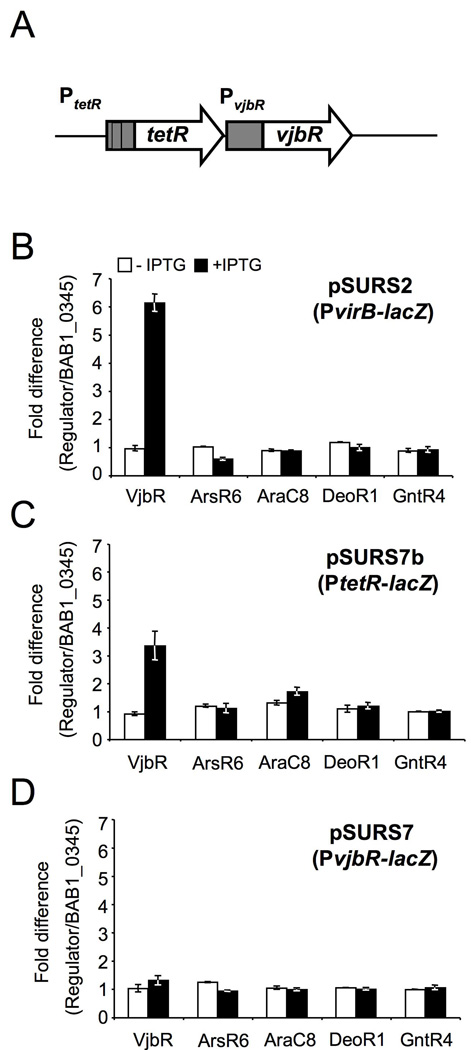
1. VjbR of B. abortus activates PvirB∷lacZ and PtetR∷lacZ in E. coli
We hypothesized that a common regulator would control expression of both the virB genes encoding the T4SS and its secreted effector proteins. Therefore, as a first step toward identification of genes that are co-regulated with the virB genes, we performed experiments to identify a direct regulator of the virB operon. In previous studies VjbR was found to be an activator of the B. abortus VirB system (Delrue et al., 2005). Further, Brucella melitensis strains lacking arsR6, araC8, deoR1 or gntR4 have reduced expression levels of virB genes, suggesting that they may also function as activators of virB expression (Haine et al., 2005). However it remained unknown whether the effect of all of these regulatory proteins on expression of virB genes is direct or is part of a regulatory cascade. In order to distinguish between these two possibilities, we reconstituted virB regulation in E. coli. A transcriptional fusion of the virB1 upstream region to lacZ (PvirB∷lacZ) was constructed and the resulting plasmid pSURS2, was introduced into E. coli BL21 lacZ (Supplementary table 1). In addition, we constructed plasmids encoding IPTG-inducible copies of vjbR, arsR6, araC8, deoR1, gntR4 or a predicted two-component response regulator that did not affect expression of virB (BAB1_0345; unpublished results). Expression of each regulator together with the PvirB∷lacZ construct into E. coli strain BL21 lacZ allowed us to determine whether the regulator of interest could affect expression of the PvirB∷lacZ construct (pSURS2). To determine whether the corresponding regulatory proteins alter expression of a PvirB∷lacZ reporter construct, we measured the β-galactosidase activity of the PvirB∷lacZ fusion in E. coli strains expressing VjbR, ArsR6, AraC8, DeoR1, GntR4, or the control protein BAB1_0345 (Figure 1 and Figure 2). Data shown in Figure 1 demonstrate that induction of vjbR expression activated transcription of the PvirB∷lacZ reporter construct on pSURS2 (Fig. 1A). Furthermore, the induction of PvirB activity correlated well with the expression of VjbR as shown by Western blotting (Figure 1A, top panel). Induction of the control protein BAB1_0345 caused a slight reduction in expression of the PvirB∷lacZ reporter, which is likely due to T7 promoter-driven overexpression of the protein, as we have observed this effect with overexpression of other proteins as well (Fig. 1A). To correct for this effect of protein overproduction on expression of the virB reporter construct, we used our negative control to normalize data presented in subsequent figures.
Figure 1.
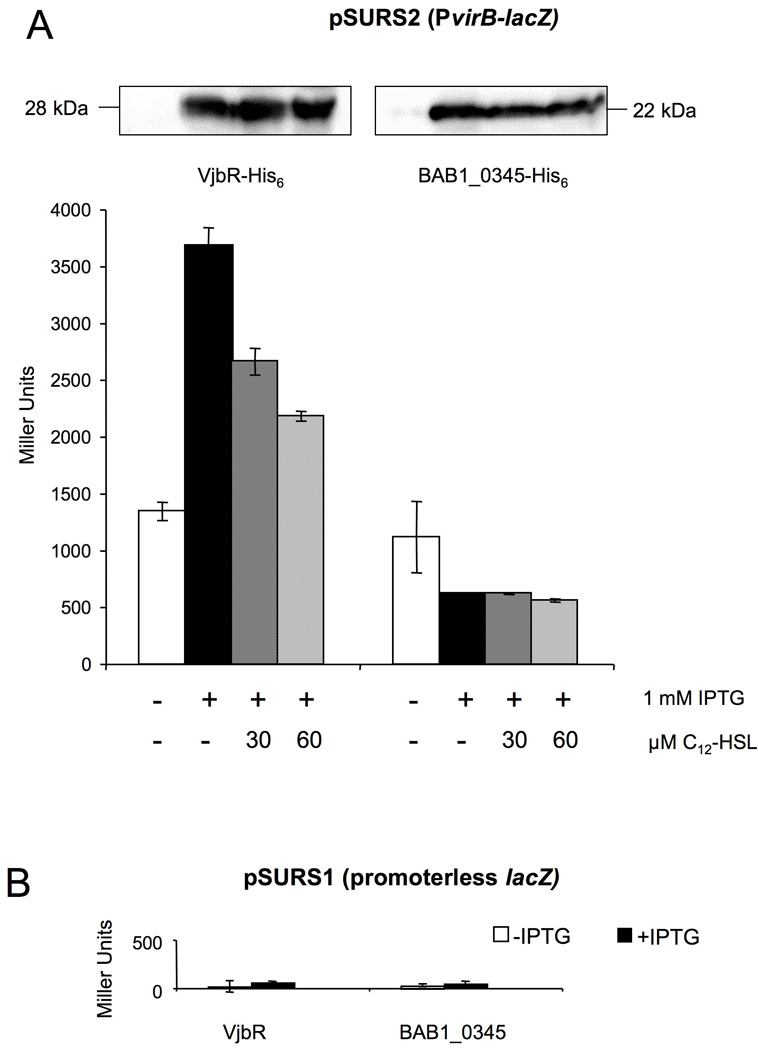
VjbR activates the promoter upstream of virB1 in E. coli.(A) Top: Western blot using anti-His antibodies showing expression of VjbR-His6 or BAB1_0345-His6 after induction with IPTG and showing that expression of VjbR-His6 is not reduced after addition of C12-HSL. Bottom: β-galactosidase activity of E. coli with pSURS2 was measured after no induction (white bars) or induction with IPTG (black bars) of VjbR-His6 and a negative control BAB1_0345-His6. Hatched bars indicate induction with IPTG and the addition of 30 µM or 60 µM C12-HSL to the culture. Values are the averages ± standard deviations of duplicate samples from a representative experiment that was repeated at least three times independently. (B) β-galactosidase activity of E. coli with empty pSURS1 vector and expressing VjbR-His6 or BAB1_0345-His6.
Figure 2.
(A) Map of the tetR-vjbR operon. (B–D) Fold difference of β-galactosidase activity of PvirB (B), PtetR (C) and PvjbR (D) in E. coli VjbR, ArsR6, AraC8, DeoR1 or GntR4 strain versus the negative control strain. White bars represent the ratio of measured Miller Units in uninduced E. coli expressing one of the regulators versus uninduced E. coli BAB1_0345. Black bars represent the ratio of measured Miller Units in IPTG induced E. coli expressing one of the regulators versus induced E. coli expressing BAB1_0345.
Since C12-homoserine lactone (C12-HSL) inhibits transcriptional activation by VjbR in Brucella (Delrue et al., 2005, Uzureau et al., 2007), we determined whether addition of C12-HSL would reduce VjbR-mediated activation of the PvirB∷lacZ reporter construct. Addition of 30 µM and 60µM C12-HSL to E. coli cultures reduced activity of PvirB in a dose-dependent manner only in the strains expressing VjbR, but not in strains expressing the negative control protein BAB1_0345 (Fig. 1A). The residual expression of the PvirB∷lacZ reporter in cultures treated with 60µM C12-HSL is likely due to the high expression level of VjbR from the T7 promoter in E. coli. Notably, the β-galactosidase activity levels observed for cells carrying the PvirB∷lacZ fusion on pSURS2 were significantly higher than the background β-galactosidase levels observed in cells carrying the promoterless control vector pSURS1 (Fig. 1B).
Only expression of vjbR, but not of arsR6, araC8, deoR1 or gntR4, activated expression of the PvirB-lacZ reporter construct in E. coli, suggesting that whereas VjbR is able to activate virB expression in the absence of other Brucella-specific factors, ArsR6, AraC8, DeoR1, GntR4 may require additional Brucella-specific gene products to activate expression of the virB genes (Fig. 2B). We therefore focused on identifying additional members of the VjbR regulon to identify proteins secreted by the B. abortus T4SS.
Members of the LuxR regulator family in other bacteria (e.g. Vibrio fischeri) are known to regulate their own expression (Shadel & Baldwin, 1991, Shadel & Baldwin, 1992). We therefore examined the effect of VjbR on its own promoter. Since vjbR is predicted to be in an operon with an upstream gene encoding a tetR family regulator (Fig. 2A), we constructed lacZ fusions to the promoter directly upstream of tetR (BAB2_0117; PtetR) and to the tetR-vjbR intergenic region (PvjbR; 2C and 2D). LacZ promoter fusions were introduced into E. coli strains expressing vjbR, arsR6, araC8, deoR1 or gntR4 or BAB1_0345 genes and induction of lacZ expression was measured using β-galactosidase assays. The results from the assays showed that VjbR activated its own expression via PtetR (Fig. 2C). Compared to the negative control BAB1_0345, induction of tetR∷lacZ expression by VjbR was approximately 3 fold higher. We also examined the regulatory effect of VjbR on the intergenic region between the tetR and vjbR genes using the same system and, although this promoter showed background activity in E. coli, no activation by VjbR was observed (Fig. 2D). Since in B. melitensis it was recently shown that VjbR regulates its own expression (Rambow-Larsen et al., 2008), these findings suggest that the promoter in the tetR-vjbR intergenic region may control VjbR expression in a VjbR-independent manner or that it may require Brucella-specific factors in addition to VjbR for transcriptional regulation.
2. VjbR-His6 binds directly to both a fragment of PvirB and a fragment of the virB1-virB2 intergenic region
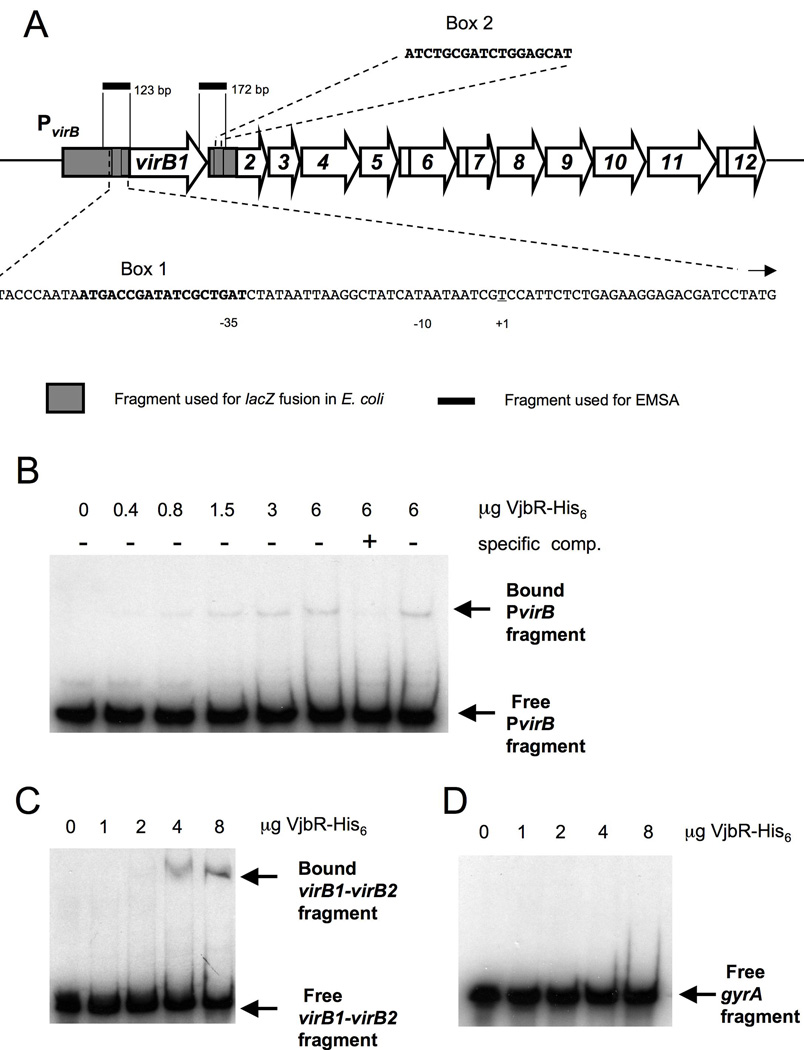
The results presented above suggested direct activation of PvirB by VjbR due to binding of this regulator to the virB promoter. To test this idea, we performed electrophoretic mobility shift assays (EMSA) using purified, His-tagged B. abortus VjbR protein and a 123 bp fragment containing the DNA region directly upstream of the virB promoter (Fig. 3). Addition of increasing amounts of VjbR-His6 to the 32P labeled PvirB fragment resulted in an increased intensity of a band with reduced electrophoretic mobility (Fig. 3B). Competition with the unlabeled PvirB probe reduced the intensity of the shifted band, and this effect was not observed when the same amount of an unlabeled and unrelated probe was added to the binding reaction. These results demonstrate a specific binding of VjbR to the upstream region of the B. abortus virB operon.
Figure 3.
(A) Schematic representation of the virB operon in the B. abortus chromosome II including PvirB and intergenic regions. The putative PvirB box 1 and box 2 that were used in the first prediction round are shown and the fragments that were used in EMSAs are indicated in black. (B–D) EMSA experiments showing specific binding of VjbR-His6 to regions directly upstream and downstream of virB1(B) VjbR-His6 binding to a 123 bp fragment of PvirB upstream of virB1. In the first lane no VjbR-His6 was added to the binding reaction, and in the subsequent five lanes VjbR-His6 was added in increasing amounts. In lanes 7 and 8 VjbR-His6 protein concentration was the same as in lane 6, but here a 100-fold excess of unlabeled specific (PvirB) or non-specific (vjbR gene) DNA fragment was added to the reaction. (C–D) VjbR-His6 binding to a 172 bp fragment of the virB1-virB2 intergenic region (C), and not to a 139 bp fragment of the gene gyrA (D) when VjbR-His6 was added in the same, increasing amounts to the binding reactions.
Since the virB1-virB2 intergenic region is also predicted to contain a promoter, we tested binding of VjbR by EMSA to a 32P labeled 172 bp DNA fragment containing the intergenic region (Fig. 3A). We found that purified VjbR-His6 bound to this region similarly as to the PvirB fragment (Fig. 3C). Addition of increasing amounts of VjbR-His6 to the fragment containing the virB1-virB2 intergenic region resulted in an increased intensity of a band with reduced electrophoretic mobility (Fig. 3C). As a control for nonspecific binding of VjbR, we used a 32P labeled 139 bp internal fragment of the housekeeping gyrA gene (Fig. 3D). No band shift was observed for the 32P labeled gyrA (BAB1_1121) fragment at any of the VjbR concentrations examined, demonstrating a specific binding of VjbR to the virB1-virB2 intergenic region. However, in E. coli, induction of vjbR expression did not increase expression of a virB1-virB2∷lacZ transcriptional fusion (pSURS3; data not shown). It is possible that for proper VjbR mediated activation of the virB1-virB2 region other Brucella specific factors are required that are not present in E. coli. Collectively, these data suggested that VjbR binds specifically to two promoter regions within the virB operon of B. abortus.
3. A conserved 18 bp sequence in PvirB is important for activation by VjbR
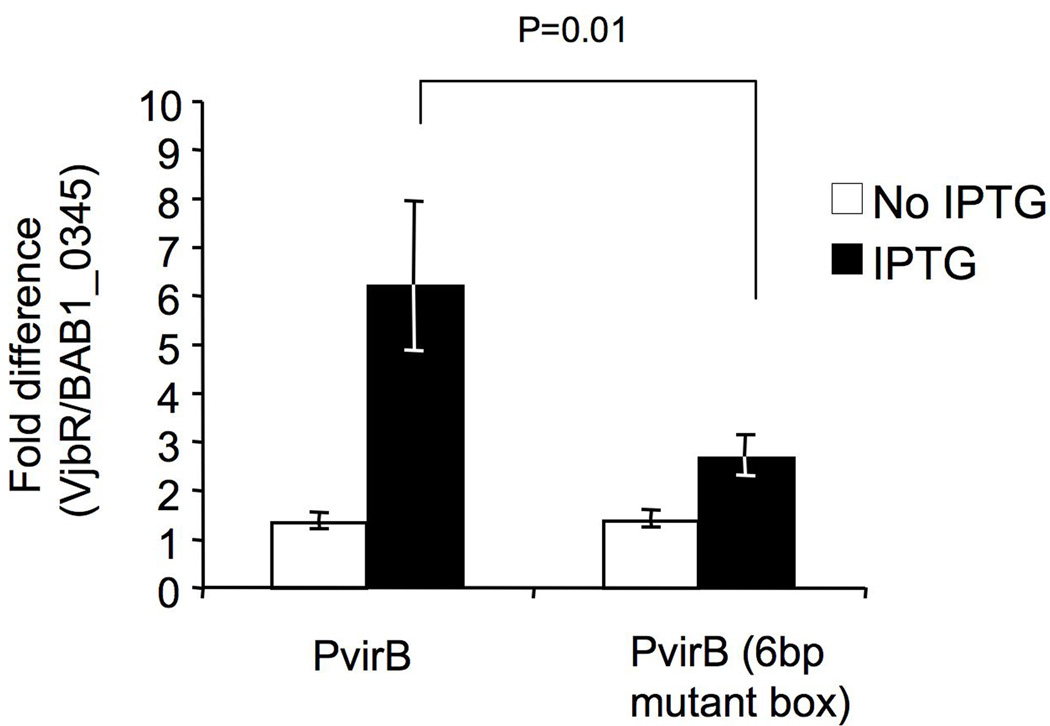
Since VjbR was found to activate PvirB and PtetR in our E. coli expression system, we examined the DNA sequence of both promoters for the presence of conserved motifs. We identified a conserved palindromic sequence of 18 bp in PvirB and 19 bp in PtetR (Fig. 3A). We also found a similar sequence in the intergenic region between virB1 and virB2 (Fig. 3A). In B. abortus PvirB the 18 bp box is positioned at −37 relative to the transcription start site. Remarkably, this box is similar to the lux box consensus sequence that has been implicated in promoter activation by the regulator TraR of A. tumefaciens (White & Winans, 2007, Pappas & Winans, 2003). To investigate whether this sequence is required for activation by VjbR, a lacZ fusion was constructed to a 460 bp fragment of PvirB in which bp 2–7 in the upstream half of the 18 bp box was substituted with a 6 bp HindIII site (TGACCG to AAGCTT), thereby disrupting the palindrome, but not the putative −35 regulatory sequence. Control of the promoter activity of this construct by VjbR was monitored in E. coli using a β-galactosidase assay, as described above. Compared to wild type PvirB, the background activity of the mutated promoter remained the same (about 1300 Miller Units) but activation by VjbR was significantly reduced (Fig. 4). Replacement of bp 7–12 or bp 13–18 of the promoter box consensus sequence by AAGCTT also reduced activation of the resulting PvirB∷lacZ fusions by VjbR (data not shown). However, these changes to the virB promoter box also resulted in an overall decrease of background promoter function, possibly because the downstream half of the box overlaps the putative −35 regulatory sequence of the promoter (Fig. 3A).
Figure 4.
VjbR-dependent expression of of PvirB∷lacZ constructs containing the native wild type (pSURS2) or mutated PvirB box (pSURS2b) in E. coli expressing vjbR. Values are presented as fold difference above E. coli expressing the negative control gene BAB1_0345. White bars represent expression levels in the absence of vjbR induction (no IPTG), and black bars represent expression after induction of vjbR expression with IPTG.
4. Many promoter regions in the Brucella genome contain a conserved virB promoter box
Since the 18 bp lux box-like sequence was required for full PvirB induction, we performed a bioinformatic search for the virB promoter box in intergenic regions of the B. suis 1330 and B. abortus 2308 genomes using the motif alignment search tool MAST (http://meme.nbcr.net/meme/)(Halling et al., 2005, Chain et al., 2005, Paulsen et al., 2002, Bailey & Gribskov, 1998). In the first search, a consensus of the three motifs in PvirB, PtetR and the virB1-virB2 intergenic region was used, resulting in the identification of 45 promoter regions containing similar motifs. A selection of 9 promoters was fused to lacZ for expression analysis in our E. coli strains containing vjbR or BAB1_0345 under control of a T7 promoter. Out of these 9 promoters we found 5 to be activated by VjbR, and the conserved motifs in these promoters were used to refine the consensus sequence for a new round of promoter searches. After repeating the promoter prediction process 3 times, a total of 144 promoters were predicted to contain the consensus 18 bp virB promoter box (Supplementary Table 2). Among genes directly downstream of these promoters, 13 are predicted to encode transcriptional regulators, including FtcR, whose expression was shown previously to be partially under control of VjbR (Leonard et al., 2007). Other genes include proteins of hypothetical function (40), enzymes of unknown specificity (22) and transport and binding proteins (11). The list also includes 14 proteins predicted to be involved in adaptation of Brucella to atypical conditions such as the intracellular environment, including RelA/SpoT, BacA and cyclic β-glucan synthetase (Arellano-Reynoso et al., 2005, Dozot et al., 2006, LeVier et al., 2000).
Based on known or predicted roles in host-pathogen interaction, 24 promoters were tested for VjbR-dependent transcriptional activation in E. coli. Transcriptional fusions of 15 of these promoters to lacZ were activated by induction of vjbR expression (Fig. 5 and Supplementary Table 3). These promoters included those upstream of two genes required for virulence in mice: fliC encoding flagellin and cgs, encoding cyclic β-glucan synthetase (Fretin et al., 2005, Arellano-Reynoso et al., 2005, Briones et al., 2001). Expression of two regulators was shown to be induced by VjbR, LysR12 (BAB2_0329) which is required for virulence of B. melitensis during infection of mice (Haine et al., 2005) and OmpR (BAB2_0762/3), predicted to encode the response regulator of the OmpR/EnvZ two-component system. In addition, virB promoter box-containing upstream regions of several uncharacterized genes were found to be activated by VjbR, including BAB2_0328, BAB1_1881, BAB1_1066, BAB1_1837, BAB1_1994, BR0951 (ORF BR0951 is not annotated in B. abortus), BAB1_1652, BRA1111, BAB1_1058, BAB2_0403 and BAB1_0604. BAB2_0880 was predicted in our first generation screen to contain an upstream virB promoter box, however we found the BAB2_0880∷lacZ transcriptional fusion not to be activated by VjbR in E. coli (Fig. 5). In the subsequent rounds of promoter searches the promoter of BAB2_0880 was no longer predicted to contain a virB promoter box, indicating that the consensus sequence predictions became more refined as we included more motifs from promoters that were activated by VjbR in E. coli.
Figure 5.
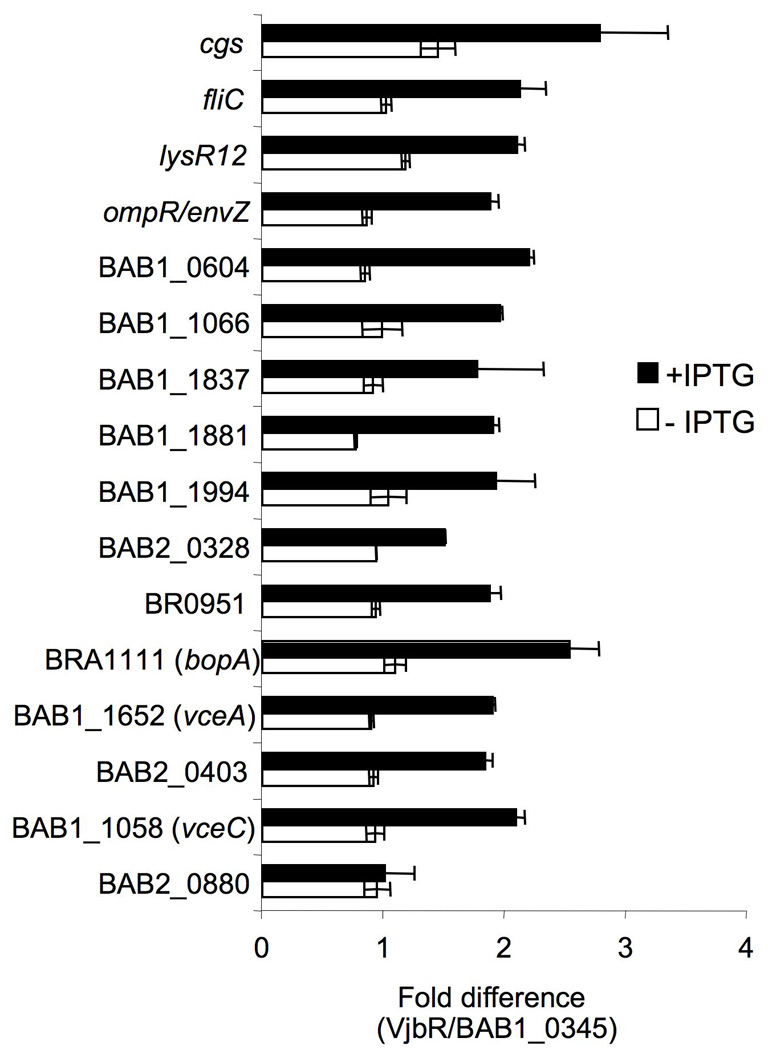
VjbR-dependent expression in E. coli of lacZ transcriptional fusions to 16 promoter regions containing a putative PvirB box. Values are presented as fold difference above E. coli expressing the negative control gene BAB1_0345. White bars represent expression levels in the absence of vjbR induction (no IPTG), and black bars represent expression after induction of vjbR expression with IPTG.
5. TEM1 β-lactamase can be used to detect translocation of T4SS effector proteins into macrophages
TEM-1 β-lactamase has been used by our lab and others (Charpentier & Oswald, 2004, Raffatellu et al., 2005, Sun et al., 2007) to demonstrate cellular translocation of Type III secretion system (T3SS) substrates, including Salmonella enterica serovar Typhi SipA and serovar Typhimurium Phase I flagellin. Since the requirements for passage through the T3SS needle and the T4SS apparatus may differ, we tested the utility of this reporter using a well characterized T4SS effector, Legionella pneumophila RalF. For these experiments, we generated translational fusions to the C terminus of TEM-1 β-lactamase (see Materials and Methods). To this end, each protein was fused to the C terminus of a modified TEM-1, in which the N-terminal Sec-dependent signal sequence was replaced by a 3×FLAG tag (Raffatellu et al., 2005). The resulting constructs were introduced into L. pneumophila Philadelphia 1 or a dotA mutant, in which the Dot/Icm T4SS is inactivated (Fig. 6). The strains expressing the TEM-1 fusion proteins were then used to infect J774 macrophages for detection of protein translocation, as has been reported previously (Raffatellu et al., 2005, Sun et al., 2007, Charpentier & Oswald, 2004). Translocation of a TEM1-RalF fusion by L. pneumophila into a host cell loaded with the fluorescent β-lactamase substrate (CCF2/AM) should lead to a shift in color of the cells from green to blue. At 4h after infection of J774 cells, we observed translocation of TEM1∷RalF into macrophages (Fig. 6A). A RalF mutant protein in which a lysine residue in the C terminus was replaced by alanine (K368A) was translocated, as reported previously, but replacement of the essential leucine residue at the C terminus by alanine (L372) strongly reduced translocation. Finally, the C-terminal 20 amino acids of RalF mediated translocation of TEM1 into J774 cells, in agreement with a previous report (Nagai et al., 2005). Figure 6B shows a blue cell infected with Lp01 expressing dsRed and TEM1∷RalF and a green cell infected with the dotA mutant expressing the same fusion. A lack of translocation of all the fusion proteins by the dotA mutant was not the result of reduced intracellular bacteria or decreased expression of the fusion proteins, as both of these were similar between Lp01 and the dotA mutant (Fig. 6C and 6D). These results showed that TEM-1 β-lactamase is a useful reporter for detection of T4SS-mediated protein translocation.
Figure 6.
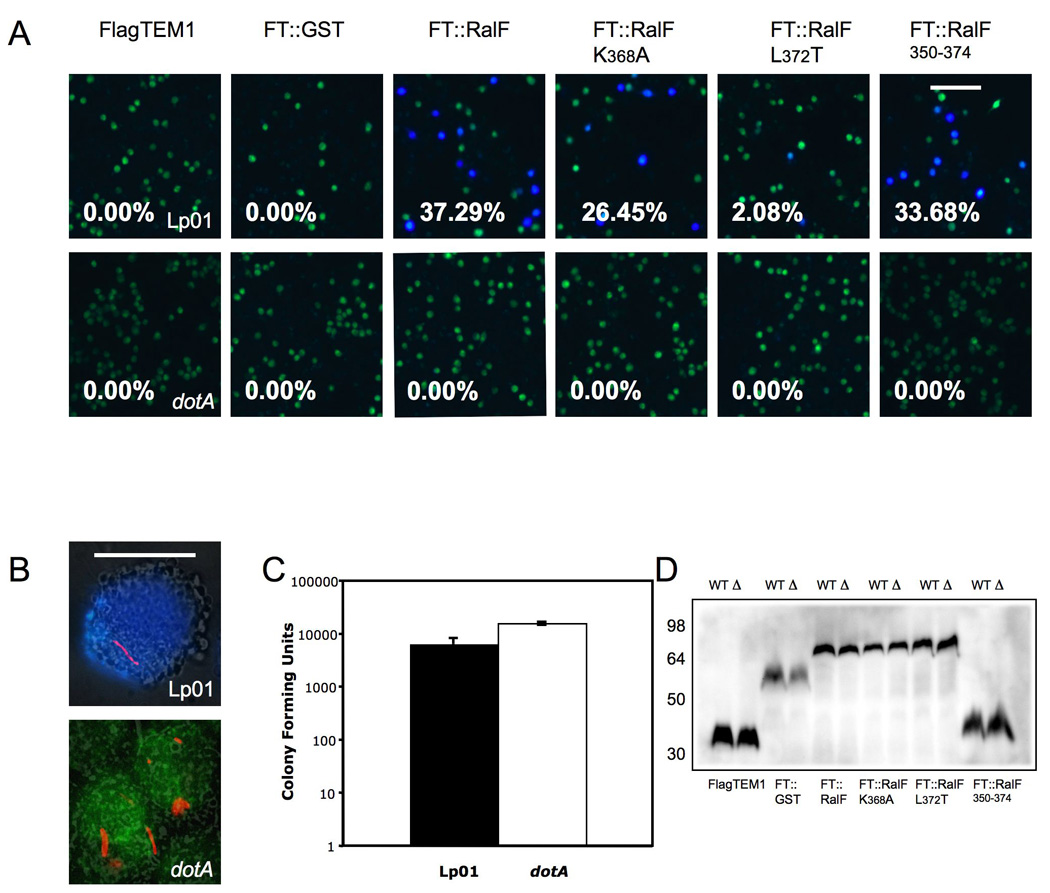
Translocation of TEM1∷RalF into J774 macrophages by L. pneumophila.
(A) J774 macrophages were infected with Lp01 (top row) or dotA (bottom row) mutant transformed with plasmids expressing FLAG-TEM-1 fusions for a total of four hours. Translocation efficiency is given as percentage of blue cells. Data is representative of three experiments that produced similar results. Bar, 100 µm. (B) Same experiment as above by using DsRed labeled Lp01 and dotA mutant bearing a plasmid expressing FT∷RalF350–374. These high magnification images were merged from the β-lactamase color channel (Blue and Green), DsRed mono channel (Red) and phase contrast control. The upper blue cell contains Lp01 expressing FT∷RalF350–374 and the bottom green cell contains the dotA mutant expressing the same fusion protein. Bar, 5 µm. (C) Number of bacteria per well. At the end of experiment cells were lysed in 0.5% Tween 20 after incubation for 90 min with 50µg/ml gentamicin. Colony forming units were determined by serial dilution and plating on BYCE plates. (D) Western blot using anti Flag, showing all β-lactamase fusions were expressed and their expression were similar between Lp01 (WT) and DotA mutant (Δ). Numbers on right indicate protein standard in kDa.
6. Translocation of BAB1_1652 (VceA) and BAB1_1058/BR1038 (VceC) into macrophages by B. abortus is dependent on the VirB T4SS
We analyzed the 144 candidate genes with the 18 bp PvirB promoter motif for features typical of translocated bacterial effector proteins. Since no predicted function was found initially for some T4SS effectors identified in other bacterial pathogens (de Felipe et al., 2005), we considered 13 hypothetical proteins from this group as candidate effectors. To determine whether these effector candidates were substrates of the T4SS, we generated translational fusions to the C terminus of TEM-1 β-lactamase, as described above for RalF (see Materials and Methods). The resulting constructs were introduced into the B. abortus wild type or a virB2 mutant (ADH3; (den Hartigh et al., 2004). The strains expressing the TEM-1 fusion proteins were then used to infect J774 macrophages for detection of protein translocation. For 10 of these constructs, we observed fewer than 0.5% blue cells, suggesting that these fusion protein are not translocated into host cells similar to what was observed for our negative control protein FLAGTEM1∷GST (Fig. 7A and 7B). However, we did observe blue cells in macrophage cultures infected with B. abortus expressing TEM1∷BAB1_1652, B. suis TEM1∷BR1038 or a TEM1 fusion to its B. abortus orthologue (TEM1∷BAB1_1058), suggesting that these proteins are translocated into macrophages by B. abortus (Fig. 7A and 7B). No blue cells were seen in macrophage cultures infected with a virB2 mutant (ADH3) expressing these fusion proteins, indicating that an intact T4SS is required for cellular translocation of TEM1∷BAB1_1652, TEM1∷BR1038 and TEM1∷BAB1_1058 (Fig. 7A–B). Western blotting of wild type and virB2 mutant B. abortus expressing TEM1∷BAB1_1652, TEM1∷BR1038 or TEM1∷BAB1_1058 showed that both strains expressed the fusion protein at equivalent levels (Fig. 7C).
Figure 7.
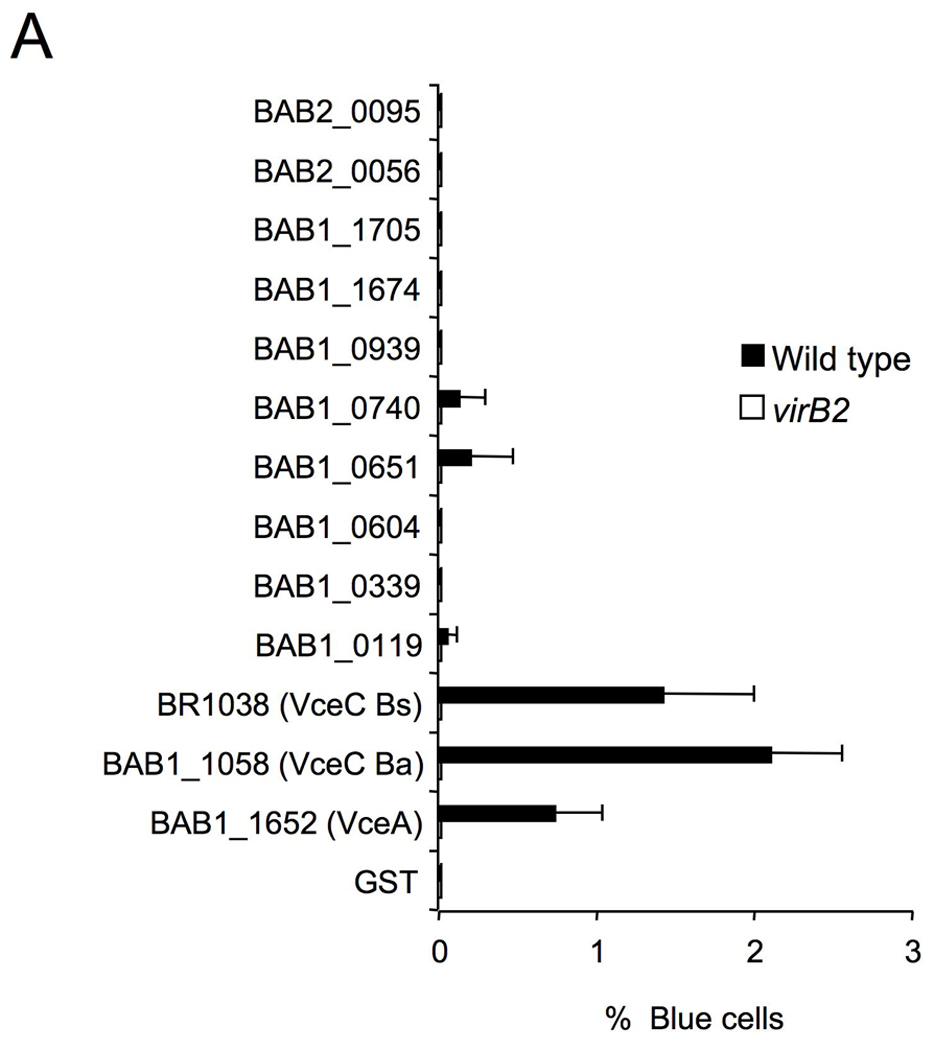
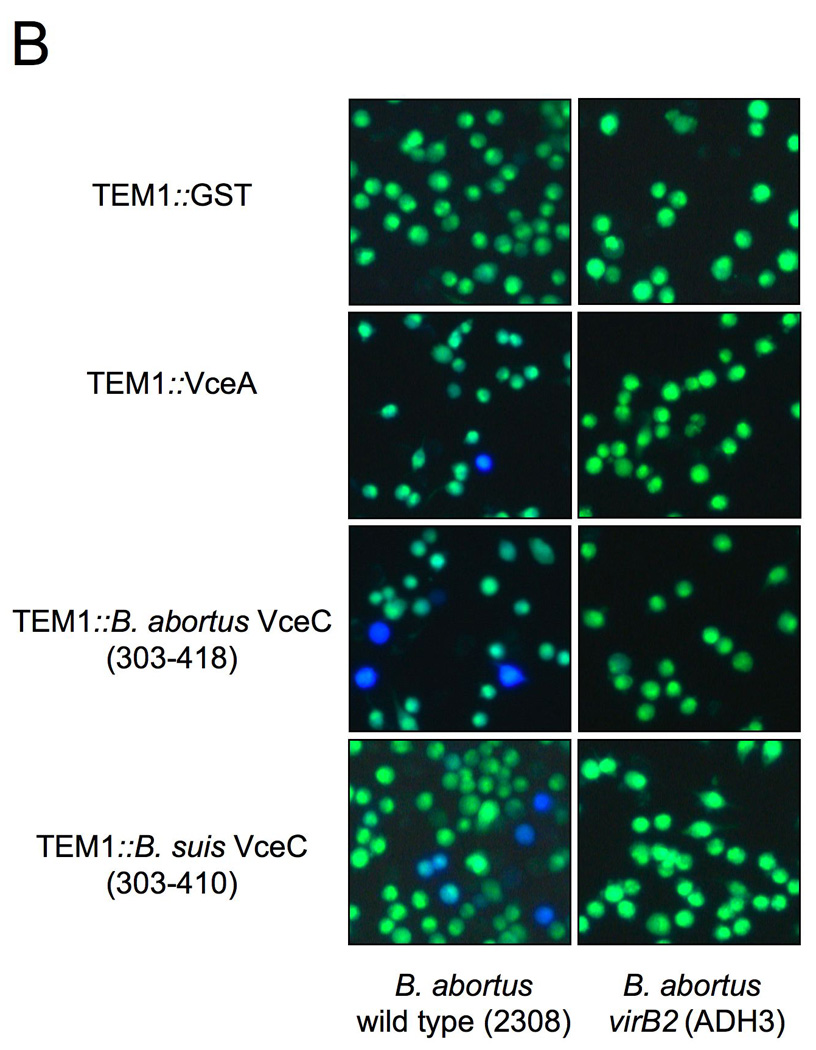
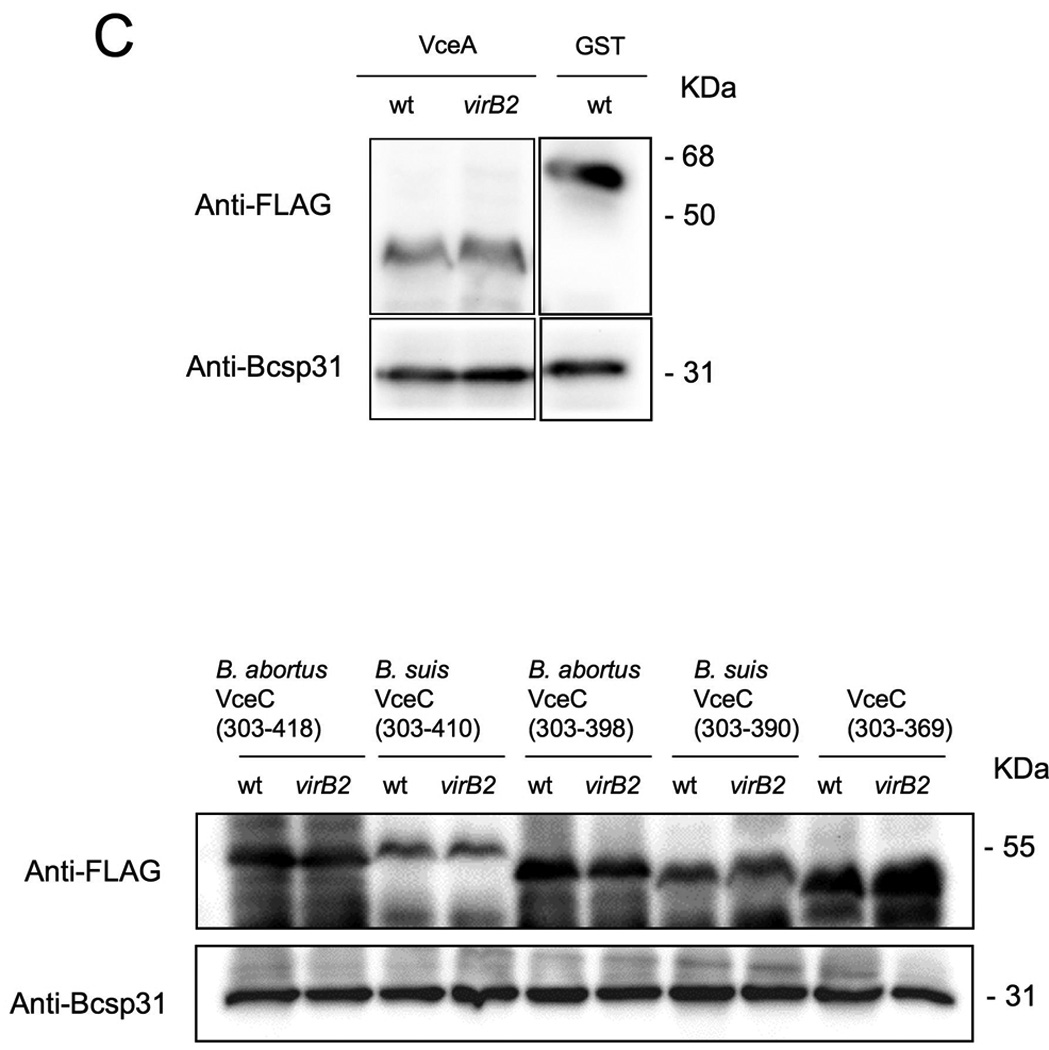
Translocation of TEM1∷VceA and TEM1∷VceC into J774 macrophages. Cytosolic translocation of β-lactamase by wild type (2308) or ΔvirB2 mutant strain (ADH3) of B. abortus was assessed by fluorescence microscopy. Cells in which translocation of the fusion protein has occurred appear blue. (A) Quantification of effector translocation. Cells from ten independent random microscope fields were counted and the percent blue cells calculated. Results shown are the mean ± SD of three independent experiments. (B) TopB. abortus 2308 containing the control fusion protein pFlagTEM1-GST, showing lack of translocation of FLAG-TEM1∷GST. Second from top to bottomB. abortus 2308 (left) or virB2 mutant (right) expressing FLAG-TEM-1 fused to B. abortus VceA or the C-terminal domain of VceC from B. abortus or B. suis. Results are from a representative individual experiment that was repeated three times independently. (C) Western blots showing equal expression levels of FLAG-TEM1 fusion proteins in B. abortus wild type and virB2 mutant. Proteins were detected using anti-FLAG antiserum (upper row). As a loading control, the blot was probed with anti-Bcsp31 (lower row).
To ensure that the lack of detectable translocation from the virB2 mutant compared to wild type B. abortus was not due to a lower number of T4SS mutant Brucella associated with the macrophages, we determined the kinetics of intracellular survival of each strain after a 90 minute treatment with gentamicin to kill extracellular bacteria (Fig. 8A). At time points up until 11h, we recovered equal numbers of B. abortus wild type and the virB2 mutant, which demonstrated that failure of the virB mutant to translocate the TEM1 fusion proteins is not due to intracellular killing of the virB mutant. Based on these findings, we designated TEM1∷BAB1_1652 as vceA (for virB-coregulated effector A), BAB1_1058 as B. abortus vceC and BR1038 as B. suis vceC.
Figure 8.
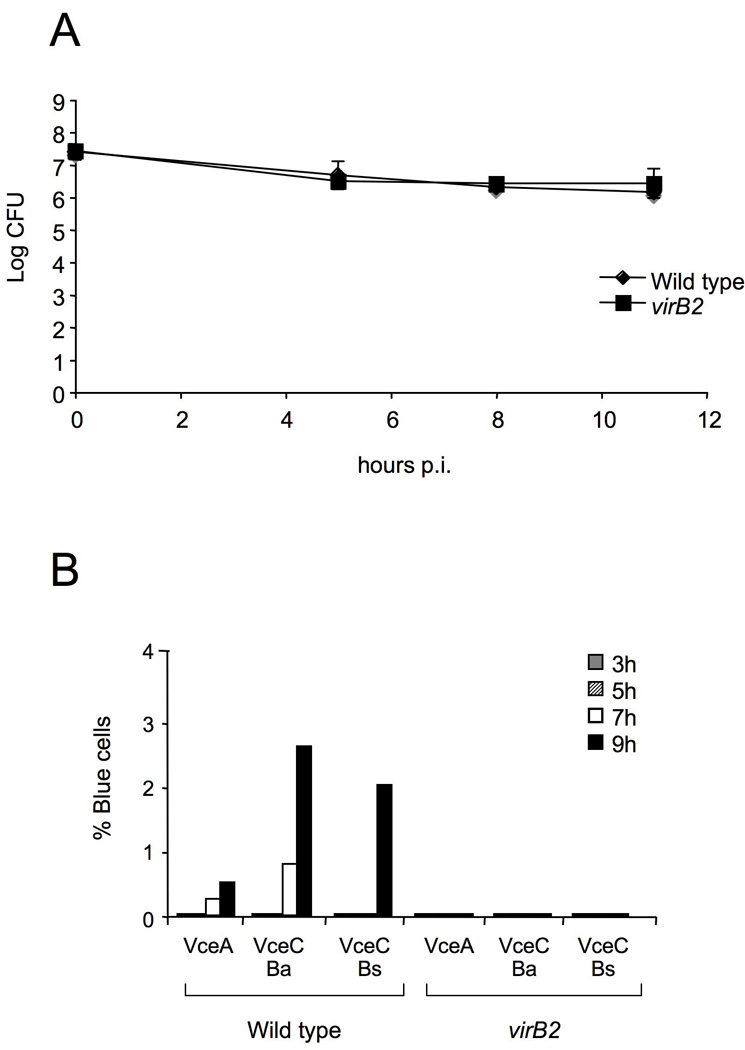
Translocation of TEM1∷VceA and TEM1∷VceC into J774 macrophages at different time points after infection. (A) Survival of wild type (2308) and virB2 mutant (ADH3) B. abortus in J774 macrophages after infection at an MOI of 500 bacteria per macrophage. (B) Cytosolic translocation of TEM1∷VceA and TEM1∷VceC by wild type (2308) or ΔvirB2 mutant strain (ADH3) B. abortus was assessed by fluorescence microscopy at 3, 5, 7 and 9 hours post infection (MOI 1:500). Cells in which translocation of the fusion protein has occurred appear blue and were quantified from independent random microscope fields from a single experiment and the percent blue cells was calculated.
Expression of the B. abortus virB genes has been shown to be induced upon phagosomal acidification and to reach its maximal expression level at 5h after infection (Sieira et al., 2004). Since we initially screened for translocation at 16h after infection, we determined the kinetics of translocation of VceA and VceC into macrophages (Fig. 8B). Translocation of VceA and VceC was first detectable at 7h after macrophage infection. These findings were consistent with a requirement for induction of the T4SS genes for translocation of the effectors into host cells.
7. VceA and VceC are conserved in all sequenced Brucella genomes
VceA is a protein of 105 amino acids that is conserved in all of the sequenced Brucella genomes. It is not well conserved in close phylogenetic relatives of Brucella spp., including Ochrobactrum anthropi, O. intermedium, Bartonella spp., or A. tumefaciens, however a conserved domain of this protein is found in sequenced genomes of some environmental strains of Alpha- and Betaproteobacteria, such as Paracoccus denitrificans and Burkholderia spp. This information did not allow us to infer a putative function for this protein.
The VceC protein of B. abortus contains 418 amino acids, with a proline rich central domain. BLASTP searches using the VceC amino acid sequence showed that an N-terminal region of approximately 100 amino acids was conserved in proteins of several α-proteobacteria including Mesorhizobium loti, Methylobacterium radiotolerans and Bartonella spp., among others. However, most of the full-length protein was specific to Brucella spp. and its close relatives, Ochrobactrum species anthropi and intermedium (unpublished genome sequence). Figure S1 shows a multiple alignment of different VceC proteins revealing a high level of conservation among VceC proteins from O. anthropi, O. intermedium and Brucella spp. (see also Table S3). A PSI-BLAST search using the conserved central proline rich region of VceC showed similarity to several eukaryotic proteins of hypothetical function containing proline rich regions. Interestingly, the C-termini of the B. suis and B. abortus VceC proteins differed, as a result of 1bp that is missing in B. suis vceC, leading to a frameshift in the C-termini of B. suis and B. canis (Figure S1). This difference was not the result of a sequencing error, as we re-sequenced these genes in our laboratory to confirm the different C termini (data not shown). The differences in the C terminus of B. abortus and B. suis variants of VceC suggest that the Type IV translocation signal may not need to be completely conserved. Based on this finding, as well as similarity of the proline-rich region of VceC to eukaryotic proteins, we chose to characterize VceC further.
8. The C terminal 20 amino acids of VceC are required for its translocation into host cells
For several Type IV effectors, including L. pneumophila RalF (Nagai et al., 2005), A. tumefaciens VirF (Vergunst et al., 2005), Bartonella henselae BepA-D (Schulein et al., 2005), and Helicobacter pylori CagA (Hohlfeld et al., 2006), a C-terminal domain is important for translocation into host cells. To determine whether the C terminus of B. abortus VceC was required for its translocation into macrophages, we generated two sets of truncated TEM1∷VceC fusions, using the B. suis and B. abortus variants (Fig. 9A): one in which the C-terminal 20 amino acids were deleted (Ba VceC303–398 and Bs VceC303–390) and one in which the C-terminal 49 (Bs VceC) or 41 (Ba VceC) amino acids were deleted (TEM1∷VceC303–369). Each of these truncations abrogated translocation into macrophages, showing that despite their divergence in the C terminus of the protein, the C-terminal 20 amino acids of both B. abortus VceC and B. suis VceC are required for VirB-dependent translocation into macrophages. Truncation of the C terminus did not reduce stability of the proteins, as shown by Western blot (Fig. 7C). Alignment of the C termini of B. abortus VceC and B. suis VceC with that of VceA revealed no obvious motif that could be involved in translocation, although both VceA and VceC had a C-terminal K-X-K-X-K/H motif reminiscent of the C-terminus of A. tumefaciens effectors (Vergunst et al., 2005).
Figure 9.
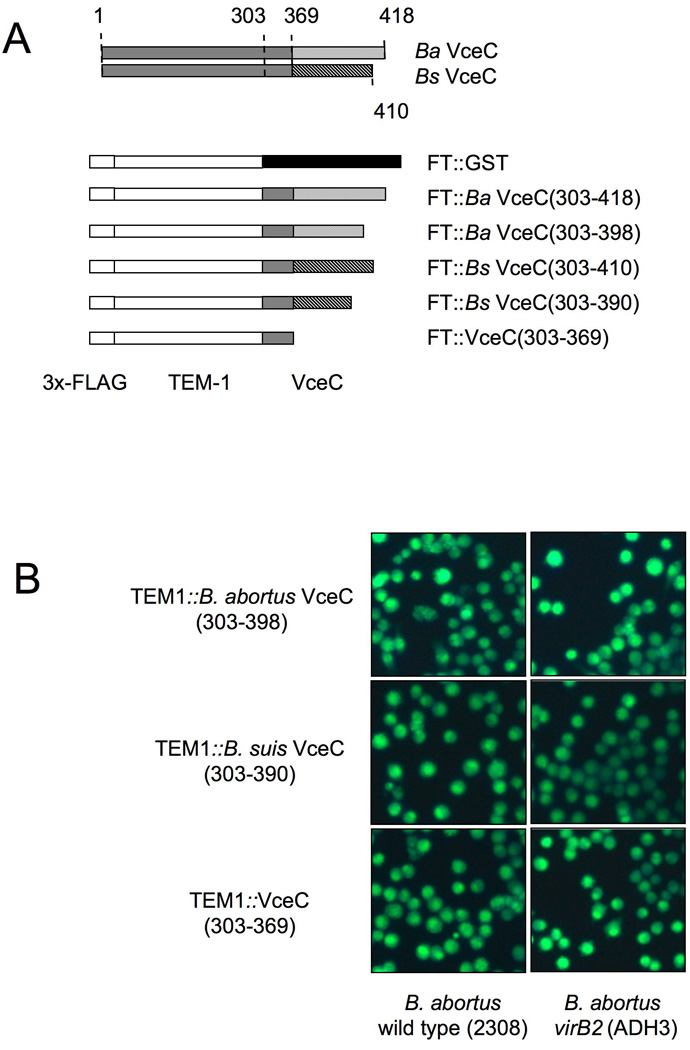
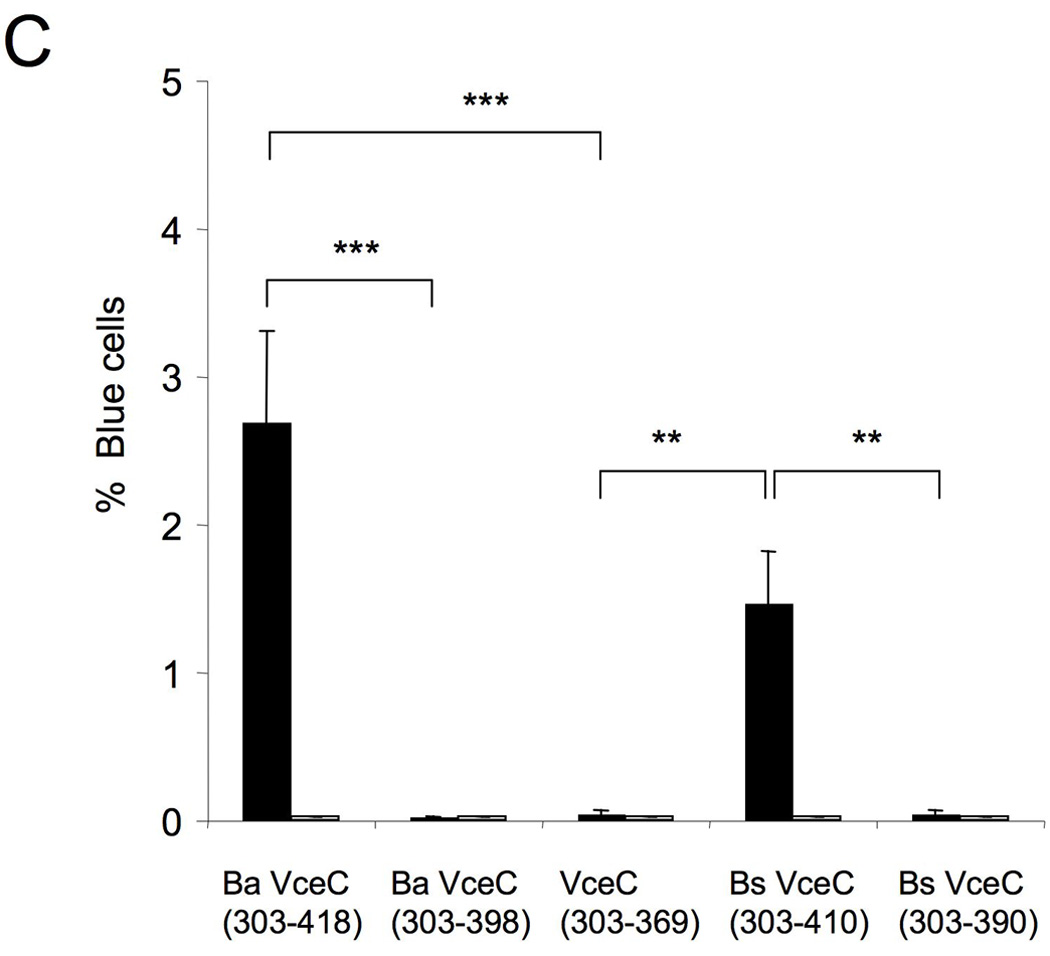
The C-terminal 20 amino acids of both B. abortus VceC and B. suis VceC are required for translocation into infected macrophages. (A). Schematic representation of VceC in B. abortus and B. suis which differ in their last 49 or 41 amino acids respectively. For translocation experiments the C-terminal 115 (B. abortus) or 107 (B. suis) amino acids of VceC and truncations lacking the last 20 amino acids were fused to TEM1. Also a truncation of VceC (303–369) lacking the entire C-terminal region that differs between VceC B. abortus and B. suis was fused to TEM1 and tested for translocation in macrophages. (B). Translocation of TEM1-VceC fusions into J774 macrophages. Cytosolic translocation of β-lactamase by wild type (2308) or ΔvirB2 mutant strain (ADH3) of B. abortus was assessed by fluorescence microscopy. (C) Quantification of effector translocation. Cells from ten independent random microscope fields were counted and the percent blue cells calculated. Results shown are the mean ± SD of three independent experiments. For statistical analysis and calculation of SD, data were logarithmically transformed and significance of differences between cultures infected with wild type and virB2 mutant B. abortus was analysed using a Student’s t test. ***, p<0.001; **, p<0.01.
9. Translocation of TEM1∷VceC results in a cytotoxic effect on J774 macrophages
Since we observed in some translocation assays a large number of blue cell “ghosts” suggestive of lysis of blue cells, we tested whether a cytotoxic phenotype may contribute to the low number of β-lactamase positive blue cells in our assays. To test this idea, we assayed for LDH release from J774 macrophages infected with B. abortus wild type or virB2 mutant expressing TEM-1 β-lactamase fusions. At the multiplicity of infection of 500 used for these assays, at 11h post infection we observed 30% cytotoxicity in cells infected with B. abortus, while cells infected with the virB2 mutant were indistinguishable from uninfected controls (Fig. 8C). This likely represents the T4SS-dependent macrophage cytotoxicity described by Pei et al using a similar multiplicity of infection (Pei et al., 2008). However strikingly we found that 60% of cells infected with B. abortus 2308 expressing the TEM1∷VceC fusion were lysed by 11h (Fig. 9C). Truncation of the C terminus of VceC by 20 or 49 amino acids led to a reduction in cytotoxicity that corresponded with the reduced ability of these fusion proteins to be translocated into host cells (Fig. 9A–C). In a second experiment, we determined the time course of lysis in cells infected with B. abortus 2308 expressing the TEM1∷VceC fusion (Fig. 10D). At 8h and 11h post infection, we observed increasing lysis of infected cells with kinetics paralleling those of blue cell appearance in the cultures (Fig. 8B). This finding suggested that under the conditions used for this assay, cells into which the TEM1∷VceC fusion is translocated may ultimately lyse, thereby limiting our ability to detect translocation of this fusion protein.
Figure 10.
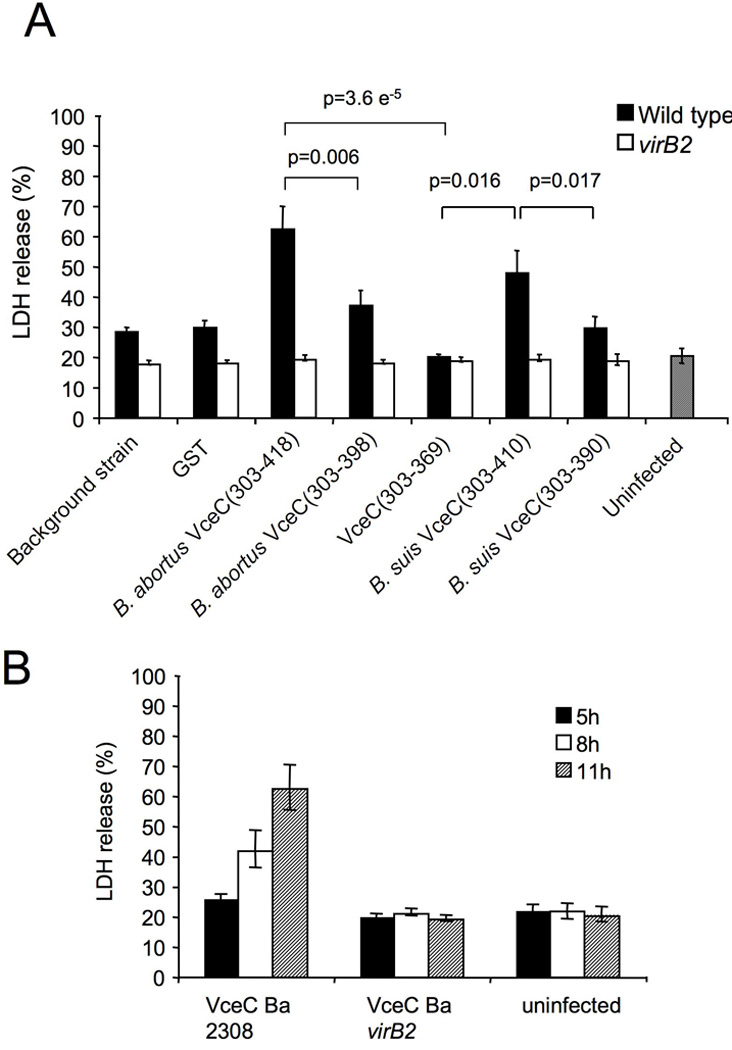
(A) Cytotoxicity assay showing % LDH release at 11 hours post infection from J774 macrophages infected at MOI 1:500 with either wild type or virB2 mutant Brucella abortus expressing TEM1-GST or TEM1-effector fusions. (B) Cytotoxicity assay showing % LDH release at 5, 8 and 11 hours post infection from J774 macrophages infected at MOI 1:500 with either wild type or virB2 mutant Brucella abortus expressing TEM1∷VceC. Results shown in A and B are the mean ± SD of three independent experiments. For statistical analysis and calculation of SD, data were analysed using a Student’s t test. ***, p<0.001; **, p<0.01; *, p<0.05.
10. The C terminus of B. suis VceC contains a translocation signal that is functional in L. pneumophila
T4S signals from some substrates, such as that of RSF1010 MobA, are able to function in heterologous systems, including Bartonella henselae (Schulein et al., 2005)and Helicobacter pylori (Hohlfeld et al., 2006), suggesting that some of these secretion signals share common features. To determine whether the C-terminal signal of VceC can also function in a heterologous system, we expressed the C-terminal 107 or 115 amino acids of B. abortus and B. suis VceC (CyaA-VceC) as a fusion to adenylate cyclase in both B. abortus and L. pneumophila. Translocation was detected by infecting CHO-FcR cells with either B. abortus or L. pneumophila expressing the fusion proteins and detection of cyclic AMP. While translocation of the CyaA-VceC fusions by B. abortus could not be detected, L. pneumophila translocated the B. suis CyaA-VceC fusion protein into the CHO-FcR cells at a level similar to that of the Dot/Icm T4SS substrate RalF (Fig. 11 A and 11B). The B. abortus CyaA-VceC fusion protein was translocated, albeit at a lower level. Translocation of both VceC fusion proteins was dependent on the Dot/Icm T4SS, since a dotA mutant did not translocate either VceC variant. Examination of the C terminus of both proteins showed that similar to RalF, B. suis VceC had a Leu residue at the −3 position, as well as the motif K-X-K-X-K directly upstream of the Leu. B. abortus VceC lacked both of these features, suggesting that they may contribute to recognition of the B. suis VceC secretion signal by the Dot/Icm T4SS (Fig. 11A).
Figure 11.
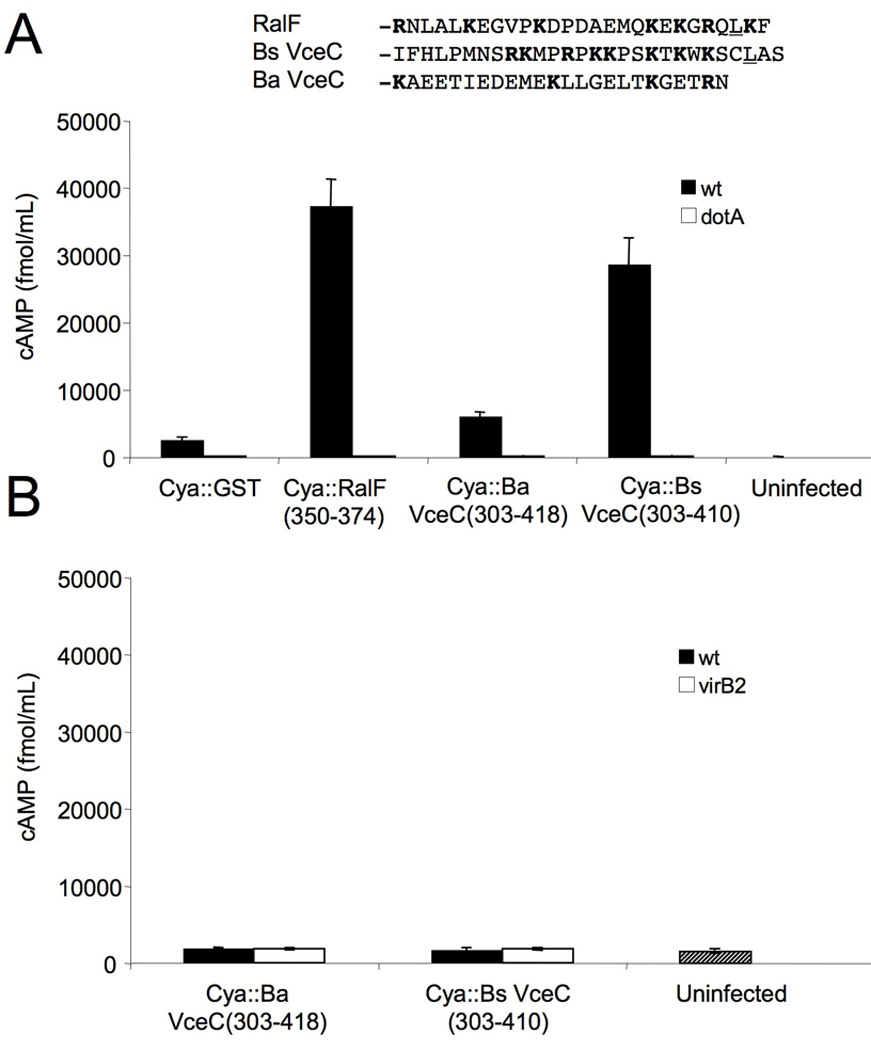
The C-terminus of B. suis VceC contains a signal that is recognized by the L. pneumophila Dot/Icm T4SS (A) Translocation of CyaA∷VceC into CHO-FcR cells by L. pneumophila wild type or dotA mutant. (B) Translocation of the same fusions by B. abortus cannot be detected. Results shown are representative of two independent experiments.
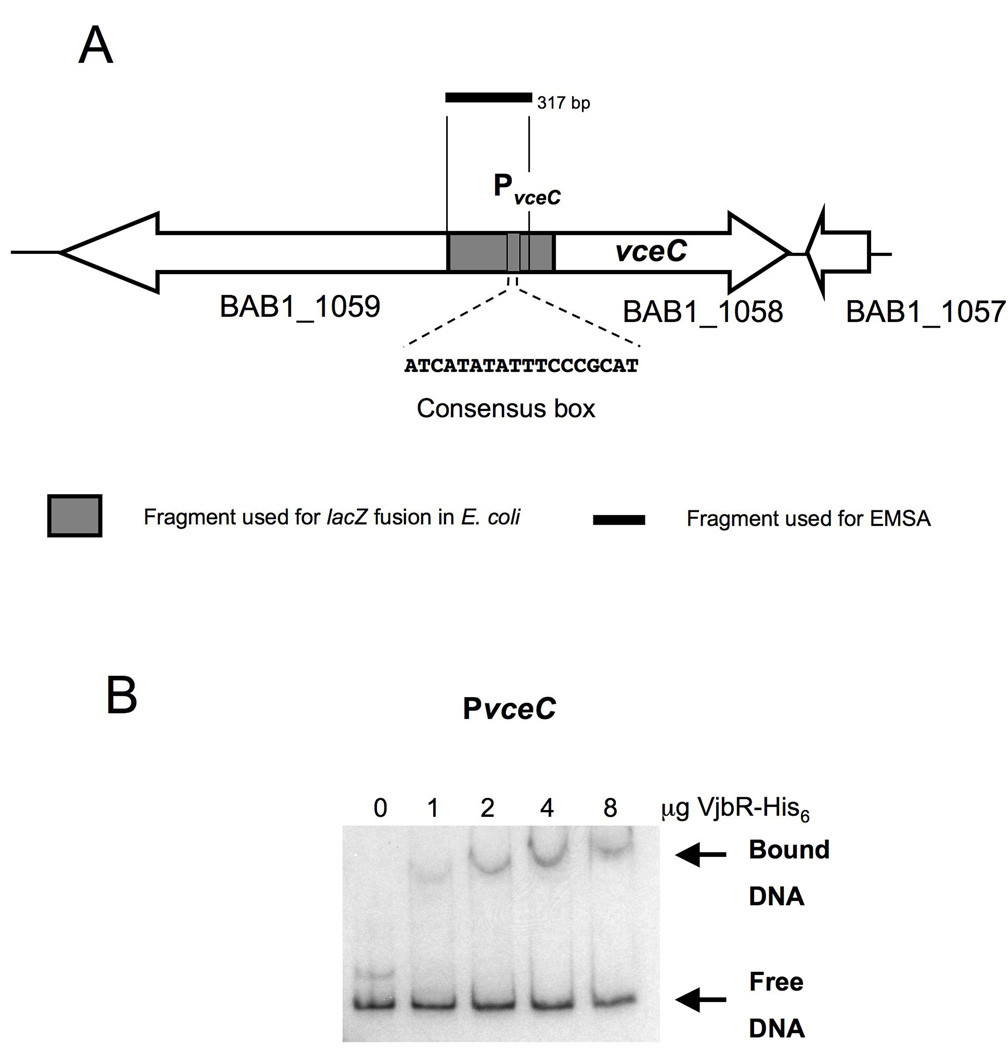
11. VjbR-His6 binds directly to PvceC
To determine whether VjbR would directly bind a promoter of a translocated effector identified in the bioinformatic screen, we performed EMSAs with PvceC in a similar manner as with the PvirB fragments. In EMSAs with PvceC we used a 32P labeled 317 bp fragment containing a DNA region upstream of the vceC start codon (Fig. 12A). Fig. 12B shows that the intensity of a band with decreased electrophoretic mobility increased as increasing amounts of purified VjbR-His6 were added to the binding reaction. These results demonstrated direct binding of VjbR to PvceC, and together with the expression analysis in E. coli (Fig. 5) strongly supported VjbR mediated co-regulation of vceC and the virB genes in B. abortus.
Figure 12.
VjbR binds to PvceC(A) Schematic representation of VceC in chromosome I of B. abortus showing the putative PvirB box and promoter fragments used for EMSA and lacZ fusions. (B) EMSA showing binding of VjbR-His6 to a32P labeled DNA fragment containing 317 bp upstream of the vceC start codon.
12. VjbR activates expression of vceC in B. abortus
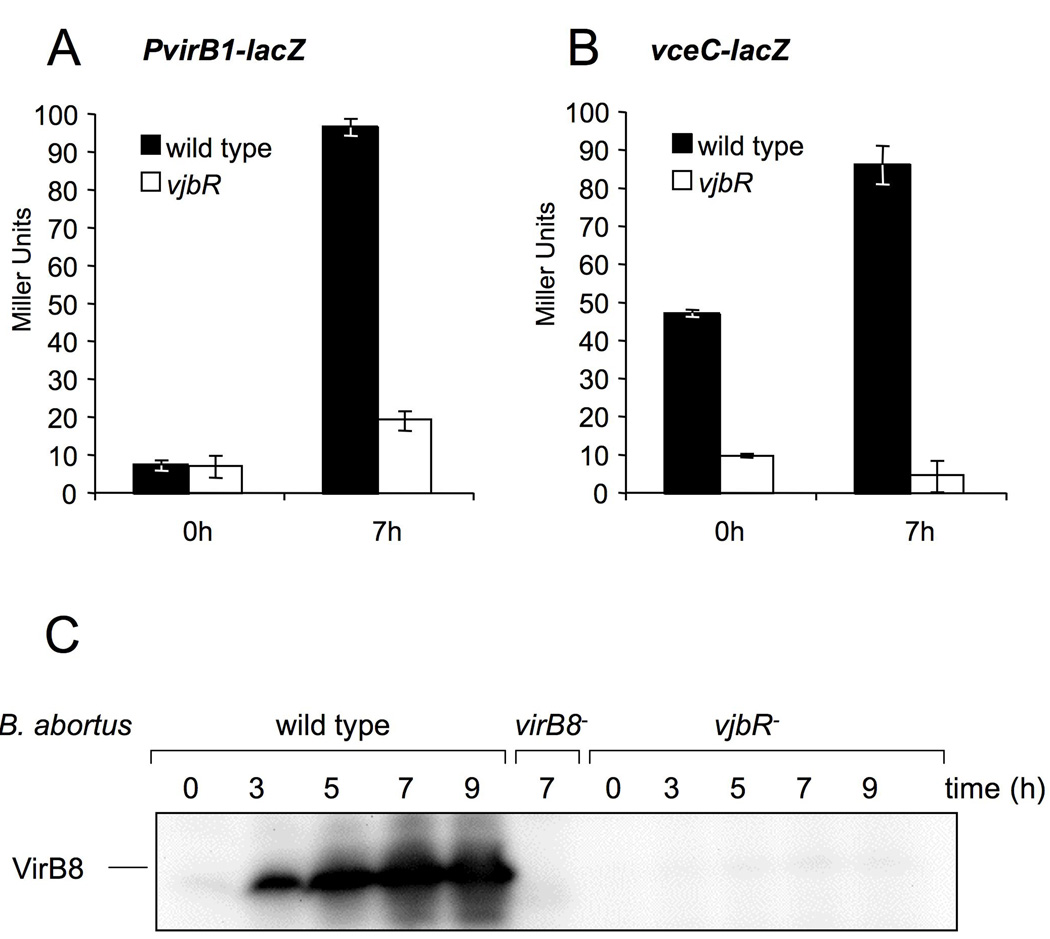
To assess the biological significance of the VjbR-dependent regulation of vceC in E. coli and of results from in vitro EMSAs, we constructed chromosomal lacZ fusions to the virB1 and vceC promoters in the B. abortus wild type (2308) and in a vjbR mutant (ADH17) background. The β-galactosidase activity of these strains was measured at several time points during growth in modified minimal E-medium at pH 5, which has been shown to induce expression of the virB genes (Kulakov et al., 1997, Patey et al., 2006). As expected, PvirB∷lacZ expression was reduced in the vjbR mutant (Fig. 13A) compared to wild type B. abortus. This was also shown by Western blot, in which the VirB8 protein levels were markedly lower in the vjbR mutant than the wild type (Fig. 13C). Similarly, our results showed that vceC∷lacZ expression was down regulated in the B. abortus vjbR mutant (ADH17; Fig. 13B). These data suggested that VjbR is required for optimal expression of both vceC and the virB operon, indicating that this activator coordinates expression of the B. abortus T4SS and one of its secreted targets.
Figure 13.
VjbR-dependent activation of virB and vceC expression in B. abortus at different time points after switching from TSB to modified minimal E-medium, pH 5. At time points 0 h and 7 h after switching media, β-galactosidase activity was measured in samples of B. abortus wild type and vjbR mutant strains containing a chromosomal virB1∷lacZ fusion (A) or vceC∷lacZ fusion (B). C. Western blot using anti-VirB8 antiserum shows protein levels of VirB8 in wild type and vjbR mutant B. abortus in samples taken at several time points after switching to modified minimal E-medium at pH 5. In each lane, the total protein equivalent to 1 × 108 CFU was loaded, as determined by measurement of OD600 for each culture.
Discussion
Type IV secretion systems are used by many Gram-negative bacteria to manipulate their host’s cells for optimal survival and multiplication in a hostile environment. In a wide variety of eukaryotic hosts such protein secretion systems therefore play a critical role in pathogenesis (Backert & Meyer, 2006). All Brucella species including the human pathogens B. abortus, B. suis and B. melitensis require a T4SS for intracellular survival and persistent infection, however to date no substrates of the T4SS have been identified (Delrue et al., 2001, den Hartigh et al., 2004, Hong et al., 2000, O’Callaghan et al., 1999, Sieira et al., 2000). We hypothesized that in order to function optimally, the T4SS should be expressed together with its secreted effectors. To test this idea, we screened for genes co-regulated with the VirB system to identify an effector of the B. abortus T4SS. The LuxR-family quorum sensing regulator VjbR, is an important activator of virB gene expression (Delrue et al., 2005), however, it is unclear from this work whether this regulation is direct or occurs as part of a regulatory cascade. Binding of VjbR to a 123 bp fragment of PvirB in an EMSA provided evidence that this regulator directly activates virB transcription. Activation of a PvirB∷lacZ transcriptional fusion by VjbR in E. coli was reduced by the quorum sensing pheromone C12-HSL, which was in accordance with recent work showing inhibition of transcriptional activation by VjbR in the presence of C12-HSL (Uzureau et al., 2007).
Other regulators of the LuxR family, such as TraR of A. tumefaciens, are known to bind a palindromic motif of 18 bp often located close to the −35 regulatory sequence of promoters (Pappas & Winans, 2003, White & Winans, 2007). We identified a structurally related palindromic motif that enabled us to identify candidate VjbR-regulated genes in B. abortus and B. suis.
The virB promoter box was found upstream of 144 B. abortus and B. suis genes. Since some of these genes are predicted to be part of operons, this finding suggested that the VjbR regulon may include more than 144 genes. VjbR regulated several genes required for virulence of Brucella, including cgs and fliC (Briones et al., 2001, Arellano-Reynoso et al., 2005, Fretin et al., 2005). The gene cgs encodes the protein required for synthesis of cyclic β-1,2-glucan, which mediates evasion of lysosomal fusion with the Brucella-containing vacuole (Briones et al., 2001, Arellano-Reynoso et al., 2005, Fretin et al., 2005) and fliC encodes flagellin, a component of the flagellum that has been shown to be required for persistent colonization of the reticuloendothelial system in mice (Briones et al., 2001, Arellano-Reynoso et al., 2005, Fretin et al., 2005). A gene with potential function in host-pathogen interactions, the bopA gene (BRA1111), was also found to contain a virB promoter box. This gene, shown to be regulated in B. melitensis by VjbR, encodes a protein with similarity to T3SS effector protein HopAN1 from the plant pathogen Pseudomonas syringae (Boch et al., 2002, Lindeberg et al., 2005, Rambow-Larsen et al., 2008). Thus, VjbR may be a global regulator of genes involved in interactions with host cells.
To determine whether the reporter TEM-1 would be useful for detection of T4S, we showed translocation of a TEM1∷RalF fusion protein by L. pneumophila (Fig. 6). Our results are in agreement with a recently published study showing translocation of several new Dot/Icm effectors designated Legs (de Felipe et al., 2008). Interestingly, the same authors were unable to detect translocation of several of the Legs using translational fusions to Bordetella pertussis adenylate cyclase (CyaA), suggesting that TEM-1 may be a more sensitive translocation reporter for at least a subset of effectors (de Felipe et al., 2005). Based on our success in demonstrating cytosolic translocation of S. enterica serovar Typhimurium flagellin into macrophages using this system (Sun et al., 2007), we chose it as a sensitive reporter assay for translocation of candidate B. abortus effectors. We assayed for translocation into host cells of 13 hypothetical proteins containing upstream virB promoter boxes. Ten hypothetical proteins were not translocated into J774 cells by B. abortus in our experiments. For BAB2_0056 and BAB2_0095 this might be caused by the fact that we constructed a fusion with TEM fused to the N-terminus of these proteins, possibly disrupting an N-terminal Sec (BAB2_0056) or TAT (BAB2_0095) secretion signal. In this respect, it should be noted that the precise route of translocation across the cell envelope of most T4SS effectors is still unknown. Therefore, it is possible that some Brucella effectors are exported to the periplasm prior to translocation into the host cell by the T4SS, as is the case for subunits of the B. pertussis pertussis toxin (Covacci & Rappuoli, 1993, Weiss et al., 1993).
Importantly, our screen identified the first effectors of the Brucella spp. T4SS, VceA and VceC. Both of these effectors were co-regulated with the virB genes via VjbR. Further, the timing of secretion (7–9h after infection of macrophages) coincided with the time at which the B. abortus virB genes encoding the T4SS apparatus are induced in intracellular bacteria (Sieira et al., 2004). The percent of cells positive for translocated VceA and VceC ranged from 0.5% to 2.5%, which is similar to translocation levels reported for the Bartonella henselae BepB, BepC and BepD secretion signals using the Cre reporter assay for translocation, a method that, similar to the TEM-1 reporter, allows quantification of the number of cells containing translocated proteins (Schulein et al., 2005). It is possible that we were able to detect translocation of TEM-1 fusions by B. abortus, but not CyaA fusions (data not shown) to these proteins, because with the low number of cells into which proteins are translocated, the signal of the few cells containing translocated protein would be diluted out by the majority of cells in a lysate that do not contain cytosolic protein. It has been reported previously, that in macrophage cultures infected with Brucella, only few cells contain replicating bacteria, while in the majority of cells, bacteria do not increase in numbers (Celli et al., 2003). Thus, it is possible that while many cells contain Brucella, only a few contain bacteria that successfully inject effector proteins and go on to replicate intracellularly.
The C terminus of VceC was required for optimal secretion, similar to what has been found for other Type IV effector proteins (Vergunst et al., 2005). VceC of B. suis had a positively charged C-terminal region, which could form a secretion signal similar to that of A. tumefaciens T4SS effector proteins (Vergunst et al., 2005). However, for VceC, a R/K-X-R/K-X-R/K motif found in the C terminus was not essential, since this motif was present in the C terminus of B. suis 1330 VceC but not B. abortus 2308 VceC, both of which were translocated with similar frequency into macrophages by B. abortus. There is evidence that domains of effector proteins other than the C terminus contribute to T4SS-dependent translocation in other organisms (Cambronne & Roy, 2007, Hohlfeld et al., 2006, Schulein et al., 2005), which could explain why the C-terminal regions of VceC proteins were not conserved between B. abortus and B. suis, yet both could be translocated into macrophages. Interestingly, B. suis CyaA-VceC was translocated into CHO-FcR cells by L. pneumophila, dependent on the Dot/Icm T4SS, which suggests the T4S signal of VceC is recognized by the Dot/Icm T4SS. Additional studies will be required to define the precise residues of the VceC translocation signal that are required for its transfer to the host cell.
In summary, this report provided the first evidence that VjbR coordinates expression of the virB T4SS with its translocated effectors. This situation is reminiscent of other T4SS, including the Dot/Icm T4SS of L. pneumophila, in which the two-component system regulators PmrA and CpxR regulate genes of the Dot/Icm T4SS as well as several Dot/Icm effector proteins (Zusman et al., 2007, Altman & Segal, 2008), and the Mesorhizobium loti R7A VirB/D4 T4SS, in which the virB and virD4 genes are coregulated with the effectors msi059 and msi061 via a VirA/VirG two-component regulatory system (Hubber et al., 2007). The discovery of new host cell targets for VceA and VceC should shed light on their role in facilitating intracellular Brucella infection.
Experimental Procedures
Bacterial strains and plasmids
The Brucella abortus and Escherichia coli strains used in this study are listed in Table S1. B. abortus 2308 was used as a wild type strain. B. abortus strains were cultured on tryptic soy agar (TSA; Difco/Becton-Dickinson, Sparks, Md.), in tryptic soy broth (TSB) with appropriate antibiotics, or in modified E-medium (Kulakov et al., 1997). E. coli strains were grown on Luria Bertani (LB) agar. Antibiotics were used at the following concentrations for E. coli and B. abortus: carbenicillin (Carb), 100 µg/ml; kanamycin (Kan), 100 µg/ml; chloramphenicol (Cm), 30 µg/ml. E. coli and B. abortus were grown at 37°C. Work with B. abortus was performed at biosafety level 3. DNA techniques were performed according to standard protocols. Restriction enzymes were purchased from New England Biolabs and primers from Operon Technologies. Wild type L. pneumophila strain Lp01 (Berger and Isberg, 1993) and dotA mutant were grown on CYE plates or in AYE broth as described previously (Feeley et al., 1979). Antibiotics were used at the following concentrations: chloramphenicol (10 µg/ml) or kanamycin (Kan; 50 µg/ml).
Construction of plasmids for LacZ fusions
To obtain a low copy number plasmid for study of B. abortus promoters in E. coli, pSURS was constructed. To this end, the P15A origin of replication together with a Cm resistance gene were amplified by PCR from pSU19 (a derivative of pACYC184) using primers pACYC184-F and pACYC184-R (Table S4). The resulting 1900 bp product was digested with PstI and SalI and ligated to a 7500 bp fragment of pRS528, containing lacZYA, which was also digested with PstI and SalI, to yield pSURS1. Promoters of interest were introduced in the 5’ region of lacZ between the BamHI and EcoRI sites of pSURS. A 463 bp fragment of PvirB was PCR amplified using primers PVirBBamH1-F and PVirBEcoR1-R, digested with BamHI and EcoRI, and ligated into pSURS1. The resulting plasmid was named pSURS2 (see Table S1 for all the pSURS plasmids constructed).
For construction of lacZ fusions in B. abortus, regions of virB1 and vceC were PCR amplified using primers virB-1F and virB874R for virB1 and VceC-Pst1-F and VceCXba1-R for vceC. These amplicons were digested with PstI and XbaI and inserted between PstI and XbaI sites in the 5’ region of lacZ of plasmid pUJ10. The resulting plasmids were introduced into B. abortus 2308 and ADH17 strains by electroporation. Recombinants carrying the plasmid integrated into the chromosome were selected on ampicillin.
β-galactosidase assays
For β-galactosidase expression assays in E. coli, BL21 lacZ− (Stratagene) was transformed with a pET103 plasmid containing the vjbR gene or ORF BAB1_0345 (a randomly selected putative transcriptional regulator of Brucella, which was used as negative control). Subsequently a pSURS plasmid containing a promoter of interest fused to lacZ was introduced into this strain. For the expression assay strains were grown overnight in LB containing 20 mM glucose. The overnight cultures were diluted 1:20 in LB with 20 mM glucose and grown for 2 h to an OD600nm of 0.4, after which expression of regulators was induced by adding IPTG to a final concentration of 1 mM to the cultures. For some experiments, N-Dodecanoyl-DL-homoserine lactone (C12-HSL from Sigma-Aldrich, Switzerland; dissolved in acetonitrile- to a final concentration of 30 µM or 60 µM) was added to inhibit virB promoter activity. Cultures were then grown for an additional 1.5 h until the OD600nm reached about 1.0.
For β-galactosidase assays in B. abortus, wild type and vjbR mutant strains containing single-copy transcriptional fusions of lacZ to the virB1 or vceC genes were grown in TSB overnight and then switched to modified minimal E-medium, pH 5.0 (Kulakov et al., 1997). Samples were then taken at different time points for β-galactosidase activity measurement and Western blotting. β-galactosidase assays in E. coli and B. abortus were performed according to the protocol developed by Miller (1972). Assays were performed at least 3 times independently for each strain. Western blotting was performed to measure VirB8 protein expression after switching to modified minimal E-medium (pH 5.0) at the same time points β-galactosidase activity was measured. Bacteria were pelleted, resuspended in 2X Laemmli sample buffer and heated at 100°C for 15 min. The total protein equivalent of 1 × 108 CFU was loaded and run on a 12% polyacrylamide gel and then transferred to a nitrocellulose membrane. The membranes were blocked in 2% non-fat skim milk powder in PBS for 1 h and probed with rabbit anti-VirB8 polyclonal antibody (1:5000), kindly provided by C. Baron (Rouot et al., 2003). Goat-anti-rabbit IgG antibody (Biorad) conjugated with horseradish peroxidase (HRP) was used (1:5000) as a secondary antibody and HRP activity was detected with a chemiluminescent substrate (Perkin-Elmer).
Purification of VjbR-His6
To generate a C-terminally His-tagged VjbR protein, the vjbR gene was PCR amplified from B. abortus 2308 DNA using the primers VjbR-F and VjbR-R. The PCR product was digested with NdeI and SalI and cloned in pET103 digested with the same enzymes. The plasmid was introduced into E. coli BL21 by electroporation. The C-terminal His-tagged fusion protein was purified as follows. A culture of BL21 pET103-vjbR was grown overnight in LB containing 10 mM glucose and carb. The culture was diluted 1:50 in LB with carb and grown for an additional 24 hours at room temperature without the addition of Isopropyl β-D-thiogalactopyranoside (IPTG), in order to obtain more correctly folded recombinant VjbR-His6 protein. Purification of VjbR-His6 was performed according to standard protocols (Qiagen).
Electrophoretic mobility shift assays (EMSA)
A 123 bp fragment upstream of the virB1 start codon was amplified from B. abortus 2308 DNA by PCR using the primers PvirB100-F and PvirB463-R and cloned into pCR2.1 to give rise to pWIL1 (Table S1). The fragment was isolated from pWIL1 by digestion with EcoRI. For EMSAs with the intergenic region virB1-virB2, a 172 bp fragment containing the putative PvirB box was PCR amplified from B. abortus 2308 DNA using primers PvirB1ig2-F and virB1369R. A 317 bp fragment of PvceC (BAB1_1058) was obtained from pSURS31 by digestion with BamHI and HindIII (PvceC in Brucella contains a HindIII site). A 139 bp fragment of the gyrA gene served as a negative control. This fragment was PCR amplified from B. abortus 2308 DNA using primers RTgyrA-F and RTgyrA-R. All fragments were purified on a 5% polyacrylamide gel. The purified fragments were 5’ labeled using T4 polynucleotide kinase (New England Biolabs) and γ-32P-dATP (GE-healthcare). Binding reactions contained 20,000 cpm (0.4 ng) of radio labeled DNA and varying concentrations of VjbR protein in a final volume of 20 µl. Furthermore, each reaction contained 10 mM Tris HCl (pH 7.4), 50 mM KCl, 1mM DTT, 6% glycerol, 0.5 mM EDTA, 50 µg/ml BSA and 50 µg/ml Poly(dI-dC) (Pierce Nucleic Acid). Samples were incubated at room temperature for 30 min and electrophoresed on a 5% non-denaturing polyacrylamide gel in 0.5x TBE. Gels were dried and exposed to an X-ray film for several hours. EMSAs were repeated at least 3 times with similar results.
Prediction of promoter regions containing a PvirB consensus box
A consensus prediction of the PvirB box was made using the online program MEME (http://meme.nbcr.net/meme/) using motifs found in the promoter regions upstream of virB1 and tetR (BAB2_0117) and in the intergenic region virB1-virB2. The resulting consensus was used to find related boxes in other promoters by searching the B. abortus intergenic nucleotide sequences (TIGR) with the online motif alignment search tool MAST (http://meme.nbcr.net/meme/). Several promoter regions identified in this first generation search were PCR amplified from B. abortus 2308 genomic DNA, cloned into pSURS and analysed as described earlier for the virB promoter for activation by VjbR. Promoters with PvirB boxes, which were found to be activated by VjbR in the E. coli system were further used to refine the box consensus sequence and the search for new promoters containing this motif. In total this refining process was performed three times leading to a fourth generation promoter prediction.
Effector translocation assays
For fusions to the C terminus of TEM-1 β-lactamase or adenylate cyclase (CyaA), genes encoding candidate effectors were PCR amplified without their start codons (see Supplementary table 5 for primers), digested with XbaI and PstI and cloned into pFlagTEM1 (Raffatellu et al., 2005) or pFlagCya, which were also digested with XbaI and PstI. pFlagTEM1 encodes a copy of TEM1 β-lactamase, in which the Sec-dependent signal sequence has been deleted and replaced with a 3xFLAG tag at the N terminus (Raffatellu et al., 2005). All plasmid constructs were checked by DNA sequencing. Flag-TEM1-effector or CyaA-effector fusion constructs were introduced into Brucella abortus 2308 and ADH3 (ΔvirB2) strains by electroporation. Expression of the fusion proteins in Brucella was confirmed by Western blot using anti-Flag antibodies (Sigma). For the translocation assay 6 × 104 J774.A1 mouse macrophages were seeded in 96-well plates and infected with B. abortus 2308 or ADH3 expressing TEM-1 fusion proteins at a multiplicity of infection of 500:1. Plates were centrifuged for 5 min at 250 × g at room temperature. Cells were incubated for 20 min at 37°C in 5% CO2 and washed 2 times with phosphate-buffered saline (PBS) to remove free bacteria. Then 0.2 ml new DMEMsup plus 1 mM IPTG were added to each well and plates incubated at 37°C in 5% CO2. After different time points after infection (3, 5, 7, 9 and 16 hours), cells were washed once with Hank’s balanced salt solution (Invitrogen) and loaded with a solution containing the fluorescent substrate CCF2/AM (Zlokarnik et al., 1998) at a final concentration of 1mM, for 1.5h at room temperature using the standard loading protocol recommended by the manufacturer (Invitrogen). Fluorescence microscopy analysis was performed inside a BSL3 facility using an Axiovert M200 (Carl Zeiss, Germany), equipped with a CCF2 filter set (Chroma Technology, Brattleboro, VT, USA). Fluorescence micrographs were captured using a Zeiss Axiocam MRC5 and Zeiss AxioVision 4.5 software.
To detect translocation of CyaA fusion proteins, CHO-FcR cells, seeded 2 × 104 in 96 well plates, were infected by opsonized B. abortus or L. pneumophila strains expressing CyaA-effector fusions. Plates were centrifuged for 5 min at 250 × g at room temperature and the cells were incubated for 60 min at 37°C in 5% CO2. After 3 washes with PBS, cells were incubated for another 5 h at 37°C in 5% CO2. Then cells were lysed in 0.1 M HCL/0.5 % Triton X-100 for 10 min at room temperature followed by heating for 10 min at 95°C. cAMP levels were determined by using the Direct cAMP Correlate-EIA Kit (Assay Designs).
To detect translocation of TEM-1 fusion proteins by L. pneumophila, pFLAGTEM1 plasmids expressing GST and RalF (full length, mutated or truncated) were introduced into L. pneumophila Lp01 wild type or dotA mutant by electroporation. To label Legionella with DsRed a constitutive promoter was amplified from pJC43 (Celli et al., 2005) and then was assembled with the dimer Tomato (dTomato) gene, which was obtained from Dr. R. Tsien (Shaner et al., 2004). The gene encoding dTomato, driven by the aphA promotor, was then cloned into pFT/RalFc (see above) to give rise to pdTFT/RalFc, which was confirmed by DNA sequencing to have all components in the correct reading frame, and introduced into L. pneumophila Lp01 wild type and dotA. For the translocation assay 1 × 105 J774.A1 mouse macrophages were seeded in 24-well plates and infected with L. pneumophila expressing TEM1 fusion proteins at a multiplicity of infection of 100:1. Plates were centrifuged for 5 min at 250 × g at room temperature. Cells were incubated for 60 min at 37°C in 5% CO2 and washed 3 times with phosphate-buffered saline (PBS) to remove free bacteria. Then 0.5 ml fresh medium containing 2 mM IPTG and 50 µg/ml gentamicin, was added to each well and plates were incubated at 37°C in 5% CO2. After 3 hours, cells were treated and processed as described above for B. abortus. To determine the bacterial loads, cells were lysed in 0.5% Tween 20 and scraped from each well. Expression of the fusion proteins to TEM-1 in L. pneumophila was confirmed by Western blot using anti-Flag antibodies (Sigma), as described above for B. abortus.
Supplementary Material
Table 1.
VjbR regulated proteins translocated into macrophages
| ORF | Name | Aa | C-terminusa |
|---|---|---|---|
| BAB1_1652 | VceA | 105 | TMKVVAGKVKRYGDGTPAKDKGHAPKN |
| BR1038 | B. suis VceC | 410 | IFHLPMNSRKMPRPKKPSKTKWKSCLAS |
| BAB1_1058 | B. abortus VceC | 418 | DEQPEDAKAEETIEDEMEKLLGELTKGETRN |
Brucella abortus/suis candidate T4SS substrates. Positively charged amino acids in the C-termini are shown in bold.
Acknowledgments
The authors would like to thank C. Baron for providing the VirB8 antiserum, R. Isberg for the dotA mutant, J. Celli for providing pJC43, R. Vance for the CHO-FcR cells and A. Bäumler, T. Rolán and C. Roux for critical comments on the manuscript. This work was funded by PHS grants AI050553 and AI065739 to RMT.
References Cited
- Altman E, Segal G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol. 2008;190:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Reynoso B, Lapaque N, Salcedo S, Briones G, Ciocchini AE, Ugalde R, Moreno E, Moriyon I, Gorvel JP. Cyclic beta-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat Immunol. 2005;6:618–625. doi: 10.1038/ni1202. [DOI] [PubMed] [Google Scholar]
- Arenas GN, Staskevich AS, Aballay A, Mayorga LS. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect Immun. 2000;68:4255–4263. doi: 10.1128/iai.68.7.4255-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol. 2002;44:73–88. doi: 10.1046/j.1365-2958.2002.02877.x. [DOI] [PubMed] [Google Scholar]
- Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O'Callaghan D. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A. 2002;99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones G, Inon de Iannino N, Roset M, Vigliocco A, Paulo PS, Ugalde RA. Brucella abortus cyclic beta-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect Immun. 2001;69:4528–4535. doi: 10.1128/IAI.69.7.4528-4535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne ED, Roy CR. The Legionella pneumophila IcmSW Complex Interacts with Multiple Dot/Icm Effectors to Facilitate Type IV Translocation. PLoS Pathog. 2007;3:e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med. 2003;198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Salcedo SP, Gorvel JP. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci U S A. 2005;102:1673–1678. doi: 10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, Garcia E. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect Immun. 2005;73:8353–8361. doi: 10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, Segal G, Shuman HA. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- Christie PJ. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerci DJ, Martinez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 2001;3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- Covacci A, Rappuoli R. Pertussis toxin export requires accessory genes located downstream from the pertussis toxin operon. Mol Microbiol. 1993;8:429–434. doi: 10.1111/j.1365-2958.1993.tb01587.x. [DOI] [PubMed] [Google Scholar]
- de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, Bonnot S, Ferooz J, Tibor A, De Bolle X, Letesson JJ. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. 2005;7:1151–1161. doi: 10.1111/j.1462-5822.2005.00543.x. [DOI] [PubMed] [Google Scholar]
- Delrue RM, Martinez-Lorenzo M, Lestrate P, Danese I, Bielarz V, Mertens P, De Bolle X, Tibor A, Gorvel JP, Letesson JJ. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 2001;3:487–497. doi: 10.1046/j.1462-5822.2001.00131.x. [DOI] [PubMed] [Google Scholar]
- den Hartigh AB, Sun Y-H, Sondervan D, Heuvelmans N, Reinders MO, Ficht TA, Tsolis RM. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect Immun. 2004;72:5143–5149. doi: 10.1128/IAI.72.9.5143-5149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozot M, Boigegrain RA, Delrue RM, Hallez R, Ouahrani-Bettache S, Danese I, Letesson JJ, De Bolle X, Kohler S. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol. 2006;8:1791–1802. doi: 10.1111/j.1462-5822.2006.00749.x. [DOI] [PubMed] [Google Scholar]
- Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, Danese I, Vandenhaute J, Tibor A, DeBolle X, Letesson JJ. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 2005;7:687–698. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- Fullner KJ. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J. Bacteriol. 1998;180:430–434. doi: 10.1128/jb.180.2.430-434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, Lestrate P, Lambert C, Letesson JJ, De Bolle X. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect Immun. 2005;73:5578–5586. doi: 10.1128/IAI.73.9.5578-5586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP, Olsen SC. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J Bacteriol. 2005;187:2715–2726. doi: 10.1128/JB.187.8.2715-2726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohlfeld S, Pattis I, Puls J, Plano GV, Haas R, Fischer W. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol. 2006;59:1624–1637. doi: 10.1111/j.1365-2958.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- Hong PC, Tsolis RM, Ficht TA. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect Immun. 2000;68:4102–4107. doi: 10.1128/iai.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber AM, Sullivan JT, Ronson CW. Symbiosis-induced cascade regulation of the Mesorhizobium loti R7A VirB/D4 type IV secretion system. Mol Plant Microbe Interact. 2007;20:255–261. doi: 10.1094/MPMI-20-3-0255. [DOI] [PubMed] [Google Scholar]
- Kulakov YK, Guigue-Talet PG, Ramuz MR, O’Callaghan D. Response of Brucella suis 1330 and B. canis RM6/66 to growth at acid pH and induction of an adaptive acid tolerance response. Res Microbiol. 1997;148:145–151. doi: 10.1016/S0923-2508(97)87645-0. [DOI] [PubMed] [Google Scholar]
- Leonard S, Ferooz J, Haine V, Danese I, Fretin D, Tibor A, de Walque S, De Bolle X, Letesson JJ. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J Bacteriol. 2007;189:131–141. doi: 10.1128/JB.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVier K, Phillips RW, Grippe VK, Roop RM, 2nd, Walker GC. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science. 2000;287:2492–2493. doi: 10.1126/science.287.5462.2492. [DOI] [PubMed] [Google Scholar]
- Lindeberg M, Stavrinides J, Chang JH, Alfano JR, Collmer A, Dangl JL, Greenberg JT, Mansfield JW, Guttman DS. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol Plant Microbe Interact. 2005;18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. Embo J. 2001;20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- Pappas KM, Winans SC. A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol Microbiol. 2003;48:1059–1073. doi: 10.1046/j.1365-2958.2003.03488.x. [DOI] [PubMed] [Google Scholar]
- Patey G, Qi Z, Bourg G, Baron C, O’Callaghan D. Swapping of periplasmic domains between Brucella suis VirB8 and a pSB102 VirB8 homologue allows heterologous complementation. Infect Immun. 2006;74:4945–4949. doi: 10.1128/IAI.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I, Seshadri R, Nelson K, Eisen J, Heidelberg J, Read T, Dodson R, Umayan L, Brinkac L, Beanan M, Daugherty S, Deboy R, Durkin A, Kolonay J, Madupu R, Nelson W, Ayodeji B, Kraul M, Shetty J, Malek J, van Aken S, Riedmuller S, Tettelin H, Gill S, White O, Salzberg S, Hoover D, Lindler L, Halling S, Boyle S, Fraser C. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc Natl Acad Sci USA. 2002;99:13148–13153. doi: 10.1073/pnas.192319099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Wu Q, Kahl-McDonagh M, Ficht TA. Cytotoxicity in macrophages infected with rough Brucella mutants is type IV secretion system dependent. Infect Immun. 2008;76:30–37. doi: 10.1128/IAI.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Meresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goni I, Moreno E, Gorvel JP. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998a;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Moreno E, Sanguedolce V, Mege JL, Gorvel JP. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun. 1998b;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Sun YH, Wilson RP, Tran QT, Chessa D, Andrews-Polymenis HL, Lawhon SD, Figueiredo JF, Tsolis RM, Adams LG, Baumler AJ. Host restriction of Salmonella enterica serotype Typhi is not caused by functional alteration of SipA, SopB, or SopD. Infect Immun. 2005;73:7817–7826. doi: 10.1128/IAI.73.12.7817-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow-Larsen AA, Rajashekara G, Petersen E, Splitter G. Putative Quorum Sensing Regulator BlxR of Brucella melitensis Regulates Virulence Factors Including the Type IV Secretion System and Flagella. J Bacteriol. 2008 doi: 10.1128/JB.01915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouot B, Alvarez-Martinez MT, Marius C, Menanteau P, Guilloteau L, Boigegrain RA, Zumbihl R, O’Callaghan D, Domke N, Baron C. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect Immun. 2003;71:1075–1082. doi: 10.1128/IAI.71.3.1075-1082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, Carena I, Dehio C. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Baldwin TO. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J Bacteriol. 1991;173:568–574. doi: 10.1128/jb.173.2.568-574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Baldwin TO. Positive autoregulation of the Vibrio fischeri luxR gene. LuxR and autoinducer activate cAMP-catabolite gene activator protein complex-independent and -dependent luxR transcription. J Biol Chem. 1992;267:7696–7702. [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieira R, Comerci DJ, Pietrasanta LI, Ugalde RA. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol Microbiol. 2004;54:808–822. doi: 10.1111/j.1365-2958.2004.04316.x. [DOI] [PubMed] [Google Scholar]
- Sieira R, Comerci DJ, Sanchez DO, Ugalde RA. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- Taminiau B, Daykin M, Swift S, Boschiroli ML, Tibor A, Lestrate P, De Bolle X, O’Callaghan D, Williams P, Letesson JJ. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect Immun. 2002;70:3004–3011. doi: 10.1128/IAI.70.6.3004-3011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs IM, De Keersmaecker SC, Fadda A, Engelen K, Zhao H, McClelland M, Marchal K, Vanderleyden J. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J Bacteriol. 2007;189:4587–4596. doi: 10.1128/JB.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzureau S, Godefroid M, Deschamps C, Lemaire J, De Bolle X, Letesson JJ. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J Bacteriol. 2007;189:6035–6047. doi: 10.1128/JB.00265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss AA, Johnson FD, Burns DL. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci U S A. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CE, Winans SC. The quorum-sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol Microbiol. 2007;64:245–256. doi: 10.1111/j.1365-2958.2007.05647.x. [DOI] [PubMed] [Google Scholar]
- Worley MJ, Ching KH, Heffron F. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien RY. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.